Abstract
5-methylcytosine (5mC) in DNA plays an important role in gene expression, genomic imprinting, and suppression of transposable elements. 5mC can be converted to 5-hydroxymethylcytosine (5hmC) by the Tet proteins. Here we show that, in addition to 5hmC, the Tet proteins can generate 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) from 5mC in an enzymatic activity-dependent manner. Furthermore, we reveal the presence of 5fC and 5caC in genomic DNA of mouse ES cells and mouse organs. The genomic content of 5hmC, 5fC, and 5caC can be increased or reduced through overexpression or depletion of Tet proteins. Thus, we identify two new cytosine derivatives in genomic DNA as the products of Tet proteins. Our study raises the possibility that DNA demethylation may occur through Tet-catalyzed oxidation followed by decarboxylation.
Although enzymes that catalyze DNA methylation process are well-studied (1), how DNA demethylation is achieved is less known, especially in animals (2, 3). A repair-based mechanism is used in DNA demethylation in plants, but whether a similar mechanism is also used in mammalian cells is unclear (3, 4). Identification of 5hmC as the 6th base of the mammalian genome (5, 6) and the capacity of Tet (ten_eleven_translocation) proteins to convert 5mC to 5hmC in an Fe(II) and alpha-ketoglutarate (α-KG)-dependent oxidation reaction (6, 7) raised the possibility that a Tet-catalyzed reaction might be part of the DNA demethylation process.
A potential 5mC demethylation mechanism can be envisioned from similar chemistry for thymine to uracil conversion (3, 8, 9) (Fig. S1A) with the Tet proteins oxidizing 5mC not only to 5hmC, but also to the aldehyde (5fC) and potentially the carboxylic acid (5caC) forms (Fig. S1B). The failure to detect such reaction products may simply be due to the limitations of the previous assay employed (6, 7). To determine if this might be the case, we synthesized 20mer DNA oligos with 5fC or 5caC in the internal C of an MspI site (10) and found that although MspI is efficient in digesting the oligo DNAs with C/5mC/5hmC in the internal C, it failed to digest the DNA containing 5fC or 5caC (Fig. S2A-B). Thus, if Tet proteins have the capacity to convert 5mC to 5fC or 5caC, these products would have evaded detection due to the inability of MspI to digest 5fC or 5caC-containing DNA. To overcome this problem, we identified and demonstrated that TaqI is capable of digesting DNA modified with 5mC, 5hmC (11), 5fC or 5caC (Fig. S2C-D).
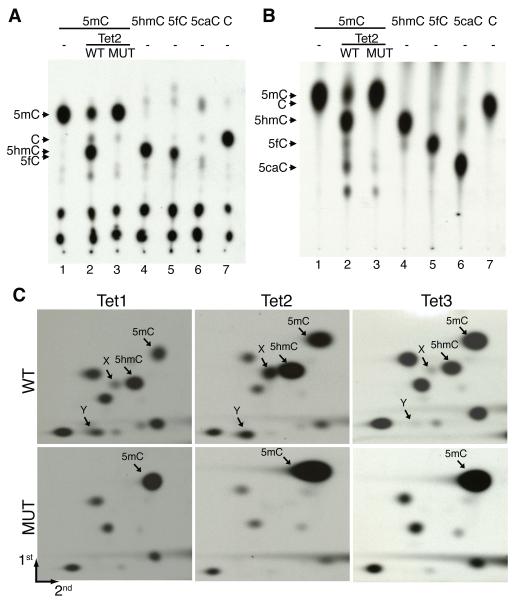
In addition to restriction enzyme, TLC conditions can also affect the detection of 5fC and 5caC. Under previous TLC conditions (7), 5hmC and 5fC have almost identical migration patterns (Fig. 1A, lanes 4 and 5) and 5caC failed to migrate (Fig. 1A, lane 6). Using a more acidic TLC buffer, all cytosine derivatives migrated (Fig. 1B, lanes 4-7). However, 5mC and C cannot be separated under this condition (Fig. 1B, lanes 1 and 7). Given that the TLC buffer used in Fig. 1A can separate C from 5mC, two-dimensional TLC (2D-TLC) using the two buffer conditions should allow for separation of cytosine and its derivatives.
Figure 1. Optimization of conditions for detection of cytosine and its 5-position modified forms by TLC.
(A)Migration of labeled C and its 5-position modified forms by TLC under the first developing buffer. Lanes 1-3 serve as controls for the migration of 5mC and 5hmC generated from DNA oligos incubated with wild-type or catalytic mutant Tet2.
