Abstract
Epigenetic deregulation of gene expression plays a role in the initiation and progression of prostate cancer (PCa). The histone methyltransferase MMSET/WHSC1 (Multiple Myeloma Set Domain) is overexpressed in a number of metastatic tumors, but its mechanism of action has not been defined. In this work, we found that PCa cell lines expressed significantly higher levels of MMSET compared to immortalized, non-transformed prostate cells. Knockdown experiments showed that, in metastatic PCa cell lines, dimethylation of lysine 36 and trimethylation of lysine 27 on histone H3 (H3K36me2 and H3K27me3, respectively) depended on MMSET expression, while depletion of MMSET in benign prostatic cells did not affect chromatin modifications. Knockdown of MMSET in DU145 and PC-3 tumor cells decreased cell proliferation, colony formation in soft agar, and strikingly diminished cell migration and invasion. Conversely, overexpression of MMSET in immortalized, non-transformed RWPE-1 cells promoted cell migration and invasion, accompanied by an epithelial to mesenchymal transition (EMT). Among a panel of EMT-promoting genes analyzed, TWIST1 expression was strongly activated in response to MMSET. Chromatin immunoprecipitation analysis demonstrated that MMSET binds to the TWIST1 locus, leading to an increase in H3K36me2, suggesting a direct role of MMSET in the regulation of this gene. Depletion of TWIST1 in MMSET-overexpressing RWPE-1 cells blocked cell invasion and EMT, indicating that TWIST1 was a critical target of MMSET, responsible for the acquisition of an invasive phenotype. Collectively, these data suggest that MMSET plays a role in PCa pathogenesis and progression through epigenetic regulation of metastasis-related genes.
Keywords: MMSET, histone methylation, epithelial-mesenchymal transition, invasion, prostate cancer
Introduction
Prostate Cancer (PCa) remains the second leading cause of cancer deaths among men in the US (1). A large burden of PCa mortality is related to advanced, metastatic disease when androgen-depletion therapy becomes ineffective. A better understanding of the mechanisms of progression of PCa and the acquisition of metastatic properties is needed to identify new therapeutic targets.
Increasing evidence shows that epigenetic changes have an important role in the initiation and progression of PCa (2–4). While aberrant DNA methylation has been the main focus of the epigenetic studies of PCa (5), recent findings suggest that chromatin remodeling and post-translational histone modifications are also important in the deregulation of gene expression in this disease (2–4). The N-terminal tails of histones are subjected to multiple reversible post-translational modifications such as methylation, acetylation, phosphorylation, ubiquitination and sumoylation (6–9). The combination of these modifications on specific residues results in changes in the conformation of the chromatin and in the binding of transcriptional cofactors, regulating gene expression. The deregulation of several enzymes that control histone modifications has been described in PCa, including histone deacetylases (HDACs) such as HDAC1, histone demethylases (HDMs) like LSD1, and histone methyltransferases (HMTs) like EZH2 (10–13). Furthermore, the global state of several histone modifications has been shown to have prognostic importance in this disease (13–15).
MMSET (WHSC1, NSD2) is a HMT that was first identified as a candidate gene for Wolf-Hirschhorn Syndrome (WHS), characterized by fetal overgrowth and malformations (16). MMSET belongs to a family of NSD (nuclear receptor SET domain) proteins, whose members possess a SET domain encoding lysine methyltransferase activity (17). Although the substrate specificity of NSD proteins was controversial (18), recently it has been narrowed down to dimethylation of histone H3 lysine 36 (19, 20), a mark commonly associated with active transcription. The MMSET gene is composed of 25 exons and undergoes alternative splicing to yield two major species: MMSET I and MMSET II. While MMSET II gives rise to the full-length protein, MMSET I is a shorter isoform that lacks the c-terminal region, encoding for a protein that does not posses the SET domain or HMT activity (16, 21, 22). The importance of MMSET in malignancy was first highlighted by characterization of the t(4;14) translocation in about 15–20% of Multiple Myeloma (MM), which fuses the MMSET gene to the immunoglobulin heavy chain promoter/enhancer, leading to dramatic up-regulation of MMSET expression (23). MMSET regulates growth, adhesion and clonogenicity of MM cell lines, and contributes to tumor maintenance in vivo (19, 24–26). Our group previously showed that in MM cell lines, MMSET produces aberrantly high global levels of H3K36me2 and low levels of H3K27me3, which correlates with an altered gene expression profile and changes in chromatin accessibility (19).
MMSET is also overexpressed in solid tumors, including breast cancer and glioblastoma (18, 27–32). In general, MMSET up-regulation was associated with aggressive tumor behavior and poor prognosis (28, 29). Two groups showed that MMSET mRNA was overexpressed in PCa relative to levels in normal tissues (29, 30). Moreover, analysis of Oncomine datasets showed that MMSET expression was elevated in Gleason grade 7–9 tumors compared to tumors at grade 6 and below, and that high level expression of this factor was associated with PCa recurrence (29). These findings suggested that MMSET may be involved in PCa pathogenesis and progression, however, the biological role of MMSET in this disease and the nature of the genes it may regulate remained to be elucidated.
