Abstract
Background
ARAP1 is an Arf-directed GTPase-activating protein (GAP) that inhibits the trafficking of epidermal growth factor receptor (EGFR) to the early endosome. To further understand the function of ARAP1, we sought to identify proteins that interact with ARAP1.
Results
Here we report that ARAP1 associates with the Cbl-interacting protein of 85 kDa (CIN85). Arginines 86 and 90 of ARAP1 and the SH3 domains of CIN85 are necessary for the interaction. We found that a mutant of ARAP1 with reduced affinity for CIN85 does not efficiently rescue the effect of reduced ARAP1 expression on EGFR trafficking to the early endosome. Reduced expression of CIN85 has a similar effect as reduced expression of ARAP1 on traffic of the EGFR. Cbl proteins regulate the endocytic trafficking of the EGFR by mediating ubiquitination of the EGFR. Overexpression of ARAP1 reduced ubiquitination of the EGFR by Cbl and slowed Cbl-mediated EGFR degradation. Reduced expression of ARAP1 accelerated degradation of EGFR but did not affect the level of ubiquitination of the receptor that was detected.
Conclusions
ARAP1 interaction with CIN85 regulates endocytic trafficking of the EGFR and affects ubiquitination of EGFR. We propose a model in which the ARAP1/CIN85 complex drives exit of EGF-EGFR-Cbl complex from a pre-early endosome into a pathway distinct from the early endosome/lysosome pathway.
Keywords: Epidermal Growth Factor Receptor, ARAP1, CIN85, endocytosis, membrane traffic
Introduction
ARAPs are a subtype of ArfGAPs (ADP-ribosylation factor GTPase-activating proteins) with three members: ARAP1, ARAP2 and ARAP3. The proteins contain a sterile-α motif (SAM) at the extreme amino-terminus, five pleckstrin homology (PH) domains, an Arf GAP and a Rho GAP domain, ankyrin repeats (ANK), and a ras-associating (RA) domain (Miura et al., 2002; Krugmann et al., 2002; Randazzo et al., 2007). Two of the PH domains contain the consensus sequence for phosphatidylinositol 3,4,5- trisphosphate (PI(3,4,5)P3) binding, and binding of PI(3,4,5)P3 stimulates ArfGAP activity (Miura et al., 2002; Yoon et al., 2006; Campa et al., 2009; I et al., 2004). ARAP1 has been implicated in the regulation of intracellular trafficking of the epidermal growth factor receptor (EGFR) (Yoon et al., 2008; Daniele et al., 2008).
EGFR, a member of the ErbB family of receptor tyrosine kinases (RTKs), is a transmembrane protein that delivers extracellular signals into cells, thereby controlling cellular functions including cell proliferation, differentiation and migration (Jorissen et al., 2003). Overexpression and/or hyperactivation of the EGFR are frequently detected in breast cancer, ovarian cancer, kidney cancer, non-small cell lung cancer, head and neck squamous cell carcinoma, and glioblastoma (Salomon et al., 1995; Klapper et al., 2000; Ono and Kuwano, 2006; Wong et al., 1992). EGFR forms a dimer upon binding ligands, including epidermal growth factor (EGF), transforming growth factor-α, and ampiregulin (Kim and Muller, 1999), resulting in kinase activation, phosphorylation of tyrosine residues in the cytoplasmic domain of the receptor itself, and receptor internalization and transport through the endocytic pathway. The strength and duration of intracellular signaling from the EGFR are controlled by the relative rates of internalization, recycling and degradation of the receptor (Yarden and Sliwkowski, 2001; Ceresa, 2006).
Endocytic trafficking of the EGFR has been extensively studied. Internalization has been reported to be dependent on clathrin as well as adaptor proteins (Goh et al., 2010; Sorkin and Goh, 2009; Mosesson et al., 2008; Sorkin and von Zastrow, 2009). EGFR is delivered to a Rab5-containing early endosome from which it is sorted into either recycling or degradative pathways. Regulation of internalization and subsequent trafficking involve the ubiquitin ligase (E3) Cbl and the adaptor protein CIN85 (Szymkiewicz et al., 2002). CIN85 contains three Src homology 3 (SH3) domains, a proline-rich (PR) region and a coiled-coil (Cc) domain. CIN85 was first identified based on its interaction with c-Cbl (Take et al., 2000) and is also known as a SH3KBP1 (SH3 domain kinase binding protein 1), Ruk (regulator of ubiquitous kinase) and SETA (SH3 domain-containing gene expressed in tumorigenic astrocytes) (Narita et al., 2001; Gout et al., 2000; Bogler et al., 2000; Borinstein et al., 2000; Dikic, 2002). CIN85 is thought to regulate endocytosis of receptor by binding to EGFR indirectly through Cbl and recruiting components of the endocytic machinery, such as endophilins, but CIN85 does not affect Cbl-mediated ubiquitination of the EGFR (Soubeyran et al., 2002).
We previously reported that EGF stimulation causes ARAP1 to rapidly and transiently associate with a pre-early endosome that contains EGFR, Rab5 and Rabaptin5, but not early endosomal antigen1 (EEA1). Reduction of ARAP1 expression by small interfering RNA (siRNA) accelerated association of the EGFR with the EEA1 compartment and the internalization and degradation of the EGFR, and attenuated EGFR signaling (Yoon et al., 2008). It has also been reported that reduction of ARAP1 expression leads to accumulation of EGFR in the late endosomes and prolonged signaling (Daniele et al., 2008). The exact site and molecular basis of ARAP1 action and the reason for the apparently conflicting results of reduced ARAP1 expression are still being explored.
To further understand the role of ARAP1 in regulation of EGFR endocytosis, we sought to identify proteins that bind to ARAP1. Here, we report the interaction of CIN85 with ARAP1. CIN85 was identified in a 2-hybrid screen. We confirmed the interaction by coimmunoprecipitation and colocalization in cells. The binding site on ARAP1 was mapped to a PxPxxR motif immediately C-terminal of the SAM domain. We found that a mutant of ARAP1 with reduced affinity for CIN85 was defective in modulating EGFR trafficking. We also found that ARAP1 reduced ubiquitination of the EGFR. We propose a model in which ARAP1 binds to CIN85 to drive exit of EGFR from the endosomal pathway prior to ubiquitination of the receptor.