(B) The same samples used in panel A were separated by TLC under the second developing buffer. With the exception of 5mC and C, all of the other forms of C can be separated under this condition.
(C) Autoradiographs of 2D-TLC analysis of samples derived from 5mC-containing TaqI 20mer oligo DNA incubated with wild-type and catalytic-deficient mutant Tet1, Tet2, and Tet3.
Using TaqI digestion and 2D-TLC (Fig. S3), we analyzed the enzymatic activity of the Tet proteins. Compared with the mutant control, incubation of the Tet1 protein with 5mC-containing substrate resulted in a decrease in the 5mC level concomitant with the appearance of a radioactive spot that correlates with 5hmC (Fig. 1C, left two panels). Two additional radioactive spots, labeled “X” and “Y” whose appearance depend on Tet1 enzymatic activity, were observed. Similarly, Tet2 and Tet3 also generated three enzymatic activity-dependent radioactive spots that were detected in Tet1-catalyzed reaction although the signal that corresponds to the “Y” spot from the Tet3 reaction is extremely weak (Fig. 1C, middle and right panels).
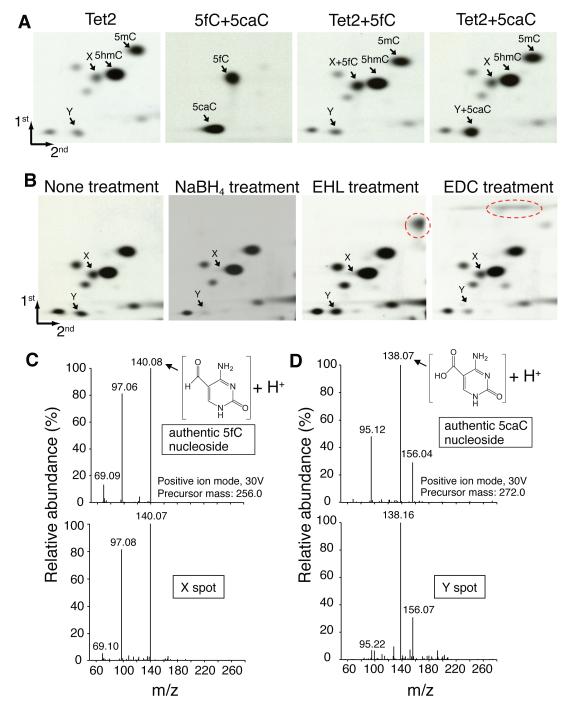
If our hypothetical model for DNA demethylation is correct (Fig. S1B), the “X” and “Y” spots are likely to be 5fC and 5caC. We compared the migration patterns of 5fC and 5caC with that of Tet2-treated 5mC-containing DNA substrates and found that the “X” and “Y” spots match 5fC and 5caC with respect to their migration (Fig. 2A, compare the first two panels). We further confirmed this by mixing radioactive 5fC (third panel) or 5caC (last panel) with the samples used in the first panel before performing 2D-TLC. To confirm the identities of the “X” and “Y” spots, we treated the Tet2-catalyzed reaction mixture with sodium borohydride (NaBH4), which resulted in the disappearance of both “X” and “Y” spots concomitant with increase in 5hmC (Fig. 2B, compare the first two panels) indicating that both are oxidation products of 5hmC, consistent with the notion that they are 5fC and 5caC.
Figure 2. Tet proteins are capable of converting 5mC to 5hmC, 5fC, and 5caC.
(A)“X” and “Y” co-migrate with 5fC and 5caC on 2D-TLC, respectively. Left panel shows the migration pattern of the Tet2 reaction mixture on 2D-TLC. The locations of 5mC, 5hmC, “X”, and “Y” are indicated. Second panel shows the locations of control 5fC and 5caC in a parallel 2D-TLC assay. Third and fourth panels contain samples used in the first panel plus radioactive 5fC or 5caC, respectively.
(B) Confirmation of the identities of “X” and “Y” by chemical treatments. Left panel shows the migration pattern of samples derived from incubation of Tet2 with the 5mC-containing TaqI 20mer oligo DNA. Second panel demonstrates that treatment of the samples used in the left panel with NaBH4. Third and fourth panels demonstrate that EHL and EDC respectively react with the formyl group of 5fC or the carboxyl group of 5caC to generate the new products indicated by the dotted circles.
(C) Mass spectrometric analysis demonstrates “X” has the same fingerprint as 5fC.