Here we report that similarly to MM, MMSET overexpression in PCa influences H3K36me2 and H3K27me3 methylation. Overexpression of MMSET in immortalized prostatic epithelial cells leads to a striking increase in migration and invasion, and changes in cell morphology and gene expression consistent with an epithelial-mesenchymal transition (EMT). These effects are mediated by the ability of MMSET to activate expression of TWIST1, a gene implicated in tumor-associated EMT and invasion (33, 34). Collectively, our data suggest that MMSET contributes to PCa progression by the aberrant epigenetic regulation of genes that drive the metastatic phenotype.
Results
MMSET Is Overexpressed in PCa Cell Lines and Influences Global Histone Methylation
MMSET mRNA was shown to be upregulated in PCa specimens (29, 30). To determine whether PCa is associated with increased expression of MMSET protein, lysates from two immortalized but non-transformed prostate epithelial cell lines (RWPE-1 and BPH1), and several PCa cell lines (LNCaP, PC-3, DU145, VCaP and 22Rv1) were subjected to immunoblot. MMSET expression was clearly upregulated in all the PCa cell lines analyzed compared to BPH1 and RWPE-1 cells (Figure 1A), with the metastasis-derived DU145 cell line displaying the highest levels.
Figure 1.
MMSET is overexpressed in PCa cell lines and influences global histone methylation. (A) Nuclear extracts from prostate cell lines were immunoblotted with the indicated antibodies, and quantified by densitometry. MMSET levels were normalized to histone H4 and represented as relative to levels in BPH1 cells. (B) RWPE-1 and BPH1 cells were infected with a control retrovirus, or viruses harboring MMSET I, MMSET II, or SET domain mutant MMSET II (Y1118A). Nuclear proteins were immunoblotted as indicated. (C–D) PCa DU145, PC-3 and 22Rv1 cell lines (C), and non-transformed RWPE-1 and BPH1 cells (D) were transfected with the indicated siRNAs. Nuclear extracts were prepared 6 days after transfection and immunoblotted with the indicated antibodies.
MMSET alters global histone methylation patterns in MM, increasing H3K36 methylation and decreasing H3K27 methylation (19). By contrast, in prostate cells, no clear correlation between the expression of MMSET and the levels of histone methylation was observed (Figure 1A). However, to determine if MMSET still had the ability to alter chromatin in PCa, gain- and loss-of-function cellular models were generated. Benign, immortalized RWPE-1 cells with low basal levels of MMSET (Figure 1A), were engineered to overexpress MMSET I, wild-type MMSET II or SET domain mutant MMSET II (Y1118A). Immunoblot analysis showed that, as in MM, overexpression of wild-type MMSET II elevated H3K36me2 and depressed H3K27me3 levels. By contrast, MMSET I, which lacks the SET domain, and mutant MMSET II (26), failed to induce a H3K36me2/H3K27me3 switch (Figure 1B). These results were confirmed in the immortalized, non-transformed BPH1 cells (Figure 1B).
To generate a loss–of–function model, PCa cell lines DU145, PC-3 and 22Rv1 were transfected with a pool of siRNAs targeting both isoforms of MMSET (siMMET pool), a C-terminal siRNA targeting MMSET II (siMMSET C-ter), or a control siRNA (scramble). Both MMSET knockdowns showed the same effect on global histone methylation patterns, a decrease in H3K36me2 and an increase in H3K27me3 (Figure 1C and Figure S1A), corroborating the results obtained above. Intriguingly, down-regulation of MMSET in the non-transformed BPH1 and RWPE-1 cell lines did not show a clear alteration of these modifications (Figure 1D and Figure S1B). Collectively, these results show that low expression of MMSET in non-transformed prostate cells does not control global levels of H3K36me2 and H3K27me3. By contrast, histone methylation in PCa cells appears to become dependent on the overexpression of this factor. This suggests that MMSET could shift patterns of gene expression in PCa and hence affect its biology.
To further quantitatively analyze the histone methylation changes observed upon MMSET manipulation, mass-spectrometric profiles were obtained. Confirming the immunoblot analysis, overexpression of the wild-type, but not mutant, MMSET in RWPE-1 cells almost doubled the amount of dimethylated lysine 36, at the expense of unmodified and mono-methylated species. Methylation of lysine 27 changed in the opposite direction, with H3K27me2/3 decreasing substantially in the presence of MMSET (Figure S2A). Similarly, MMSET knockdown in DU145 cells reduced H3K36me2 and increased levels of H3K27me2 and H3K27me3, at the expense of H3K27me1 (Figure S2B). Additionally, MMSET knockdown in non-transformed RWPE-1 and BPH1 cells had little effect on H3K27me2/3 and H3K36me2/3 levels, confirming the results obtained by immunoblot (Figure S3).