Results
ARAP1 interacts with CIN85 in mammalian cells
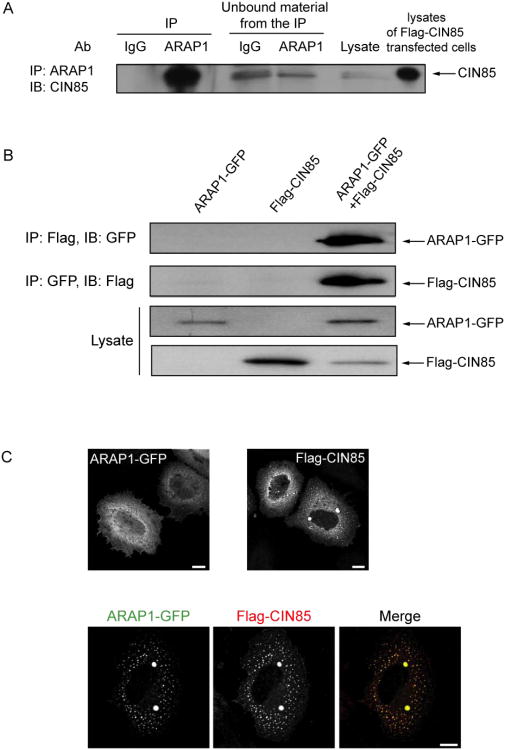
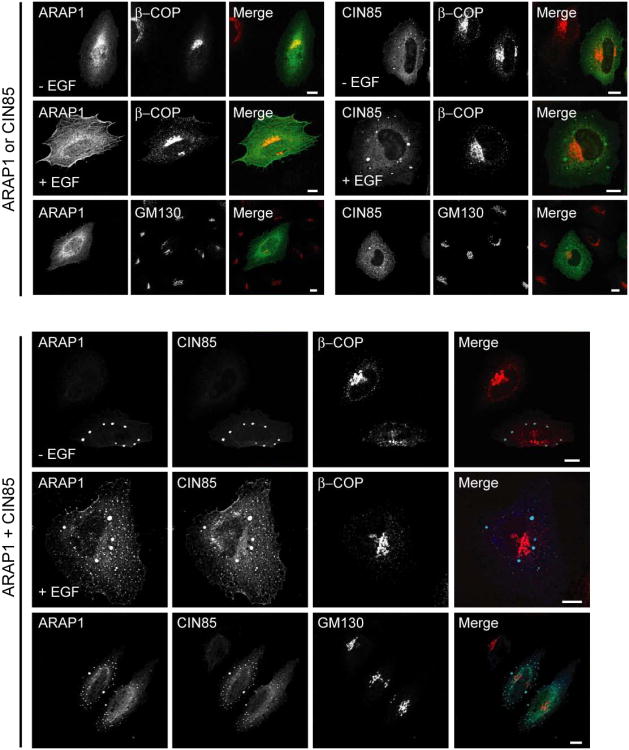
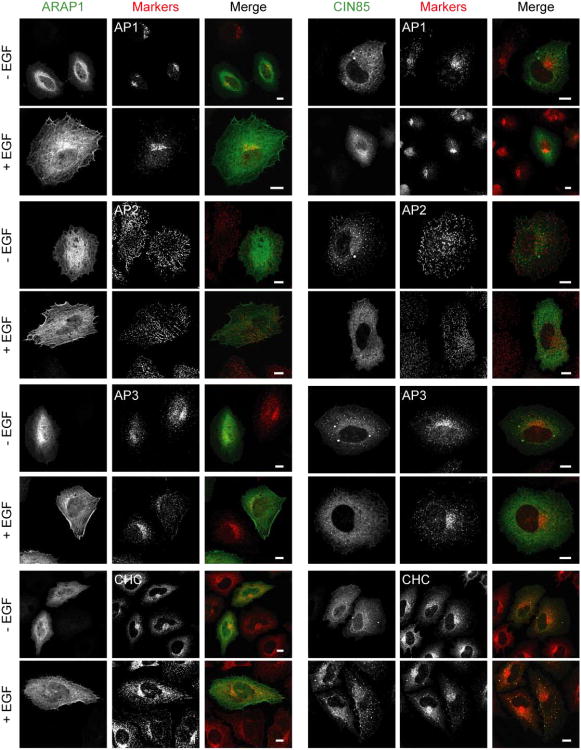
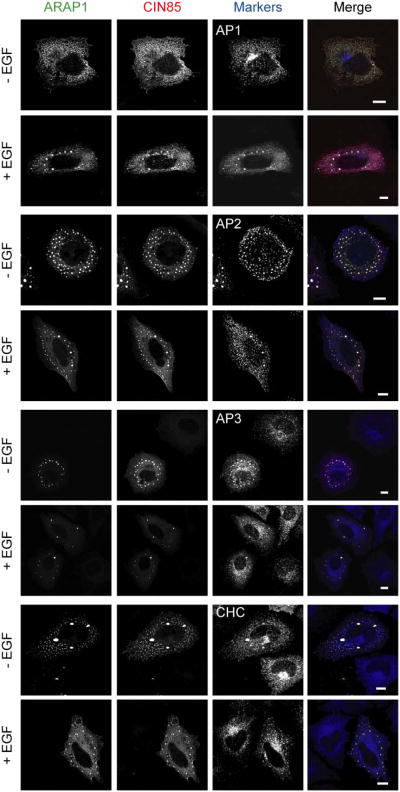
To further understand the role of ARAP1 in regulation of EGFR endocytosis, we identified interacting proteins of ARAP1 using the yeast two-hybrid system. Screens of human spleen and thymus cDNA libraries with two baits derived from ARAP1 gave rise to 7 positive clones, one of which encoded the three SH3 domains of CIN85 (Fig. 4A and Table 1). To confirm that ARAP1 associates with CIN85 in mammalian cells, proteins were immunoprecipitated using rabbit anti-ARAP1 antibody (Ab). Proteins in the precipitates and remaining in solution (“unbound material” in Fig. 1A) were detected by immunoblotting. We found that CIN85 co-immunoprecipitated with ARAP1 using anti-ARAP1 Ab, but was not precipitated with the control IgG (Fig. 1A). We estimate that approximately 50% of the CIN85 was in complex with ARAP1 based on the loss of CIN85 from solution after the specific immunoprecipitation. To further analyze ARAP1 binding to CIN85, we transfected HeLa cells with plasmids encoding GFP fused to ARAP1 (ARAP1-GFP) or Flag epitope fused to CIN85 (Flag-CIN85). ARAP1-GFP was present with Flag-CIN85 in immunoprecipitates obtained with an Ab to the FLAG epitope and Flag-CIN85 was in the precipitate with ARAP1-GFP when immunoprecipitating with an anti-GFP Ab (Fig. 1B). We examined the distribution of ARAP1-GFP and Flag-CIN85 in the cells because there is no commercial CIN85 Ab available that is suitable for immunofluorescence staining of endogenous proteins. Although endogenous ARAP1 associated with the Golgi apparatus and with the endocytic compartment in cells treated with EGF (Miura et al., 2000; Yoon et al., 2008), overexpressed ARAP1-GFP was primarily distributed diffusely throughout the cell with some perinuclear accumulation, consistent with association with the Golgi apparatus (Fig. 1C left upper panel). Flag-CIN85 had a perinuclear distribution in some cells but was mostly found associated with dot-like structures, with several large puncta that are likely artifacts of overexpression (Fig. 1C right upper panel). In cells coexpressing ARAP1 and CIN85, the proteins colocalized in the large, likely artifactual punctate structures, as well as with the small puncta, with loss of perinuclear assocated ARAP1 (Fig. 1C lower panels). To identify the punctate structures, we used organellar markers. First, we determined if overexpressed ARAP1 and CIN85 associate with the Golgi apparatus. ARAP1 had limited colocalization with β-COP in the perinuclear region (Fig. 2 left upper panel). CIN85, either alone or together with ARAP1, did not associate with the Golgi markers, β-COP and GM130 (Fig. 2 right upper panel and bottom panel). Treatment of cells with EGF did not affect the distribution of coexpressed ARAP1 and CIN85 (Fig. 2). Next, we determined if ARAP1 and CIN85 colocalized with adaptor protein complexes, AP1, AP2, and AP3, and clathrin, components of clathrin-coated vesicles, which are involved in endocytic membrane trafficking (Robinson and Bonifacino, 2001). ARAP1 partially colocalized with AP1, AP3 and clathrin in the perinuclear region but not AP2. We did not detect colocalization of CIN85 with AP1, AP2 or AP3 although limited colocalization with clathrin-positive puncta was detected in cells treated with EGF (Fig. 3A). However, in cells expressing both ARAP1 and CIN85, puncta containing ARAP1 and CIN85 also contained all three adaptor protein complexes and clathrin. The compartment containing both ARAP1 and CIN85 was not affected by EGF (Fig. 3B). Although we could not clearly identify the structures as known organellar structures, the colocalization of ARAP1 with CIN85 together with the immunoprecipitation results support the conclusion that CIN85 and ARAP1 interact in cells. Furthermore, ARAP1 and CIN85 coexpression appeared to cause collapse of the endocytic compartments into a single, artifactual compartment, similar to the mixing of the TGN and endocytic compartments observed on treating wells with brefeldin A (Lippincott-Schwartz et al., 1991).
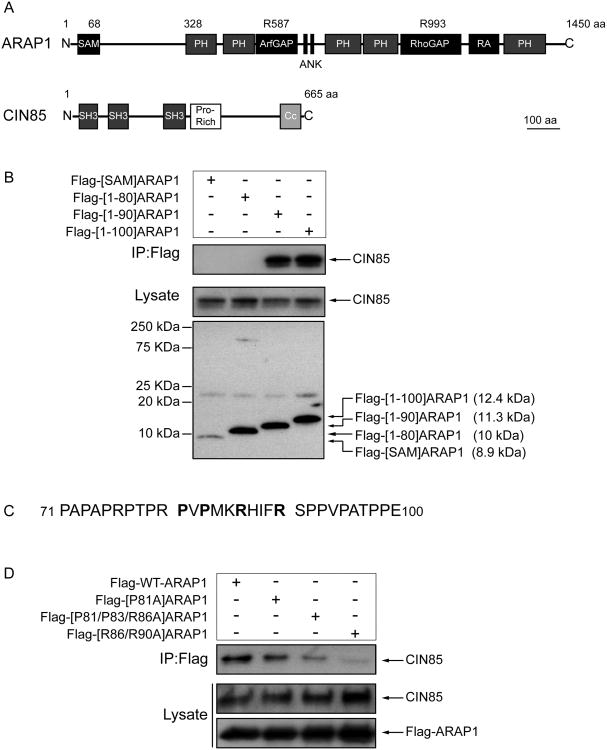
Figure 4. Identification of CIN85 binding sites in ARAP1.
(A) Schematics of ARAP1 and CIN85. Abbreviations: SAM, sterile alpha-motif domain; PH, pleckstrin homology domain; ArfGAP, Arf GTPase-activating domain; Ank, ankyrin repeat; RA, Ras associating domain; SH3, Src homology 3 domain; Pro-Rich, proline-rich motif; Cc, coiled-coil domain. (B) Association of CIN85 with truncation mutants of ARAP1. HeLa cells were transiently transfected with plasmids directing expression of the indicated mutants of Flag-ARAP1. Lysates were immunoprecipitated using an antibody to the Flag epitope and immunoblotted with anti-CIN85 or anti-Flag antibodies. (C) Amino acids 71 to 100 of ARAP1. D. Effect of point mutations in ARAP1 on association with CIN85. Flag-ARAP1 with the indicated point mutations was immunoprecipitated to analyze its interaction site with endogenous CIN85.
Table 1.
Interacting proteins of ARAP1 using the yeast two-hybrid system.
| Prey | Baits (ARAP1) | |
|---|---|---|
| CIN85 | c-Cbl-interacting protein of 85 kDa | [1-100]ARAP1 |
| mSHIP | SH2-containing inositol 5-phosphatase 2 | [1-100]ARAP1 |
| KCTD12 | potassium channel tetramerisation domain containing 12 | [300-562]ARAP1 |
| PSD3 | pleckstrin and Sec7 domain containing 3 | [300-562]ARAP1 |
| TP53BP1 | p53-binding protein 1 | [300-562]ARAP1 |
| TES | testin, isoform 1 | [300-562]ARAP1 |
| mWAPL | wings apart-like protein | [300-562]ARAP1 |
Figure 1. Association of ARAP1 with CIN85 in mammalian cells.