(D) Mass spectrometric analysis demonstrates “Y” has the same fingerprint as 5caC.
O-ethylhydroxylamine hydrochloride (EHL) and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) react with formyl and carboxyl groups to generate oximes and amides, respectively (12, 13) (Fig. S4). To determine the migration patterns of the reaction products, we performed reactions using standard 5fC and 5caC, and separated the products by 2D-TLC, establishing the migration pattern for oxime (Fig. S4A) and amide (Fig. S4B). Similar EHL treatment of the Tet2 reaction mixture specifically converted the “X” spot to a new spot that co-migrated with oxime (Fig. 2B, compare panels 1, 3 and Fig. S4A). In contrast, EDC treatment specifically converted the “Y” spot to a new signal that co-migrated with amide (Fig. 2B, compare panels 1, 4, and Fig. S4B). To unequivocally define the identities of “X” and Y”, we employed mass spectrometry. Having established the mass spectrometry fingerprints of standard 5fC and 5caC (Fig. 2C, D, top panels), we extracted the “X” and “Y” spots and subjected them to mass spectrometric analysis. The “X” spot shows the same major fragment ions as that of 5fC, while the “Y” spot shows the same major fragment ions as that of 5caC. Collectively, 2D-TLC co-migration, chemical treatment, and mass spectrometry fingerprints demonstrate that Tet proteins not only can convert 5mC to 5hmC, but also can further oxidize 5hmC to 5fC and 5caC.
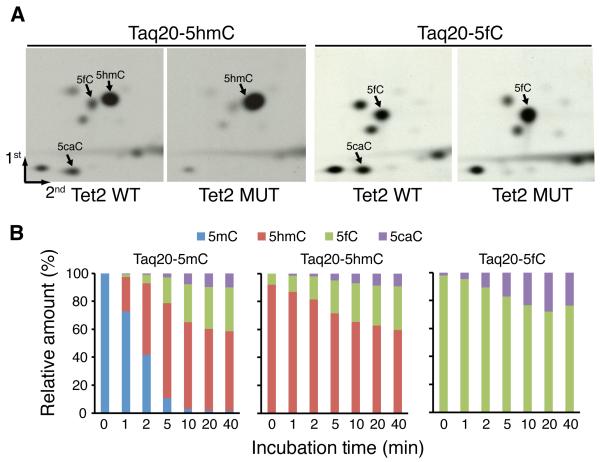
To determine if Tet proteins can use 5hmC or 5fC-containing DNA as substrates, 20mer DNA oligos with either 5hmC or 5fC in the TaqI site were incubated with Tet proteins. 2D-TLC analysis demonstrated that incubation with wild-type Tet proteins, but not the catalytic mutants, resulted in a decrease in the level of 5hmC/5fC concomitant with the appearance of 5fC and 5caC, or 5caC (Figs. 3A, S5) suggesting that Tet proteins can act upon 5hmC and 5fC-containing substrates. However, the 5caC signal generated by Tet3 is extremely weak.
Figure 3. Kinetic analysis of Tet2 using 5mC, 5hmC, and 5fC-containing oligo DNAs.
(A)Autoradiographs of 2D-TLC analysis of samples derived from 5hmC, or 5fC-containing TaqI 20mer DNA oligos incubated with wild-type or catalytic-deficient mutant Tet2.
(B) Relative percentage of 5mC, 5hmC, 5fC, and 5caC at different time points after incubation of Tet2 proteins with 5mC, 5hmC, or 5fC-containing TaqI 20mer DNA oligos.
We used a quantitative mass spectrometric assay to rule out the possibility that 5fC and 5caC are generated as a side reaction by Tet proteins. We generated a standard curve for each of the cytosine derivatives by mixing different amounts of each 5mC, 5hmC, 5fC, and 5caC followed by LC-MS (Fig. S6). We then quantified the cytosine derivatives at different time points after incubating Tet2 with 5mC, 5hmC, or 5fC-containing DNA substrates. Quantification of the relative amount of the substrate and the various products during the reaction process demonstrated that the reaction plateaued after 10 min of incubation regardless whether 5mC, 5hmC, or 5fC-containing TaqI 20mer DNA is used as a substrate (Fig. 3B). The reaction plateaued in 10 min due to the inactivation of the Tet2 enzyme during the incubation (Fig. S7).