Loss of MMSET Expression Affects Cell Growth, Invasion and Migration
To evaluate the biological role of MMSET overexpression in PCa cell lines, we analyzed the effect of MMSET knockdown on anchorage-dependent and independent proliferation, migration and invasion. Down-regulation of MMSET produced a modest decrease in the proliferation of the DU145 cell line (Figure 2A). MMSET knockdown yielded both an increase in the number of apoptotic cells (Figure S4A), and a modest reduction in the percentage of cells in S-phase (Figure S4B). Loss of MMSET had a larger effect on anchorage-independent growth, reducing the number of colonies formed in soft agar by a factor of three (Figure 2B). This assay is considered a measure of tumorigenicity, and suggests that MMSET is necessary to maintain the transformed phenotype of PCa cells. DU145 is a metastatic PCa cell line, and therefore displays conspicuous migratory and invasive properties. MMSET loss in this cell line led to a striking decrease in migration through a Boyden chamber (Figure 2C), and invasion into Matrigel (Figure 2D–E), suggesting that MMSET up-regulation may be relevant to the metastatic phenotype of PCa cells. All results were corroborated using the PC-3 metastasis-derived cell line (Figures S4 and S5).
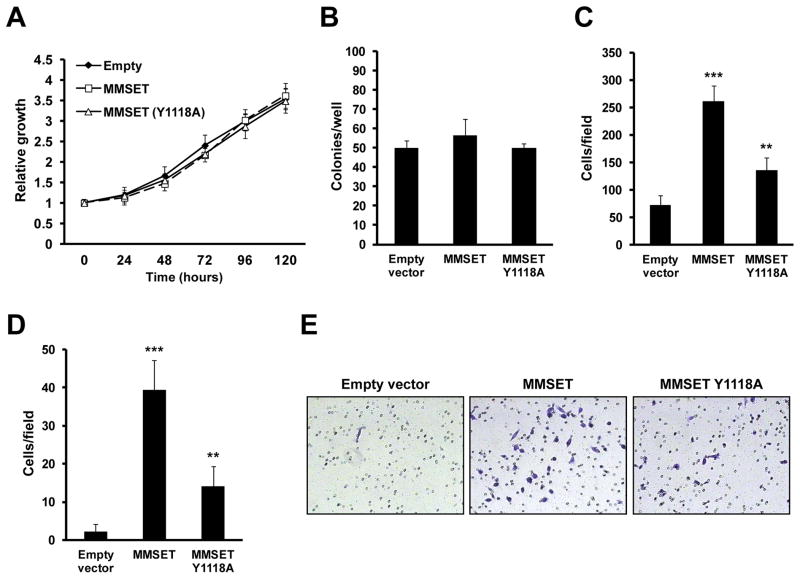
Figure 2.
MMSET knockdown decreases anchorage-dependent and independent proliferation, migration, and invasion of metastatic DU145 PCa cells. (A) Proliferation curves of DU145 cells transfected with the indicated siRNAs. Proliferation was measured by MTT conversion. Absorbance values are represented relative to time 0 (mean +/− SD of 3 independent biological replicates; *p<0.05). (B) Soft-agar colony formation. Cells were transfected in biological triplicates with the indicated siRNAs. The mean colony number per well +/− SD is presented (**p<0.01). (C–D) Quantification of one of three representative, independent migration (C) and invasion (D) experiments. The mean number of migrating or invading cells per field +/− SD of is presented (**p<0.01). (E) Representative fields of invasion assays (200x magnification).
MMSET Overexpression in Non-Transformed Cells Promotes Migration and Invasion
Using the gain-of-function model in RWPE-1 cells, we evaluated whether MMSET up-regulation in benign cells could confer aspects of the tumor phenotype. Overexpression of the wild-type or SET domain mutant MMSET did not enhance the proliferation of these cells (Figure 3A), and did not stimulate the formation of colonies in soft agar (Figure 3B). However, overexpression of MMSET in RWPE-1 cells strongly promoted migration and invasion (Figure 3C–E). The MMSET mutant induced this phenotype to a lesser degree than wild-type MMSET (Figure 3C–E), suggesting that the HMT activity of MMSET is an important factor for its biological activity. These data suggest that while MMSET may not transform cells on its own, it may play a role in disease progression and metastasis.
Figure 3.
Overexpression of MMSET promotes invasion and migration in non-transformed RWPE-1 cells. (A) Proliferation curves of RWPE-1 cells infected with a control retrovirus or a virus harboring wild-type or SET domain mutant MMSET. Proliferation was measured by MTT conversion. Absorbance values (mean +/− SD of three independent experiments) are represented as relative to time 0. (B) Soft-agar colony formation. Cells were infected in biological triplicates with retroviruses described in (A), and seeded in soft agar. The mean colony number per well +/− SD is presented. (C–D) Quantification of one of three representative, independent migration (C) and invasion (D) experiments. The mean number of cells per field +/− SD is presented (**p<0.01; ***p<0.001). (E) Representative fields of invasion assay (200x magnification).