(A) Coimmunoprecipitation of endogenous ARAP1 and endogenous CIN85. Endogenous ARAP1 from HeLa cell lysates was immunoprecipitated with rabbit anti-ARAP1 antibody (1153) and immunoblotting was used to detect endogenous CIN85 in the precipitate and that remaining in solution after precipitation (“unbound material from the IP”). Lysates of cells expressing recombinant proteins were used as positive controls for blotting; however, the lysates used for IP did not contain overexpressed recombinant protein. Rabbit IgG was used as a control antibody for immunoprecipitation. (B) Coimmunoprecipitated of Flag-CIN85 with ARAP1-GFP. Cells were transiently transfected plasmids for the expression of Flag-CIN85 and/or ARAP1-GFP. Immunoprecipitation and immunoblotting using antibodies to the Flag and GFP were performed as described in (A). (C) Colocalization of ARAP1 and CIN85. HeLa cells were transfected with plasmids encoding ARAP1-GFP (left upper panel), Flag-CIN85 (right upper panel) or both (lower panels). The cells were immunostained with an antibody to Flag and visualized by immunofluorescence. Scale bar, 10 μm.
Figure 2. Subcellular localization of exogenous ARAP1 and CIN85 in the cells.
HeLa cells with ARAP1-GFP and/or Flag-CIN85 were serum starved and stimulated with 100 ng/ml of EGF for 10 min at 37°C. The indicated proteins were detected by immunofluorescence microscopy. In upper panel, ARAP1 or CIN85: green, β-COP or GM130: red, in lower panel, ARAP1: green, CIN85: blue and β-COP or GM130: red. Scale bar, 10 μm.
Figure 3. Effect of EGF on the distribution of recombinant ARAP1 and/or CIN85 with endosomal-markers.
Serum starved HeLa cells with ARAP1-GFP (in the left panel of A) or Flag-CIN85 (in the right panel of A) or both (B) were stimulated with 100 ng/ml of EGF for 10 min at 37°C and examined by immunofluorescence microscopy. In Fig. 3A, ARAP1 or CIN85: green, markers: red. In Fig. 3B, ARAP1: green, CIN85: red and markers: blue. Scale bar, 10 μm.
Structural determinants of CIN85-ARAP1 binding
The bait used for the yeast two-hybrid screen that identified CIN85 was the N-terminal 100 amino acids of ARAP1. To define the specific regions of ARAP1 involved in CIN85 interaction, a panel of ARAP1 deletion mutants was used in immunoprecipitation experiments. Endogenous CIN85 coprecipitated with Flag-[1-90]-and Flag-[1-100]ARAP1, but not Flag-[1-80] or Flag-[SAM]ARAP1, indicating that amino acids 81-90 of ARAP1 are necessary for binding to CIN85 (Fig. 4B). This part of ARAP1 contains a PxPxxR motif (see Fig. 4C) similar to the previously identified consensus binding site for the SH3 domains of CIN85 (Kurakin et al., 2003; Kowanetz et al., 2003). To further analyze ARAP1 regions required for binding to CIN85, we mutated proline or arginine to alanine in this 81-90 amino acid portion of ARAP1 and found that Flag-[R86/90A]ARAP1 and Flag-[P81/P83/R86A]ARAP1 bound CIN85 less efficiently than wild type ARAP1. Mutating P81 alone had a small effect (Fig. 4D).
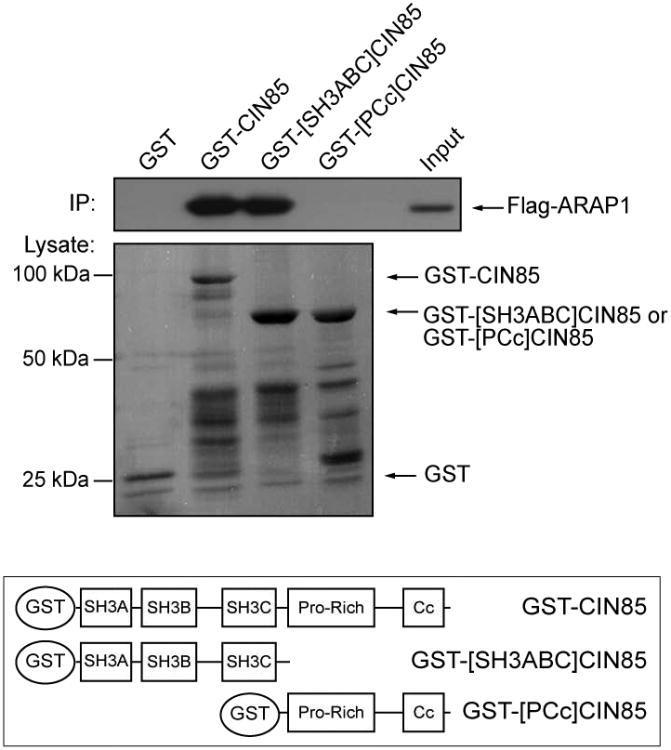
To map the regions of CIN85 responsible for binding to ARAP1, lysates of HeLa cells transfected with Flag-ARAP1 were incubated with GST fusion proteins encoding full length, the three SH3 domains (SH3ABC), or the proline-rich motif and coiled-coil domain (PCc) of CIN85. The SH3ABC protein bound to ARAP1 as efficiently as did full length CIN85, whereas no interaction was detected with the proline-rich motif and coiled-coil domain of CIN85 or with GST (Fig. 5).
Figure 5. ARAP1 interacts with the SH3 domains of CIN85.
Lysates of HeLa cells expressing Flag-ARAP1 were incubated with GST or GST fused to full length-, the three SH3 domains of CIN85 (SH3ABC-) or the proline-rich motif and coiled-coil domain of CIN85 (PCc) immobilized on glutathione Sepharose beads. After incubation, the beads were washed and associated ARAP1 was visualized by immunoblotting using anti-Flag antibody. The amount of GST-fusion proteins present was determined by Ponceaus S staining. The lower panel is a schematic CIN85 and truncation mutants of CIN85 used in the experiment.
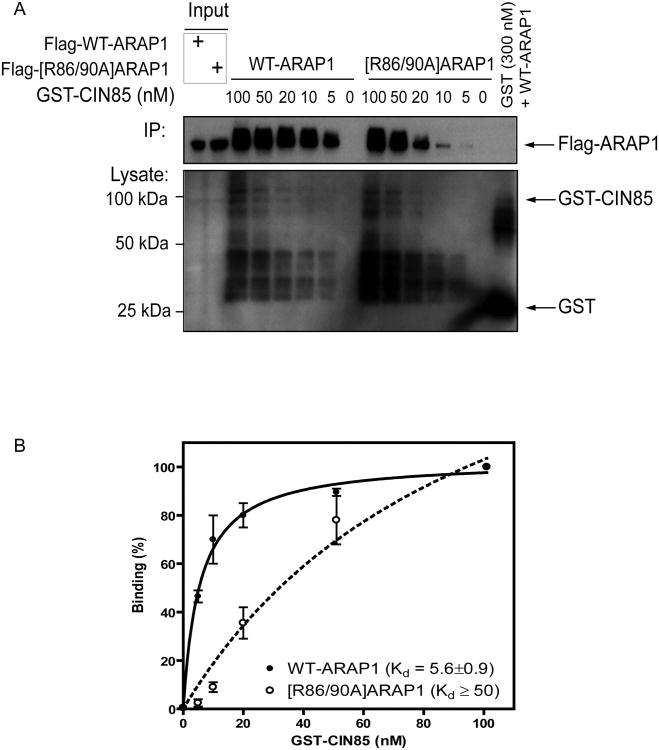
To determine relative affinities between CIN85 and ARAP1 recombinant proteins, we titrated GST-fused full length CIN85 into lysates of HeLa cells transfected with either Flag-ARAP1 or Flag-[R86/90A]ARAP1 at a fixed concentration (0.3 mg). GST was incubated with Flag-ARAP1 as a negative control (Fig. 6A). The association of CIN85 with ARAP1 was analyzed by immunoblotting using anti-Flag Ab. The signal was quantified using densitometry (Fig. 6B). Half maximal binding of ARAP1 was achieved with 5.6 ± 0.9 nM GST-CIN85. Because of uncertainties related to the stability and folding of the GST-CIN85, this value is nominal, and does not represent a Kd, but is useful for the purpose of comparison. Similar levels of binding of [R86/90A]ARAP1 required eight times more GST-CIN85, indicating the affinity was at least eight-fold less (Fig. 6B). Based on these data, we conclude that binding of CIN85 and ARAP1 involves at least one of the SH3 domains of CIN85 and arginines 86 and 90 of ARAP1.
Figure 6. Determination of ARAP1/CIN85 binding affinity.
GST-CIN85 was titrated into a reaction with a fixed concentration of cell lysate (0.3 mg) containing either Flag-ARAP1 or Flag-[R86/90A]ARAP1. GST (10 μg) was incubated with Flag-ARAP1 as a negative control. The primary data from one experiment is shown in panel A. A summary of two independent experiments in which signal was quantified by densitometry is shown in panel B. The error bars are the SEM.