During this period, Tet2 is able to convert more than 95% of the 5mC to 5hmC (~60%), 5fC (~30%), and 5caC (5%), but it can only convert about 40% or 25% when 5hmC or 5fC-contianing DNA was used as a substrate (Fig. 3B). From this data we calculated the initial reaction rate of Tet2 for 5mC, 5hmC, and 5fC-containing substrates to be 429 nM/min, 87.4 nM/min, and 56.6 nM/min, respectively (Fig. S8). Although Tet2 has a clear preference for the 5mC-containing DNA substrate, its initial reaction rate for 5hmC and 5fC-containing substrate is only 4.9-7.6 fold lower. The fact that there is clear accumulation of 5fC and 5caC when 5mC is used as a substrate (Fig. 3B, top panel) strongly suggests that Tet-catalyzed iterative oxidation is likely a kinetically relevant pathway.
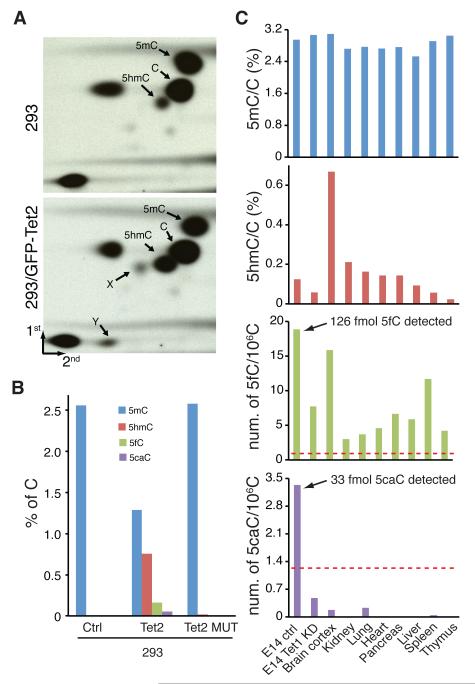
To determine whether Tet-catalyzed iterative oxidation of 5mC can take place in vivo, we transfected a mammalian expression construct containing the Tet2 catalytic domain fused to GFP into HEK293 cells. After FACS sorting, genomic DNA of GFP positive cells was analyzed for the presence of 5hmC, 5fC, and 5caC by 2D-TLC (Fig. S3). Compared with the untransfected control, cells expressing Tet2 not only have increased 5hmC levels, but also contain two additional spots (Fig. 4A), which correspond to 5fC and 5caC, respectively. In addition, we quantified the genomic content of 5hmC, 5fC, and 5caC following the procedure depicted in Fig. S9A (14). After establishing the retention times for each of the cytosine derivatives on HPLC (Fig. S9B, top panel), nucleosides derived from genomic DNA were subjected to the same HPLC conditions for fractionation. Fractions A and B (Fig. S9B) that have the same retention times as that of 5caC and 5hmC or 5fC were collected. Mass spectrometry analysis demonstrates that both 5fC and 5caC are detected in the genomic DNA of cells overexpressing Tet2 (Fig. S10A). By comparison to the standard curves (Fig. S11A), overexpression of wild-type Tet2, but not a catalytic mutant, increased the genomic content of 5hmC, 5fC and 5caC (Fig. 4B).
Figure 4. 5fC and 5caC are present in genomic DNA and their abundance is regulated by Tet proteins.
(A)Genomic DNA prepared from either HEK293 cells or HEK293 cells expressing a GFP-tagged Tet2 were digested with TaqI, end labeled with T4 polynucleotide kinase, digested with DNase I and phosphodiesterase I, and analyzed by 2D-TLC.
(B) Mass spectrometric quantification of genomic content of 5mC, 5hmC, 5fC and 5caC relative to cytosine in HEK293 cells expressing GFP-tagged wild-type or a catalytic mutant Tet2.
(C)Mass spectrometric quantification of genomic content of 5mC, 5hmC, 5fC and 5caC relative to cytosine in mouse ES cells, Tet1 knockdown ES cells, and mouse organs. Shown are averages of two biologically independent experiments. The red dotted lines indicate the limits for accurate quantification, which are 0.8 5fC/106C and 1.2 5caC/106C in 20 μg of genomic DNA.