MMSET Induces EMT
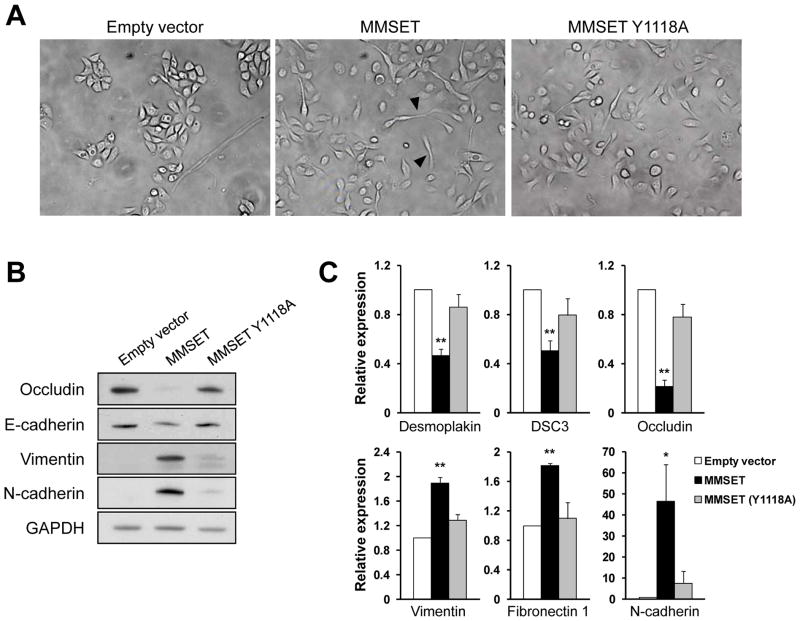
The EMT is a developmental process in which epithelial cells acquire characteristics of mesenchymal cells. Aspects of the EMT are present in advanced cancers, and contribute to tumor progression by facilitating tumor invasion and dissemination (35). RWPE-1 cells show epithelial-like growth, forming tightly packed colonies (Figure 4A-left). Overexpression of MMSET in these cells changed this pattern to more scattered growth (Figure 4A-middle), with some cells displaying fibroblast-like cytoplasmic elongations. The hallmark of EMT is the loss of epithelial markers and the neoexpression of mesenchymal markers. Accordingly, RWPE-1 cells overexpressing MMSET demonstrated decreased levels of the epithelial markers occludin and E-cadherin, and a gain of the mesenchymal markers vimentin and N-cadherin, at the protein (Figure 4B), as well as mRNA level (Figure 4C), suggesting that MMSET regulates the transcription of these genes.
Figure 4.
MMSET regulates EMT. (A) RWPE-1 cells were infected with control retrovirus or a virus harboring wild-type or SET domain mutant MMSET. Cytoplasmic fibroblast-like elongations are indicated with arrowheads (400x magnification). (B) Total protein extracts from RWPE-1 cells described in (A) were immunoblotted as indicated. (C) The expression of mesenchymal (vimentin, fibronectin 1, N-cadherin) and epithelial (desmoplakin, DSC3, occludin) markers was analyzed by real time PCR in cells described in (A). Gene expression normalized to GAPDH, from three independent experiments (+/−SD), is presented relative to that observed in cells transduced with control retrovirus (*p<0.05; **p<0.01).
We next determined whether knockdown of MMSET affected the phenotype of DU145 metastatic PCa cells. In response to MMSET depletion, these cells changed their mode of growth from a scattered pattern to the formation of more tightly packed colonies (Figure S6A). This effect was accompanied by a down-regulation of vimentin, and an up-regulation of occludin and E-cadherin at the protein (Figure S6B) and mRNA level, again, highlighting the role of transcriptional regulation by MMSET (Figure S6C). Collectively, the data suggest that MMSET depletion in PCa cells leads to loss of mesenchymal characteristics and promotes a more epithelial state.