ARAP1 association with CIN85 does not depend on EGF
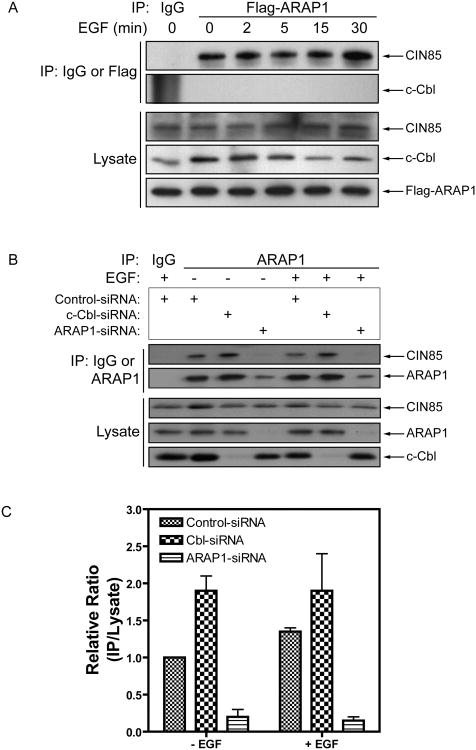
CIN85 binds to activated EGFR through the E3 ligase Cbl to facilitate endocytosis and degradation (Petreli et al., 2002; Soubeyran et al., 2002; Haglund et al., 2002; Mosesson et al., 2003). To determine if ARAP1 is in the complex with CIN85 and Cbl and if EGF affected ARAP1 association with CIN85, Flag-ARAP1 was immunoprecipitated using mouse anti-Flag-M5 Ab or control mouse IgG and then CIN85 and Cbl content of the precipitate was measured using immunoblotting. No change in CIN85 association with ARAP1 was detected upon EGF stimulation and Cbl did not bind to ARAP1 (Fig. 7A).
Figure 7. Effect of ARAP1 on c-Cbl binding to CIN85.
(A) ARAP1 binds to CIN85 constitutively but does not bind to Cbl. HeLa cells were transiently transfected with a plasmid encoding Flag-ARAP1, serum-starved and then treated with 25 ng/ml EGF. ARAP1 was immunoprecipitated with an anti-Flag antibody and CIN85 and Cbl in the precipitates were detected by immunoblotting using specific antibodies against CIN85 or Cbl. Mouse IgG was used as a negative control for the immunoprecipitation. Data are representative of three independent experiments. (B) The effect of reduced Cbl expression on the formation of ARAP1/CIN85 complexes. ARAP1 in the lysate of HeLa cells transfected with the indicated siRNA was immunoprecipitated using anti-ARAP1 antibody. Proteins in the precipitate were detected by immunoblotting. (C) Quantification of the ARAP1/CIN85 complexes. The signal was quantified by densitometry from two independent experiments. The data are presented the relative ratio of ARAP1/CIN85 immune complexes to total cellular CIN85. The error bars are the SEM.
The absence of Cbl in the ARAP1 precipitate suggests that Cbl/CIN85 and ARAP1/CIN85 complexes are mutually exclusive. We determined if reducing Cbl expression, using siRNA, affected formation of ARAP1/CIN85. ARAP1-siRNA was included as a control. We found that reduced Cbl expression increased the amount of ARAP1/CIN85 complex recovered (Fig. 7B and C). These data support the idea that ARAP1 and Cbl compete for binding to CIN85.
CIN85 does not affect ARAP1 GAP activity
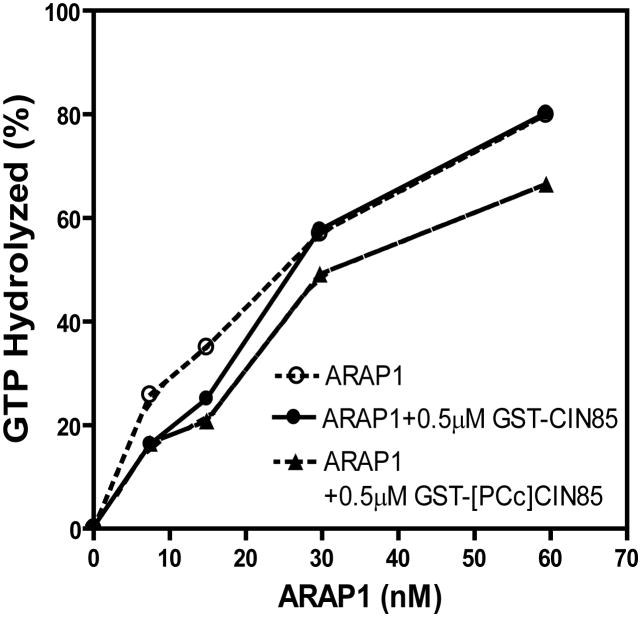
The effect of GST-CIN85 on the enzymatic activity of full length purified recombinant ARAP1 was examined. In these experiments, relative enzymatic power was determined by measuring the amount of ARAP1 required to convert 50% of a fixed amount of Arf5•GTP to Arf5•GDP. GST-CIN85 and GST-[PCc]CIN85 had no effect on ArfGAP activity of ARAP1 (Fig. 8).
Figure 8. Effect of CIN85 binding on ArfGAP activity of ARAP1.
GAP activity was determined as previously described (Campa et al., 2009). Purified WT-ARAP1 were titrated into a reaction mixture containing 0.5 μM myrArf5[α32P]GTP, 500 μM LUVs with 0.5 μM of GST-CIN85, or GST-[PCc]CIN85. The LUVs were composed of 40% phosphatidylcholine, 25% phosphatidylethanolamine, 15%phosphatidylserine, 7% PtdIns, 2.5% PtdIns(4,5)P2, 0.5% PtdIns(3,4,5)P3 and 10% cholesterol.
Effect of ARAP1 and CIN85 on EGF traffic
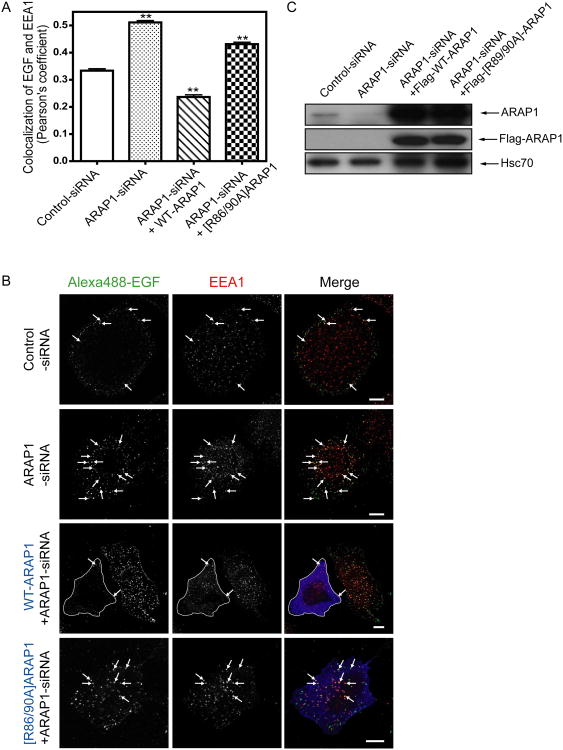
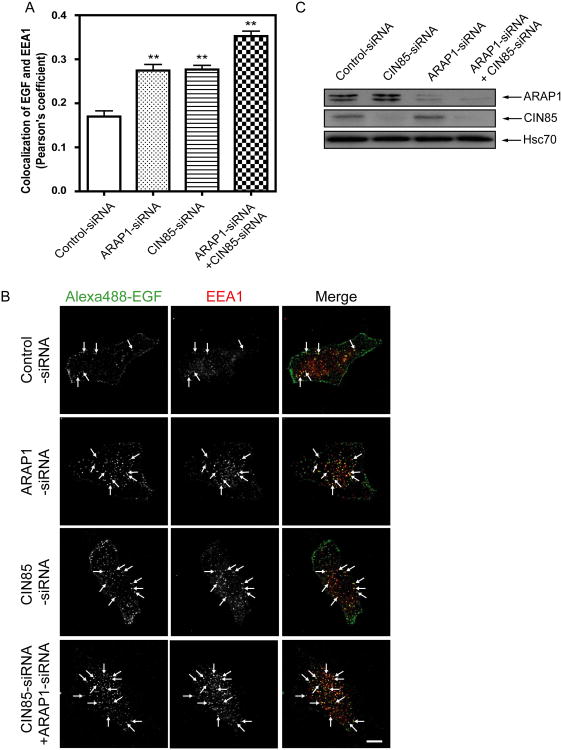
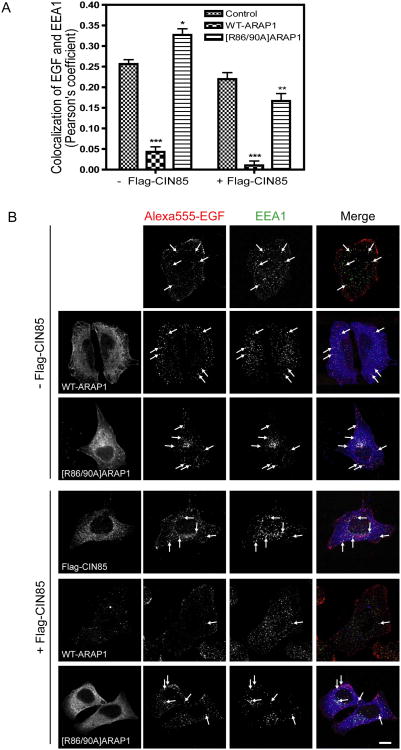
ARAP1 overexpression has been found to slow the association of EGF-EGFR with the EEA1 compartment and reduction of ARAP1 expression accelerated association of EGF-EGFR with the EEA1 compartment and degradation of the receptor (Yoon et al., 2008; Campa et al., 2009). To investigate the role of its association with CIN85 by ARAP1, we analyzed the intracellular trafficking of Alexa488-EGF to the EEA1-positive endosomes in cells transfected with control-siRNA or ARAP1-siRNA. As we had previously found, decreased ARAP1 expression increased association of EGF with the EEA1-positive endosomes. Rescue with WT-ARAP1 reversed the effect of reducing endogenous ARAP1 expression. [R86/90A]ARAP1, which has decreased binding to CIN85, was less effective than WT-ARAP1 (Fig. 9A and B). We also examined the association of EGF and the EEA1-positive endosomes in cells with control-siRNA, ARAP1-siRNA, CIN85-siRNA or both ARAP1-siRNA and CIN85-siRNA. Decreased CIN85 expression had a similar effect as reduced ARAP1 expression, with increased association of Alexa488-EGF with the EEA1-positive endosome. Reduced expression of both ARAP1 and CIN85 resulted in association of EGF and the EEA1-positive endosomes to a greater extent than that observed with the reduced expression of either protein alone (Fig. 10 A and B). We also examined the effect of overexpression of ARAP1 together with CIN85 on association of EGF and EEA1-positive endosomes. Cells were transfected with empty vector, ARAP1 or [R86/90A]ARAP1 with or without Flag-CIN85. Expression of ARAP1 alone reduced the association of EGF with the EEA1-positive compartment. CIN85 by itself had a small effect and increased the already large effect of ARAP1 to a small extent. [R86/90A]ARAP1 alone slightly increased the association of EGF with the EEA1-positive compartment. CIN85 expression reversed this effect (Fig. 11A and B).