Next, we asked whether 5fC and 5caC are present in genomic DNA under physiological conditions. Using a similar approach as that used for the genomic DNA of Tet2-overexpressing HEK293, we show that not only 5hmC, but also 5fC and 5caC are present in the genomic DNA of mouse ES cells (Fig. S10B). To quantify the genomic content of 5hmC, 5fC and 5caC in mouse ES cells, we generated standard curves for each of the 5mC derivatives at low concentrations and determined the limit of detection for 5fC and 5caC to be 5 fmol and 10 fmol, respectively (Fig. S11). We then quantified the genomic content of these cytosine derivatives in mouse ES cells to be about 1.3×103 5hmC, 20 5fC, and 3 5caC in every 106 C (Fig. 4C, Table S1). Knockdown of Tet1 reduced the genomic content of 5hmC, as well as 5fC and 5caC (Fig. 4C) indicating that Tet1 is at least partially responsible for the generation of these cytosine derivatives. The presence of 5fC is not limited to ES cells as similar analysis also revealed their presence in genomic DNA of major mouse organs (Fig. 4C). However, 5caC can be detected with confidence only in ES cells (Fig. 4C, S10B).
Here we demonstrate that the Tet family of proteins have the capacity to convert 5mC not only to 5hmC, but also to 5fC and 5caC in vitro. In addition, we provide evidence for the presence of 5fC in the genomic DNA of mouse ES cells and organs and for the presence of 5caC in moue ES cells. We note that a similar study failed to detect their existence in genomic DNA of mouse organs (15) likely due to the differences in the detection limits between the two studies (pmol vs fmol). The Tet-catalyzed oxidation reaction is reminiscent of the thymine hydroxylase catalyzed conversion of thymine to iso-orotate (8, 9) (Fig. S1) raising the possibility that 5mC demethylation could be potentially achieved through a process similar to the conversion of thymine to uracil, which is achieved by conversion of thymine to iso-orotate followed by decarboxylation by the iso-orotate decarboxylase (8, 9). Although this hypothetic pathway for DNA demethylation is simple and appealing, the enzyme that is capable of decarboxylating 5caC-containing DNA has yet to be identified. Until such an enzyme is identified, we cannot rule out the possibility that the Tet family enzymes act together with other putative DNA demethylation pathways, such as the base excision DNA repair (BER) pathway. Indeed, recent studies have provided some supporting evidence for such a possibility (16, 17).
Supplementary Material
Acknowledgments
We thank Qisheng Zhang for suggestion of the NaBH4 experiment; Chun-Xiao Song for help in oligo purification. This work was supported by NIH grants GM68804 (Y.Z.), GM071440 (C.H.), P42ES5948 and P30ES10126 (J.A.S.). S.I. is a research fellow of the Japan Society for the Promotion of Science. Y.Z. is an Investigator of the Howard Hughes Medical Institute.
References and notes
- 1.Goll MG, Bestor TH. Annu Rev Biochem. 2005;74:481. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 2.Ooi SK, Bestor TH. Cell. 2008 Jun 27;133:1145. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Wu SC, Zhang Y. Nat Rev Mol Cell Biol. 2010 Sep;11:607. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gehring M, Reik W, Henikoff S. Trends Genet. 2009 Feb;25:82. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Kriaucionis S, Heintz N. Science. 2009 May 15;324:929. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tahiliani M, et al. Science. 2009 May 15;324:930. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito S, et al. Nature. 2010 Jul 18;466:1129. [Google Scholar]
- 8.Neidigh JW, Darwanto A, Williams AA, Wall NR, Sowers LC. Chem Res Toxicol. 2009 May;22:885. doi: 10.1021/tx8004482. [DOI] [PubMed] [Google Scholar]
- 9.Smiley JA, Kundracik M, Landfried DA, Barnes VR, Sr., Axhemi AA. Biochim Biophys Acta. 2005 May 25;1723:256. doi: 10.1016/j.bbagen.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Dai Q, He C. Org Lett. 2011 Jul 1;13:3446. doi: 10.1021/ol201189n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang LH, Farnet CM, Ehrlich KC, Ehrlich M. Nucleic Acids Res. 1982 Mar 11;10:1579. doi: 10.1093/nar/10.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kukushkin VY, Pombeiro AJL. Coordination Chemistry Reviews. 1999;181:147. [Google Scholar]
- 13.Williams A, Hill SV, Ibrahim IT. Anal Biochem. 1981 Jun;114:173. doi: 10.1016/0003-2697(81)90470-x. [DOI] [PubMed] [Google Scholar]
- 14.Boysen G, et al. J Chromatogr B Analyt Technol Biomed Life Sci. 2010 Feb 1;878:375. doi: 10.1016/j.jchromb.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Globisch D, et al. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortellino S, et al. Cell. 2011 Jul 8;146:67. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo JU, Su Y, Zhong C, Ming GL, Song H. Cell. 2011 Apr 29;145:423. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.