TWIST1 Mediates MMSET-Induced EMT, Migration and Invasion
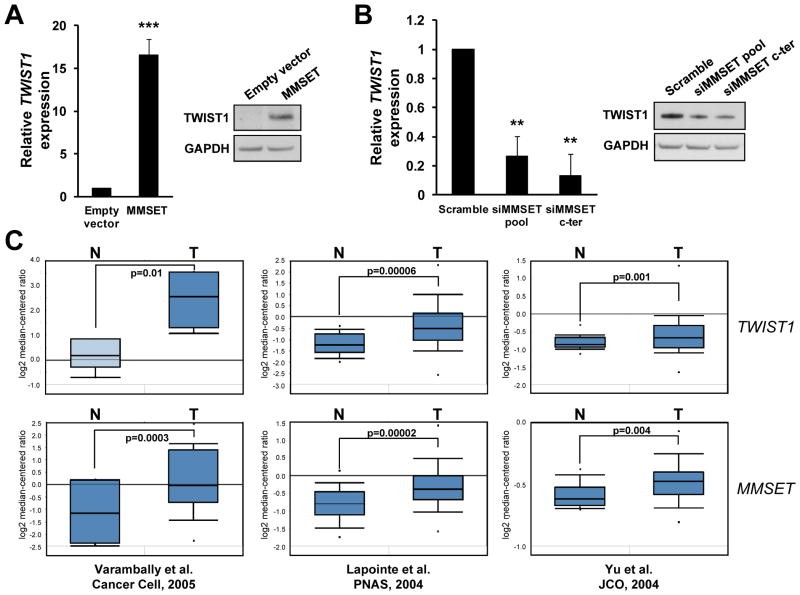
To ascertain how MMSET promotes EMT, we determined the expression of EMT-promoting genes in both the gain- and the loss-of-function models (Figure S7). Among the genes analyzed, only TWIST1 was strongly regulated by MMSET in both systems, increasing upon MMSET overexpression in RWPE-1 cells and decreasing after knockdown in DU145 cells (Figure 5A-B and Figure S7). TWIST1 is one of the core EMT-promoting genes, and a key factor responsible for metastasis in different tumors (33, 34, 36). Reanalysis of gene expression profiles from benign and malignant prostate (37–39), showed an increased expression of MMSET and TWIST1 in PCa (Figure 5C). Moreover, each data set showed a significant positive correlation between the expression of MMSET and TWIST1 (Varambally: r=0.72, p<0.05; Lapointe: r=0.84, p<0.0001; Yu: r=0.3, p<0.0001), suggesting that their expression increase in parallel in the transition from benign prostatic epithelium to PCa (Figure 5C). To further corroborate the regulation of TWIST1 by MMSET, we analyzed its expression in MM, the system where MMSET up-regulation and function has been best described. In agreement with MM cell line expression profiles (40) (GSE6205), we found that cells harboring the t(4;14) translocation had significantly higher levels of TWIST1 than those without MMSET rearrangements (Figure S8A). Moreover, in t(4;14) myeloma cells, shRNA mediated depletion of MMSET depressed TWIST1 levels, while repletion of MMSET in a t(4;14) cell line in which the rearranged MMSET locus was disrupted (KMS11-TKO), raised TWIST1 levels (Figure S8B). In a set of primary myeloma specimens categorized by the nature of their underlying chromosomal abnormalities (41) (GSE13591), those in the TC4 group, characterized by t(4;14), had significantly greater expression of TWIST1 than other subgroups of myeloma (Figure S8C). Collectively, this data suggests that MMSET promotes TWIST1 expression both in PCa and MM.
Figure 5.
MMSET regulates TWIST1 expression in PCa. (A) RWPE-1 cells were infected with control retrovirus or virus containing MMSET. TWIST1 expression was analyzed by real time PCR, normalized to GAPDH and presented as relative to that obtained in control cells (mean +/− SD from three independent experiments; ***p<0.001). Total protein extracts from these cells were immunoblotted as indicated. (B) DU145 cells were transfected with the indicated siRNAs, and analyzed as in (A); (**p<0.01). (C) MMSET and TWIST1 mRNA expression in normal (N) and malignant (T) prostate specimens was extracted from deposited gene expression datasets (37–39), analyzed in Oncomine and presented as box and whisker plots. Statistically significant differences are indicated with the p-value.
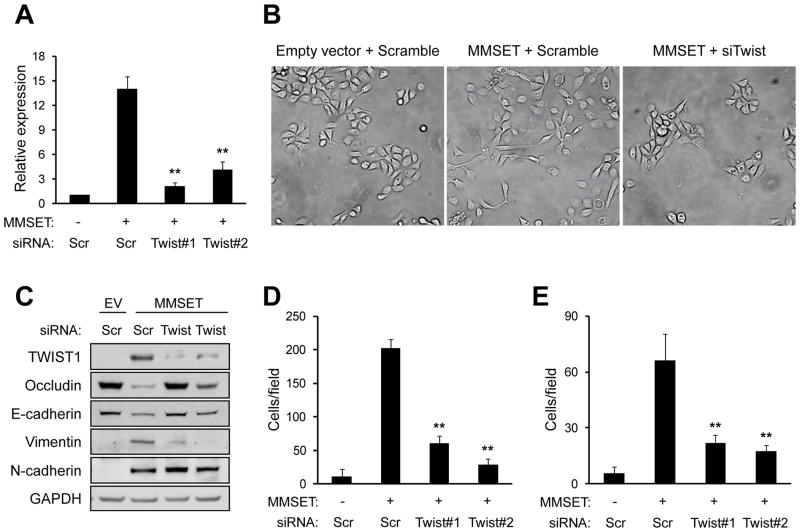
To determine whether TWIST1 was responsible for the ability of MMSET to induce cell migration, invasion and EMT, we simultaneously overexpressed MMSET and depleted TWIST1 in RWPE-1 cells. While expression of MMSET led to a >12 fold increase of TWIST1 mRNA levels, simultaneous treatment of cells with siRNA directed against TWIST1 largely blocked the accumulation of this factor (Figure 6A, 6C). The spindle-like morphology induced by overexpression of MMSET (Figure 6B-center) was reverted by TWIST1 knockdown, producing cells with an epithelial, polygonal morphology (Figure 6B-right). Loss of TWIST1 expression also reversed the ability of MMSET to induce the expression of vimentin, and blocked MMSET-mediated repression of E-cadherin and occludin (Figure 6C). N-cadherin was not decreased upon TWIST1 knockdown, suggesting its expression may be directly regulated by MMSET or another intermediary factor. Lastly, knockdown of TWIST1 in MMSET-overexpressing RWPE-1 cells significantly impaired MMSET-stimulated cell migration and invasion (Figure 6D–E), indicating that TWIST1 is a key target gene for the biological activity of MMSET.