Figure 9. Effect of ARAP1 on association of EGF with EEA1 compartment.
Endogenous ARAP1 was reduced with siRNA and was replaced with either Flag-ARAP1 or Flag-[R86/90A]ARAP1 by expression from a siRNA resistant plasmid. Cells were serum starved, incubated with 1 μg/ml of AlexaFluor 488-labeled EGF (in DMEM supplemented with 20 mM HEPES and 2 mg/ml BSA) for 30 min on ice, washed and then transferred to 37°C for 10 min before fixation. The cells were immunostained with mouse anti-EEA1 antibody and rabbit anti-Flag antibody. Images were acquired with an LSM510-meta laser scanning confocal microscope. Colocalization of fluorescent EGF and EEA1 quantified using the Imaris program by determining Pearson's coefficients for 31 cells. Results of the analysis are shown in the BAR graph in panel A. **p < 0.01, significant difference as compared with the control-siRNA (one way ANOVA with Bonferroni post test). Representative images used for the analysis are shown in panel B. The outlined cell is transfected with Flag tagged WT-ARAP1. Arrows indicate colocalization of fluorescence EGF and EEA1. Alexa488-EGF: green and EEA1: red. Expression levels of ARAP1 determined by immunoblotting using anti-ARAP1 antibody or anti-Flag antibody are shown in panel C. Anti-Hsc70 antibody was used as a protein loading control.
Figure 10. Effect of reduction of CIN85 and ARAP1 expression on EGF association with the EEA1-positive endosome.
ARAP1 and CIN85 expression were reduced using siRNA. EGF association with the EEA1-positive compartment was determined as described in Figure 9. Panel A shows quantification of colocalization of fluorescence EGF and EEA1 measured as Pearson's coefficients for 20 cells for each condition in two independent experiments. ** p<0.01 (one way ANOVA with Bonferroni post test). The error bars are the SEM. Representative images are shown in panel B. Arrows indicate colocalization of fluorescence EGF and EEA1. Alexa488-EGF: green and EEA1: red. Immunoblotting to determine the protein expression levels using indicated antibodies is shown in panel C.
Figure 11. The effect of overexpression of ARAP1 and CIN85 on association of EGF with the EEA1-positive compartment.
Cell expressing ARAP1-GFP, [R86/90A]ARAP1-GFP and Flag-CIN85 were treated and analyzed as described in Figures 9 and 10. Pearson's coefficients were determined for 20 cells at 10 min after stimulation with fluorescent EGF. *, ** and ***p<0.05, 0.01 and 0.001, respectively (two way ANOVA with Bonferroni post test). Panel A is the graph summarizing data for 2 independent experiments. The error bars are the SEM. Representative immunostained cells are shown in panel B. ARAP1: blue, Alexa555-EGF: red and EEA1: green. Scale bar, 10 μm.
Effect of ARAP1 on ubiquitination of the EGFR
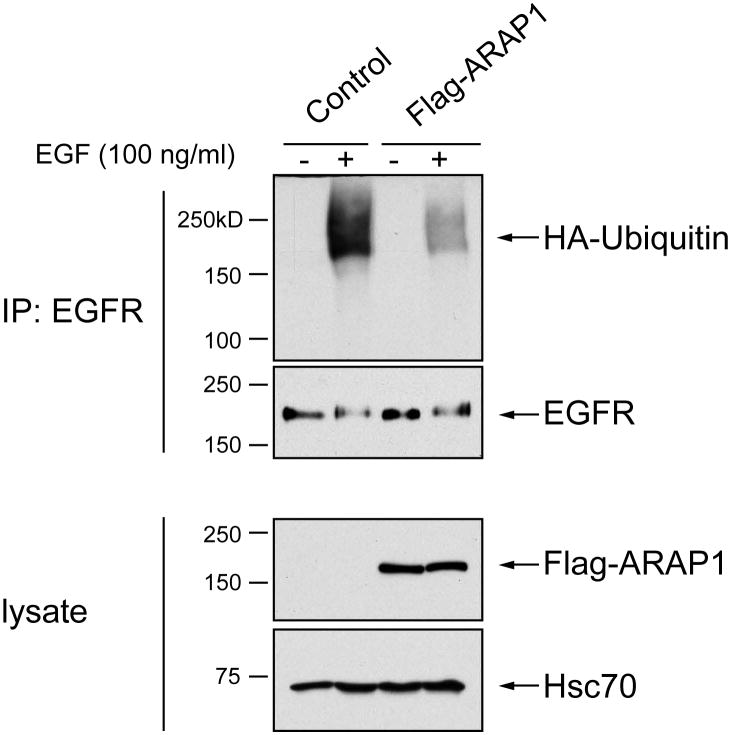
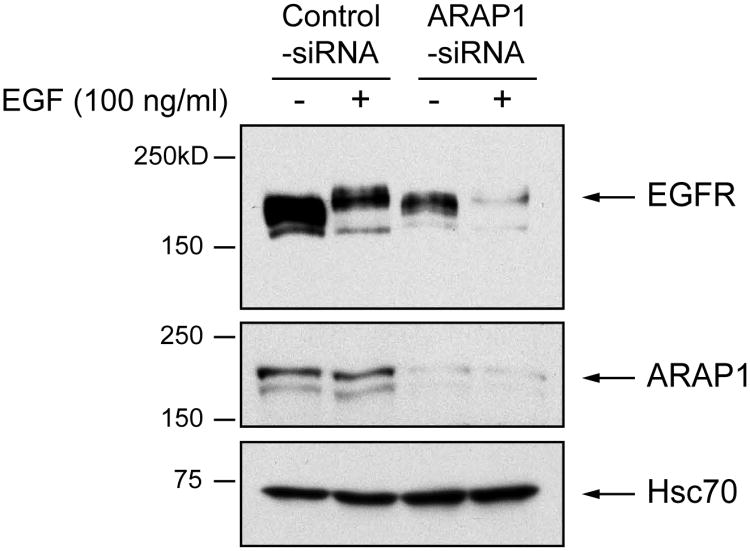
Because of the importance of ubiquitination of EGFR for trafficking by the Cbl/CIN85 pathway, we examined the effect of ARAP1 on ubiquitination of the receptor. We first examined the effect of increased expression of ARAP1. Cells were also transfected with plasmids directing expression of HA-ubiquitin. The cells were lysed after a 10 min treatment with EGF. The amount of ubiquitinated receptor was determined by immunoblot. We found that cells expressing recombinant ARAP1 had less ubiquitinated receptor than controls (Figure 12). We also examined the effect of reduced expression of ARAP1, achieved by using siRNA. As previously reported, EGFR degraded more quickly in cells with decreased ARAP1 expression (Figure 13); however, once normalized for total receptor, we did not observe difference in the extent of ubiquitination (data not shown).
Figure 12. Effect of ARAP1 on EGFR ubiquitination.
HeLa cells were transiently transfected with HA-tagged ubiquitin and either empty vector (control) or Flag-ARAP1. Cells were split into duplicate plates, serum starved and then treated with 100 ng/mL EGF (+) or an equivalent volume of water (−). EGFR was immunoprecipitated with anti-EGFR Ab and immunoblotted for HA-ubiquitin and EGFR, indicated on the right.
Figure 13. Effect of reduced ARAP1 expression on EGFR degradation.