Figure 6.
TWIST1 expression mediates MMSET-induced EMT, migration and invasion. (A) RWPE-1 cells infected with control retrovirus (MMSET-) or MMSET-harboring retrovirus (MMSET+), were transfected with siRNAs targeting TWIST1, or a control siRNA (Scr). TWIST1 expression was analyzed by real time PCR, normalized to GAPDH, and presented relative to that observed in cells transduced with control retrovirus (mean +/− SD from three independent experiments; **p<0.01). (B) Photomicrographs of cells described in (A) were taken 4 days after transfection (400x magnification). (C) Total protein extracts from cells described in (A) were immunoblotted with the indicated antibodies. (D–E) Quantification of one of three representative, independent migration (D) and invasion (E) experiments of cells described in (A). The mean number of migrating or invading cells per field +/−SD is presented (**p<0.01).
Additionally, to determine whether reduction of TWIST1 expression upon MMSET knockdown in DU145 cells could explain the decrease in the invasive phenotype observed, TWIST1 itself was downregulated. TWIST1 knockdown recapitulated the effects of MMSET downregulation, decreasing migration and invasion, and inducing a more epithelial phenotype (Figure S9). These results, in accordance with those of Kwok and colleagues (33), indicate a major role of TWIST1 in controlling the invasive phenotype of DU145 cells, and suggest that TWIST1 mediates the effect of MMSET on these cells.
MMSET Binding Across the TWIST1 Locus Correlates with Levels of H3K36me2
Since MMSET regulated the expression of TWIST1 in all the systems analyzed, we determined whether MMSET could directly bind TWIST1. ChIP analysis in RWPE-1 cells overexpressing MMSET showed MMSET binding across the TWIST1 locus. MMSET binding was accompanied with an increase in H3K36 dimethylation (Figure 7A). ChIP analysis in DU145 cells also showed MMSET-binding across the TWIST1 locus that decreased upon MMSET knockdown, in parallel with a decrease in H3K36me2 (Figure 7B). These results suggest that TWIST1 can be directly regulated by MMSET. To confirm the specificity of the regulation of this locus, ChIP analysis was performed on Snail gene (SNAI1), another EMT-promoting gene whose expression was unaffected by MMSET. MMSET binding to this locus was increased in response to MMSET up-regulation, however, an increase in H3K36me2 enrichment was not observed. Similarly, upon MMSET knockdown in DU145 cells, MMSET binding decreased, however, this was not accompanied by a reduction in H3K36me2 (Figure S10). Together, these results show that MMSET binding is not sufficient to stimulate gene expression, and suggests that the presence of other transcriptional regulators may also be required. Moreover, the ability of MMSET to methylate histones may be related to its ability to activate gene expression.
Figure 7.
MMSET binding across the TWIST1 locus correlates with high levels of H3K36me2. Chromatin from RWPE-1 cells infected with control retrovirus or a virus expressing MMSET (A), or from DU145 cells transfected with the indicated siRNAs (B), was immunoprecipitated with anti-MMSET or anti-H3K36me2 antibodies. The purified DNA was analyzed by real time PCR using primers amplifying regions across the TWIST1 locus. Results are presented as percentage of total input DNA precipitated. The average +/− SD of three independent experiments is presented (*p<0.05; **p<0.01).
Discussion
The pathogenesis and progression of PCa and other malignancies can be understood, in part, by the interplay between genetic and epigenetic changes that lead to altered patterns of gene expression. Most cases of PCa harbor genetic anomalies such as the TMPRSS2–ERG fusion gene, deletion of PTEN or other genomic rearrangements (42), and point mutations in genes such as TP53, KRAS and EGFR (www.sanger.ac.uk/genetics/CGP/cosmic/). These genetic alterations may be compounded by epigenetic anomalies mediated by altered expression of histone methyltransferases such as EZH2 and MMSET, the histone demethylase LSD1, and histone deacetylases (10–13). In general, expression of these enzymes is increased in the transition from benign epithelia to localized neoplasm to metastatic malignancy, suggesting that epigenetic alterations play a role in cancer progression. Accordingly, we found that the major effect of MMSET on PCa cell biology was on invasion and migration.