HeLa cells were transfected with the indicated siRNA, split into duplicate plates, serum starved then treated with 100 ng/mL of EGF (+) or the equivalent volume of water (−). A total of 20 μg was immunoblotted for the expression of EGFR, ARAP1 and Hsc70, as indicated.
Discussion
We previously reported that ARAP1 functions in the regulation of endocytic trafficking of the EGFR (Yoon et al., 2008). We found that upon EGF treatment of cells, ARAP1 rapidly and transiently associated with a pre-early endosome that has EGFR, Rab5 and Rabaptin5, but not EEA1. ARAP1, however, affected the rate of association of EGFR with the EEA1 compartment and EGFR degradation. Here, in work designed to elucidate the molecular mechanism linking ARAP1 and the regulation of EGFR endocytosis, we have examined the role of ARAP1 interaction with CIN85, which was identified in a yeast two-hybrid screen for ARAP1 binding partners. Binding of CIN85 to ARAP1 was confirmed in mammalian cells and the binding site was mapped. Overexpression of CIN85 increased the effect of overexpressed ARAP1 on EGF trafficking to the EEA1 compartment and a mutant of ARAP1 deficient in CIN85 binding had a diminished effect on EGF trafficking. Some of our results are at odds with the literature, including the finding that reduction of CIN85 expression enhanced trafficking to the EEA1 compartment. We conclude that ARAP1 functions with CIN85 to affect endocytic trafficking and we speculate on models that can explain that CIN85 accelerates endocytosis, as reported in some studies, but the absence of CIN85 also leads to accumulation of endocytosed EGF in the EEA1 compartment.
Our data support the idea that ARAP1 and CIN85 form a complex that selectively affects the endocytic compartment. ARAP1 is a GTPase activating protein for Arf•GTP. Maintenance of both the Golgi and endocytic compartment depends on Arf•GTP. Blocking formation of Arf•GTP with the fungal metabolite brefeldin A causes mixing of Golgi, ER-to-Golgi intermediate compartment and endoplasmic reticulum proteins and, separately, mixing of transGolgi network and endocytic compartment proteins (Lippincott-Schwartz et al., 1989; Lippincott-Schwartz et al., 1991). Here, coexpression of CIN85 and ARAP1 did not affect the Golgi apparatus but did cause mixing of transGolgi and endocytic proteins, consistent with reduced Arf•GTP levels within the TGN-endocytic pathway. Thus, the compartment we observe is likely an artifact of overexpression and, although it does provide some information about the effects of CIN85 on ARAP1, the morphological data do not provide information about the specific site of action of the complex within the endocytic compartment that would explain effects on EGF traffic.
We first considered the possibility that our results could be explained by ARAP1 competing with another protein for binding to CIN85. One binding partner of CIN85 is the E3, Cbl. Cbl binds to activated EGFR and recruits CIN85. It is thought that the complex of Cbl and CIN85 is necessary for endocytosis (Haglund et al., 2002; d'Azzo et al., 2005; Di Fiore et al., 2003; Levkowitz et al., 1998; Levkowitz et al., 1999). We found no association of ARAP1 and Cbl and that, reduction of Cbl increases the amount of ARAP1/CIN85 complex consistent with mutually exclusive complexes of ARAP1/CIN85 and Cbl/CIN85. Based on these results, we hypothesized that ARAP1 may compete with Cbl for CIN85, thereby slowing traffic in the endocytic pathways toward degradation; however, the simple model of competition is not adequate to explain all of our results. First, reduction of CIN85 had the same effect as reduction of ARAP1: if ARAP1 were competing with Cbl for CIN85, these manipulations should have opposite effects. Second, ARAP1 also affects ubiquitination of EGFR, which occurs independently of CIN85 binding to Cbl (Soubeyran et al., 2002).
The effect of reducing CIN85 highlights the complexities of EGFR trafficking. CIN85 binding to Cbl has been implicated in the internalization step. If this is true, then reducing CIN85 should slow targeting to the EEA1-positive compartment, not increase accumulation in this compartment as we observed. More recently, CIN85 in complex with Dynamin was found to affect trafficking from the early to late endosome (Schroeder et al., 2010). In this case, reduction of CIN85 may cause an accumulation of EGFR in the EEA1-positive compartment. We do not, however, have evidence that ARAP1 is required for this step of trafficking.
One model that might explain the results is that the ARAP1/CIN85 complex has a positive role in directing EGFR from the pre-early endosome to a recycling pathway, away from the pathway to the EEA1-positive compartment/degradative pathway and CIN85/dynamin promotes early endosome to late endosome traffic (Schroeder et al., 2010). It is not clear at what point in endocytic trafficking that ubiquitination occurs relative to the ARAP1 positive endosome, but it may be a relatively late event in trafficking of the receptor. In the model, ARAP1/CIN85 could remove endocytosed EGFR from the pre-early endosome to a rapid recycling compartment prior to ubiquitination. ARAP1, at the same time, could sequester CIN85 from dynamin, thereby slowing early to late endosome traffic and ubiquitination that may occur at this step. This interpretation can explain the result in which ARAP1 overexpression slows movement into the EEA1-positive compartment, reduced ARAP1 expression increases association with EEA1, because it accelerates entry into the compartment, and reduced CIN85 increases association with the EEA1 compartment, because it inhibits exit from the compartment. Another possible explanation for the effect of ARAP1 on ubiquitination of EGFR is related to the finding that ARAP1 activates Cdc42 (Miura et al., 2002). Cdc42 has been reported to sequester c-Cbl by binding Cool1/βPIX, which prevents Cbl-mediated ubiquitination and down-regulation of the EGFR (Wu et al., 2003; Feng et al., 2006). We have not yet examined the effect of the ARAP1/CIN85 complex on cellular levels of Cdc42•GTP.
EGF stimulation increases association of Cbl and CIN85, which is dependent on tyrosine phosphorylation of the C-terminal region of Cbl (Soubeyran et al., 2002). In contrast, the ARAP1/CIN85 complex was not affected by EGF treatment; however, we cannot exclude that ASAP1/CIN85 association is regulated. Because of the limitations of available antibodies, the experiments were performed with overexpressed epitope tagged proteins. The concentrations relative to endogenous proteins might mask interactions. Also, we did not examine whether pathways that can attenuate or potentiate the EGF signaling pathway affected the ARAP1/CIN85 complex, which is a focus of our future work.
Conclusion
In summary, the results of this study support the hypothesis that ARAP1 association with CIN85 regulates endocytic trafficking of the EGFR and may regulate ubiquitination of EGFR.
Materials and methods
Plasmids
A cDNA encoding full-length ARAP1 with a Flag epitope (DYKDDDDK) fused to the N-terminus was cloned into Xho I and Not I sites of the pCI vector (Promega) as described previously (Yoon et al, 2008). Expression vectors for Flag-tagged [SAM], [1-80], [1-90] and [1-100]ARAP1 were generated by inserting a stop codon at the position of amino acid 69, 81, 91, and 101, respectively. The cDNA fragments encoding full length ARAP1 was cloned into the Nhe I and Age I sites of the mammalian expression vector pEGFP-N1 (Clontech). Point mutations in the ARAP1 (P81A, P81/P83/R86A, R86/R90A) were introduced with the QuickChange® II XL kit (Stratagene) according to the manufacturer's protocol. All constructs were confirmed by DNA sequencing. Plasmids for expression of Flag-CIN85 in mammalian cells and for the expression of GST fused full length-, the three SH3 domains (SH3ABC)-, and the proline-rich motif and coiled-coil domain (PCc) of CIN85 in bacterial cells were generous gifts from Dr. Ivan Dikic (Goethe University, Frankfurt, Germany) (Soubeyran et al., 2002).
Yeast Two-Hybrid Screening
Yeast two-hybrid screening was carried out at Myriad Genetics (Salt Lake City, UT) using the SAM or PH1-PH2 domains ofARAP1 (1-100 or 300-562 amino acids of ARAP1, respectively) as a bait with a mating-based method. The corresponding cDNA for SAM or PH1-PH2 domain was cloned into pGBT.superB creating an open reading frame for ARAP1 fragments fused to the GAL4 DNA-binding domain. The bait plasmid was introduced into Myriad's ProNet yeast strain PNY200 (MATα ura3-52 ade2-101 trp1-901 his3-Δ200 leu2-3112 gal4Δ gal80Δ). The bait yeast cells were allowed to mate with Myriad's ProNet MATa yeast cells, BK100 (MATα ura3-52 trp1-901 his3-Δ200 leu2-3112 gal4Δ gal80Δ GAL2-ADE2 LYS2∷GAL1-HIS3 met2∷GAL7-lacZ) containing two independent cDNA libraries from human spleen and thymus. After mating, at least 5 million diploid yeast cells were obtained from each library and selected on His- and Ade-lacking medium. The auxotrophy is suppressed if the bait and prey proteins interact. The prey plasmids were isolated from the positive colonies, and the interaction was confirmed by expression of third reporter gene (lacZ). cDNAs in the positive prey plasmids were sequenced.