While overexpression of MMSET in MM was associated with a profound shift in global histone methylation levels (19), MMSET overexpression in PCa cell lines was associated with a more subtle difference. Specifically, depletion of MMSET depressed H3K36me2 levels and elevated H3K27me3 only in metastatic PCa cells and not in benign prostatic epithelial cells. This suggests that in benign prostate cells, other enzymes, including NSD1 and NSD3, may regulate H3K36me2 levels. These proteins are structurally similar to MMSET and can also catalyze dimethylation of H3K36 (43, 44). Upon MMSET overexpression in PCa cells lines, the levels of these modifications become dependent on MMSET. DU145 cells also depend on continued MMSET expression to maintain mesenchymal identity, optimal growth rate, growth in soft agar, and invasive phenotype. Together, this suggests that MMSET overexpression may alter the set of genes exhibiting H3K36me2 or H3K27me3, changing gene expression and facilitating PCa progression. Further definition of the gene networks controlled by MMSET in PCa will require integration of gene expression and ChIP-Seq datasets.
Why MMSET only regulates the expression of certain genes remains unknown. Despite a global change in histone modifications in myeloma cells, MMSET overexpression only regulates several hundred genes (19). In regard to genes affecting EMT, MMSET bound both the TWIST1 and SNAI1 loci, however, an increase in H3K36 methylation was only observed for the TWIST1 locus, correlating with TWIST1 mRNA up-regulation. Similarly, in myeloma cells, MMSET overexpression only affected the expression of TWIST1 and not SNAI1 (Figure S7B and data not shown). The ability of MMSET to alter TWIST1 expression in model cell systems is clinically relevant. Elevated MMSET levels in PCa and t(4;14)-associated myeloma, are correlated with increased TWIST1 expression but no change in SNAI1 (Figure 5C, S7C and data not shown). The factors that determine specific gene activation by MMSET such as promoter architecture, basal state of gene expression or three-dimensional chromatin configuration remain to be determined.
A number of studies show that MMSET is particularly upregulated in tumors of advanced grade and stage (23, 28, 29, 45). MMSET was unable to promote colony formation by immortalized benign prostate cells but strongly induced an invasive phenotype. Accordingly, in MM, MMSET regulates, among others, genes involved in cell adhesion and cell motility (19, 24, 25), processes usually deregulated during tumor progression. Collectively, these results suggest than MMSET acts in advanced stages malignancy, favoring tumor progression. Whether MMSET also acts in tumor initiation, transforming cells in collaboration with other genetic lesions remains to be elucidated.
MMSET-mediated acquisition of the invasive phenotype in prostate cells requires TWIST1. TWIST1 is a transcription factor that promotes invasion and metastasis in a variety of cancers, through the induction of EMT (33, 34, 36). TWIST1 is highly expressed in up to 90% of PCa, and its up-regulation in prostate cells results in an invasive phenotype (33). Interestingly, TWIST1 expression was also high in t(4;14)+ MM cell lines and patient specimens. MM cells do not show an epithelial phenotype, therefore, how TWIST1 expression alters the biology of MM remains to be determined. High expression of TWIST1 in solid tumors confers resistance to apoptosis induced by a chemotherapy drugs such as taxol or cisplatin (33, 46). This suggests that the aggressive biological behavior, poor response to therapy and poor prognosis of tumors displaying high MMSET levels might be mediated by anti-apoptotic effects of TWIST1. TWIST1 expression also links EMT to self-renewal, promoting the acquisition of stem cell-like characteristics (47). Accordingly, inactivation of MMSET by small molecular inhibitors, which have already been developed for other HMTs such as G9a (48, 49) and DOT1L1 (50), may represent a novel therapeutic maneuver in advanced cancers. We have shown here and previously that siRNA-mediated depletion of MMSET reverses global epigenetic changes and alteration in expression of specific genes such as TWIST1 (19). Further analysis of gene expression changes mediated by MMSET in MM and PCa may yield important biomarkers for the activity of putative inhibitors.
This work adds to the growing amount of evidence highlighting the importance of epigenetic deregulation in PCa. Our results emphasize the role that MMSET may have in PCa progression, through its regulation of EMT and invasion. Although TWIST1 is clearly an important gene that mediates MMSET effects, the relevance of other genes and pathways cannot be ruled out. The specific regulation of H3K36 and H3K27 methylation by MMSET in metastatic PCa, and the strong biological effects promoted by this factor, highlight the importance of considering MMSET as a therapeutic target for the treatment of PCa.
Materials and Methods
Cell Culture
Human prostate cell lines were obtained from the ATCC (Manassas, VA) and grown in RPMI-1640 (DU145, PC-3, LNCaP, 22Rv1, BPH1) or DMEM (VCaP) medium, supplemented with 10% fetal bovine serum. Non-neoplastic, immortalized human prostatic epithelial RWPE-1 cells (ATCC) were cultured in defined Keratinocyte-SFM (Gibco, Grand Island, NY) supplemented with recombinant epidermal growth factor (EGF) and bovine pituitary extract (BPE) (Gibco).