Antibodies
Monoclonal anti-EEA1 Ab was purchased from BD Biosciences. Monoclonal anti-M5-Flag and polyclonal anti-Flag Abs were from Sigma-Aldrich. Polyclonal Abs against CIN85 and Cbl and monoclonal anti-Hsc70 were purchased from Santa Cruz Biotechnology. Mouse anti-CIN85 Ab was from Millipore and Rabbit anti-CIN85 Ab was from ProteinTech Group, Inc. Affinity purified polyclonal Ab against ARAP1 have been previous described (Miura et al., 2000). Rabbit anti-GFP and Alexa 488, 594, 647-conjugated donkey anti-mouse or rabbit secondary Abs were from Invitrogen. Anti-mouse or rabbit IgG were purchased from Jackson Immunoresearch. Horseradish-peroxidase-conjugated anti-mouse and anti-rabbit IgG was from Bio-Rad.
Cell Culture and Transfection
HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) at 37°C with 5% CO2. Cell transfection was performed using Lipofectamine 2000 reagent (Invitrogen) following the manufacturer's instructions for both plasmid DNA and siRNA. For RNAi assays, 19-nucleotide siRNA duplexes with 3′-UU overhangs specific for the 3′-untranslated region (3′-UTR) (5′-GUUCAGACCUCUUGGCCCAUU-3′) of human ARAP1 (GenBank® accession number, NM_139181) and c-Cbl siRNA (siGenome SMART pool human c-Cbl (GenBank® accession number, NM_005188, Human CBL #M-003003-01, a mixture of four different siRNAs) were purchased from Dharmacon. CIN85 siRNA were from Qiagen (SI00141316, human SH3KBP1, GenBank® accession number, NM_031892) and the sequence was 5′-AAG ACT GTT ACC ATA TCC CAA-3′. siCONTROL non-targeting siRNA no. 2 (Dharmacon) was used as a negative control. For rescue experiments, cells were first transfected with siRNA using Lipofectamin2000 reagent. 48 h after transfection, cells were transfected with plasmid encoding Flag-ARAP1 and incubated an additional 24 h.
Confocal Microscopy
HeLa cells were plated onto fibronectin (15 μg/ml) coated coverslips for 6 h in OPTi-MEM media and then the cells were fixed in 2% formaldehyde in PBS for 10 min at room temperature. For uptake assays, HeLa cells were plated on glass coverslips in DMEM media containing 10% FBS for overnight and were serum-starved for 1 h at 37°C in pre-warmed uptake media (DMEM containing 20 mM HEPES, pH7.4 and 1% BSA). The cells were incubated with 1 μg/ml of Alexa488- or 555-EGF, a complex of biotinylated EGF with an AlexaFluor 488 or 555 streptavidin obtained from Invitrogen, on ice for 30 min, washed once in ice-cold PBS and then placed in pre-warmed uptake media for 10 min at 37°C. After incubation, cells were placed on ice and incubated in acidic PBS (pH 3.5) for 2 min on ice to remove fluorescent EGF from the cell surface. The cells were washed two times with ice-cold PBS, fixed and processed for immunofluorescence staining using monoclonal anti-EEA1 Ab. Images were captured with a Zeiss LSM510-Meta confocal microscope equipped with a 63×/NA 1.4 oil DIC (Plan-Apochromat) (Carl Zeiss Inc.). The acquired images were processed with Adobe Photoshop. To analyze the colocalization between fluorescent EGF and EEA1, Pearson's coefficients were determined for 20 to 31 cells under the same thresholds using Imaris ×64 software (Bitplane Scientific Software).
Immunoprecipitation
To detect interactions between endogenous ARAP1 and CIN85 proteins, HeLa cells were lysed in lysis buffer which contains 50 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 1% Triton X-100 and complete EDTA-free protease inhibitor (Roche) and incubated with 2 μg of affinity purified polyclonal anti-ARAP1 Ab for 2 h at 4 °C with rotating. Rabbit IgG was used as a negative control. Immune complexes were precipitated by overnight incubation with protein A/G agarose beads (Santa Cruz Biotechnology). After washing the beads with lysis buffer, the immune complexes were eluted in Laemmli sample buffer (Bio-Rad), resolved by 6% SDS-PAGE, analyzed by immunoblotting using specific Abs and detected by SuperSignalWest Dura Extended Duration Substrate (Thermo Scientific). To detect interactions from the cells transiently transfected with plasmids encoding epitope tagged proteins, lysates were incubated with anti-Flag M2 beads (Sigma-Aldrich) for 2 h or polyclonal anti-GFP Ab for 3 h with gamma-bind beads (GE Healthcare) and washed with lysis buffer. The bound proteins were eluted in Laemmli sample buffer and analyzed by SDS-PAGE and immunoblotting.
GST binding assay
20 μg of GST or GST fusion proteins were absorbed on glutathione-Sepharose beads (GE Healthcare), incubated with the lysates (0.5 mg) for 2 h at 4 °C with rotating and washed with 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 5 % glycerol, 0.5 % Triton X-100 and complete EDTA-free protease inhibitor (lysis buffer) for 3 times. The bound proteins were eluted by boiling in Laemmli sample buffer, separated by SDS-PAGE, and processed for immunoblotting with mouse anti-M5-Flag Ab. For determining relative binding affinity, a fixed amount of lysate (0.3 mg) was incubated with the indicated concentrations of GST or GST-CIN85 for 2 h at 4 °C. Flag-ARAP1 in the precipitated beads was detected by immunoblotting using anti-Flag Ab. The amount of GST-fusion proteins was determined by Ponceaus S staining. The signal was quantified using densitometry.
Ubiquitination and Degradation
To assess the effect of ARAP1 expression on EGFR ubiquitination, HeLa cells were transiently transfected with 1 μg of pMT123 HA-ubiquitin plasmid along with 3 μg of either empty pCI or pCI-Flag-ARAP1 plasmid, as described above using Lipofectamine 2000. After 24 h, cells were split and equally divided into two replicate plates. At 36 h post-transfection, cells were starved for 8 h, and one plate was treated with 100ng/mL EGF for 10 min, while the other served as control. Cells were lyzed in lysis buffer (50 mM Tris; pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate and 0.1% SDS supplemented with complete protease inhibitor). A total of 250 μg of whole cell lysate was incubated with 500 ng of monoclonal anti-EGFR (Ab-3) Ab (Calbiochem) overnight at 4°C. Immunoprecipitation was then blotted in duplicate, one for HA using anti-HA-peroxidase (3F10, Roche) to assess ubiquitination, the other for EGFR precipitation using polyclonal anti-EGFR Ab (Cell Signaling).
To assess the effect of ARAP1 reduction on EGFR degradation, HeLa cells were transiently transfected with either non-targeting siRNA or siRNA targeting endogenous ARAP1, as described above. At 48 h post-transfection, cells were split equally into duplicate plates and, at 72 h post-transfection, starved for 8 h then one treated with 100ng/mL EGF for 10 min, the other to serve as an untreated control. Cells were lysed as described and 20 μg of whole cell lysates was immunoblotted for EGFR using anti-EGFR Ab (Cell Signaling), ARAP1 using affinity purified polyclonal Ab, as previously described and Hsc70 using mouse anti-Hsc70 Ab.
Acknowledgments
This work was supported by the National Cancer Institute, National Institutes of Health Intramural Program, Bethesda, MD.
References
- Bogler O, Furnari FB, Kindler-Roehrborn A, Sykes VW, Yung R, Huang HJ, Cavenee WK. SETA: a novel SH3 domain-containing adapter molecule associated with malignancy in astrocytes. Neuro Oncol. 2000;2:6–15. doi: 10.1093/neuonc/2.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borinstein SC, Hyatt MA, Sykes VW, Straub RE, Lipkowitz S, Boulter J, Bogler O. SETA is a multifunctional adapter protein with three SH3 domains that binds Grb2, Cbl, and the novel SB1 proteins. Cell Signal. 2000;12:769–779. doi: 10.1016/s0898-6568(00)00129-7. [DOI] [PubMed] [Google Scholar]
- Campa F, Yoon HY, Ha VL, Szentpetery Z, Balla T, Randazzo PA. ARAP1 binds phosphatidylinositol 3, 4, 5-triphosphate and regulates ArfGAP activity independently of recruitment to the plasma membranes. J Biol Chem. 2009;284:28069–28083. doi: 10.1074/jbc.M109.028266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceresa BP. Regulation of EGFR endocytic trafficking by rab proteins. Histol Histopathol. 2006;21:987–993. doi: 10.14670/HH-21.987. [DOI] [PubMed] [Google Scholar]
- d'Azzo A, Bongiovanni A, Nastasi T. E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic. 2005;6:429–441. doi: 10.1111/j.1600-0854.2005.00294.x. [DOI] [PubMed] [Google Scholar]
- Daniele T, Di Tullio G, Santoro M, Turacchio G, De Matteis MA. ARAP1 regulates EGF receptor trafficking and signaling. Traffic. 2008;9:2221–2235. doi: 10.1111/j.1600-0854.2008.00823.x. [DOI] [PubMed] [Google Scholar]
- Di Fiore PP, Polo S, Hofmann K. When ubiquitin meets ubiquitin receptors: a signaling connection. Nat Rev Mol Cell Biol. 2003;4:491–497. doi: 10.1038/nrm1124. [DOI] [PubMed] [Google Scholar]
- Dikic I. CIN85/CMS family of adaptor molecules. FEBS J. 2002;529:110–115. doi: 10.1016/s0014-5793(02)03188-5. [DOI] [PubMed] [Google Scholar]
- Feng Q, Baird D, Peng X, Wang J, Ly T, Guan JL, Cerione RA. Cool-1 functions as an essential regulatory node for EGF receptor- and Src-mediated cell growth. Nat Cell Biol. 2006;8:945–956. doi: 10.1038/ncb1453. [DOI] [PubMed] [Google Scholar]
- Goh LK, Huang FT, Kim W, Gygi S, Sorkin A. Multiple mechanisms collectively regulate clathrin-mediated endocytosis of the epidermal growth factor receptor. J Cell Biol. 2010;189:871–883. doi: 10.1083/jcb.201001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout I, Middleton G, Adu J, Ninkina NN, Drobot LB, Filonenko V, Matsuka G, Davies AM, Waterfield M, Buchman VL. Negative regulation of PI 3-kinase by Ruk, a novel adaptor protein. EMBO J. 2000;19:4015–4025. doi: 10.1093/emboj/19.15.4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout I, Middleton G, Adu J, Ninkina NN, Drobot LB, Filonenko V, Matsuka G, Haglund K, Schmidt MH, Wong ES, Guy GR, Dikic I. Sprouty2 acts at the Cbl/CIN85 interface to inhibit epidermal growth factor receptor downregulation. EMBO Rep. 2005;6:635–641. doi: 10.1038/sj.embor.7400453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Shimokawa N, Szymkiewicz I, Dikic I. Cbl directed monoubiquitination of CIN85 is involved in regulation of ligand-induced degradation of EGF receptors. Proc Natl Acad Sci USA. 2002;99:12191–12196. doi: 10.1073/pnas.192462299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- I ST, Nie Z, Stewart A, Najdovska M, He H, Randazzo PA, Lock P. ARAP3 is transiently tyrosine phosphorylated in cells attaching to fibronectin and inhibits cell spreading in a RhoGAP-dependent manner. J Cell Sci. 2004;117:6071–6084. doi: 10.1242/jcs.01526. [DOI] [PubMed] [Google Scholar]
- Jorrissen RN, Walker F, Pouliot N, Garrett TP, Ward CW, Burgess AW. Epidermal growth factor receptor: mechanisms of activation and signaling. Exp Cell Res. 2003;284:31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- Kim H, Muller WJ. The role of the epidermal growth factor receptor family in mammary tumorigenesis and metastasis. Exp Cell Res. 1999;253:78–87. doi: 10.1006/excr.1999.4706. [DOI] [PubMed] [Google Scholar]
- Klapper LN, Kirschbaum MH, Sela M, Yarden Y. Biochemical and clinical implications of the ErbB/HER signaling network of growth factor receptors. Adv Cancer Res. 2000;77:25–79. [PubMed] [Google Scholar]
- Kowanetz K, Terzic J, Dikic I. Dab2 links CIN85 with clathrin-mediated receptor internalization. FEBS Lett. 2003;554:81–87. doi: 10.1016/s0014-5793(03)01111-6. [DOI] [PubMed] [Google Scholar]
- Krugmann S, anderson KE, Ridley SH, Risso N, McGregor A, Coadwell J, Davidson K, Eguinoa A, Ellson CD, Lipp P, Manifava M, Ktistakis N, Painter G, Thuring JW, Cooper MA. Identification of ARAP3, a novel PI3K effector regulating both Arf and Rho GTPases by selective capture on phosphoinositide affinity matrices. Mol Cell. 2002;9:95–108. doi: 10.1016/s1097-2765(02)00434-3. [DOI] [PubMed] [Google Scholar]
- Kurakin AV, Wu S, Bredesen DE. Atypical recognition consensus of CIN85/SETA/Ruk SH3 domains revealed by target-assisted iterative screening. J Biol Chem. 2003;278:34102–34109. doi: 10.1074/jbc.M305264200. [DOI] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon WY, Beguinot L, Geiger B, Yarden Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, Lipkowitz S, Yarden Y. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Yuan L, Tipper C, Amherdt M, ORCI L, Klausner RD. Brefeldin-A's Effects on Endosomes, Lysosomes, and the TGN Suggest a General Mechanism for Regulating Organelle Structure and Membrane Traffic. Cell. 1991;67:601–616. doi: 10.1016/0092-8674(91)90534-6. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid Redistribution of Golgi Proteins Into the ER in Cells Treated with Brefeldin-A - Evidence for Membrane Cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Jacques KM, Stauffer S, Kubosaki A, Zhu K, Hirsch DS, Resau J, Zheng Y, Randazzo PA. ARAP1: a point of convergence for Arf and Rho signaling. Mol Cell. 2002;9:109–119. doi: 10.1016/s1097-2765(02)00428-8. [DOI] [PubMed] [Google Scholar]
- Mosesson Y, Shtiegman K, Katz M, Zwang Y, Vereb G, Szollosi J, Yarden Y. Endocytosis of receptor tyrosine kinase is driven by monoubiquitylation, not polyubiquitylation. J Biol Chem. 2003;278:21323–21326. doi: 10.1074/jbc.C300096200. [DOI] [PubMed] [Google Scholar]
- Mosesson Y, Mills GB, Yarden Y. Derailed endocytosis: an emerging feature of cancer. Nat Rev Cancer. 2008;8:835–850. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- Narita T, Amano F, Yoshizaki K, Nishimoto N, Nishimura T, Tajima T, Namiki H, Taniyama T. Assignment of SH3KBP1 to human chromosome band Xp22.1 → p21.3 by in situ hybridization. Cytogenet Cell Genet. 2001;93:133–134. doi: 10.1159/000056966. [DOI] [PubMed] [Google Scholar]
- Ono M, Kuwano M. Molecular mechanisms of epidermal growth factor receptor (EGFR) activation and response to gefitinib and other EGFR-targeting drugs. Clin Cancer Res. 2006;12:7242–7251. doi: 10.1158/1078-0432.CCR-06-0646. [DOI] [PubMed] [Google Scholar]
- Petrelli A, Gilestro GF, Lanzardo S, Comoglio PM, Migone N, Giordano S. The endophilin–CIN85–Cbl complex mediates ligand-dependent downregulation of c-Met. Nature. 2002;416:187–190. doi: 10.1038/416187a. [DOI] [PubMed] [Google Scholar]
- Randazzo PA, Inoue H, Bharti S. ArfGAPs as regulators of the actin cytoskeleton. Biol Cell. 2007;99:583–600. doi: 10.1042/bc20070034. [DOI] [PubMed] [Google Scholar]
- Robinson MS, Bonifacino JS. Adaptor-related proteins. Curr Opin Cell Biol. 2001;13:444–453. doi: 10.1016/s0955-0674(00)00235-0. [DOI] [PubMed] [Google Scholar]
- Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol. 1995;19:183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- Schroeder B, Weller SG, Chen J, Billadeau D, McNiven MA. A Dyn2-CIN85 complex mediates degradative traffic of the EGFR by regulation of late endosomal budding. EMBO J. 2010;29:3039–3053. doi: 10.1038/emboj.2010.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2009;315:683–696. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PubMed] [Google Scholar]
- Sorkin A, von Zastrow M. Endocytosis and signaling: intertwining molecular networks. Nat Rev Mol Cell Biol. 2009;10:609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubeyran P, Kowanetz K, Szymkiewicz I, Langdon WY, Dikic I. Cbl-CIN85-endphilin complex mediates ligand-induced downregulation of EGF receptors. Nature. 2002;416:183–187. doi: 10.1038/416183a. [DOI] [PubMed] [Google Scholar]
- Szymkiewicz I, Kowanetz K, Soubeyran P, Dinarina A, Lipkowitz S, Dikic I. CIN85 participates in Cbl-b-mediated down-regulation of receptor tyrosine kinases. J Biol Chem. 2002;277:39666–39672. doi: 10.1074/jbc.M205535200. [DOI] [PubMed] [Google Scholar]
- Take H, Watanabe S, Takeda K, Yu ZX, Iwata N, Kajigaya S. Cloning and Characterization of a Novel Adaptor Protein, CIN85, That interacts with c-Cbl. Biochem Biophys Res Commun. 2000;268:321–328. doi: 10.1006/bbrc.2000.2147. [DOI] [PubMed] [Google Scholar]
- Thien CB, Walker F, Langdon WY. RING finger mutations that abolish c-Cbl-directed polyubiquitination and downregulation of the EGF receptor are insufficient for cell transformation. Mol Cell. 2001;7:355–365. doi: 10.1016/s1097-2765(01)00183-6. [DOI] [PubMed] [Google Scholar]
- Wong AJ, Ruppert JM, Bigner SH, Grzeschik C, Humphrey PA, Bigner DS, Vogelstein B. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci USA. 1992;89:2965–2969. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WJ, Tu S, Cerione RA. Activated Cdc42 sequesters c-Cbl and prevents EGF receptor degradation. Cell. 2003;114:715–725. doi: 10.1016/s0092-8674(03)00688-3. [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signaling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- Yoon HY, Lee JS, Randazzo RA. ARAP1 regulates endocytosis of EGFR. Traffic. 2008;9:2236–2252. doi: 10.1111/j.1600-0854.2008.00839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HY, Miura K, Cuthbert EJ, Davis KK, Ahvazi B, Casanova JE, Randazzo PA. ARAP2 effects on the actin cytoskeleton are dependent on Arf6-specific GTPase-activating-protein activity and binding to RhoA-GTP. J Cell Sci. 2006;119:4650–4666. doi: 10.1242/jcs.03237. [DOI] [PubMed] [Google Scholar]