Gain of Function and Loss of Function Models
RWPE-1 cells were transduced with retroviral vectors harboring MMSET I, MMSET II wild-type or MMSET II mutant isoforms. Retroviruses were generated by transfection of 293T cells with plasmids previously described (19) and FuGENE6 (Roche Applied Science, Indianapolis, IN). RWPE-1 cells were sorted using the DSRed (MMSET II wild-type and mutant) or green fluorescent protein (MMSET I) markers. For MMSET and TWIST1 knockdown, cells were transfected with 20 nM siRNA, using Lipofectamine RNAiMAX (Invitrogen, Grand Island, NY). siRNAs target sequences are included in Table S1. A scrambled siRNA (D-001206-14, Dharmacon, Lafayette, CO) was used as a control.
Immunoblotting
For extraction of total proteins, cells were disrupted in 1% NP-40 lysis buffer (140 mM NaCl, 10 mM Tris-HCl pH 8, 1% NP-40) supplemented with proteinase inhibitors (Roche). Nuclear proteins were extracted using the Nuclear Complex Co-IP Kit (Active Motif, Carlsbad, CA). Proteins were electrophoretically separated, blotted and detected using enhanced chemiluminescence. Primary antibodies were: MMSET (described in reference (19)), H3K36me2 (Millipore, Billerica, MA; 07-369), H3K27me3 (Millipore, 07-449), pan-H4 (Abcam, Cambridge, MA; Ab7311), N-cadherin (BD Transduction Laboratories, San Jose, CA; 610920), Vimentin (Thermo Scientific, Pittsburgh, PA; MS-129), Occludin (Santa Cruz, Santa Cruz, CA; sc-5562), E-cadherin (Cell Signaling, Danvers, MA; 4065), TWIST1 (Active Motif; 61097), and GAPDH (Millipore, MAB374). Secondary antibodies were horseradish peroxidase-conjugated donkey anti-rabbit and sheep anti-mouse IgG (GE Healthcare Life Sciences, Piscataway, NJ).
cDNA Preparation and Real Time PCR
Total RNA was extracted using the RNeasy kit (Qiagen, Valencia, CA). Reverse transcription was performed using 2 μg of total RNA and the iScript Advanced cDNA Synthesis Kit (BioRad, Hercules, CA). Real time PCR determinations were performed using the Lightcycler 480 SYBR Green I Master reagent (Roche) on the Lichtcycler 480 II (Roche). Primers used are listed in Table S2.
Cell Proliferation and Soft Agar Assays
DU145, PC-3 and RWPE-1 cells (500, 1,000 and 500 cells per well, respectively) were seeded in 96-well plates in 6 replicates, and proliferation was measured at 24-hour intervals by MTT conversion (Sigma-Aldrich, St. Louis, MO). Anchorage-independent proliferation was determined by plating DU145, PC-3 and RWPE-1 cells (5,000, 3,000 and 10,000 cells, respectively) in 6-well plates, with a bottom layer of 0.6% agar and a top layer of 0.3% agar containing the cells. After an 8-day incubation, colonies were visualized using MTT reagent.
Cell Cycle and Apoptosis Analysis
Cell cycle distribution and the percentage of apoptotic cells were measured by flow cytometric analysis (BD LSR II). For cell cycle determinations, cells were incubated with BrdU (5-bromo-2-deoxyuridine) for 30 minutes. BrdU incorporation was analyzed using the APC BrdU Flow Kit (BD Pharmigen). Apoptosis was measured using the Annexin V-Cy5 Apoptosis Detection Kit (Biovision, Mountain View, CA).
Migration and Invasion Assays
Migration assays were performed using polycarbonate membranes with 8 μm pores in 24-well chemotaxis chambers (Costar/Corning, Lowell, MA). DU145, PC-3 (5 × 104 cells) or RWPE-1 cells (1 × 105 cells) were loaded into the top of each chamber in 150 μl of serum-free RPMI or Keratinocyte-SFM without EGF and BPE. The lower chamber was filled with 500 μl of RPMI containing 10% FBS for DU145 and PC-3, or Keratinocyte-SFM with EGF and BPE for RWPE-1 cells. After a 24 hour-incubation, cells that migrated to the lower surface of the membrane were fixed with 100% methanol and stained with 5% crystal violet. Cells were counted in 2 different inserts, considering 4 random fields per insert. Invasion assays were performed using 24-well Matrigel invasion chambers (BD Biocoat, BD Biosciences), as above.
Histone Preparation and Mass Spectrometry Analysis
Histones were recovered from cells using sulfuric acid extraction, chemically derivatized using propionic anhydride, and digested with trypsin, as described previously (51). The peptide containing both K27 and K36 (K27SAPATGGVKKPHR40) was one of the peptides liberated from histone H3.1 and H3.2 during this process. All peptides were analyzed with a nano LC-MS system consisting of a Dionex UltiMate 3000 coupled to a Thermo Fisher Scientific TSQ Quantum QQQ mass spectrometer. Transitions for selected reaction monitoring (SRMs) were developed and data were analyzed using Skyline software (v1.1; MacCoss Lab, University of Washington) (52). Fifteen out of all 16 possible combinatorial methylations in H3K27-K36 peptide, except H3 K27me3-K36me3, were measured in 15 channels, each of them consisting of 3 unique transitions for that particular methylation form. Individual K27 or K36 relative methylation levels were calculated by combining corresponding channels.
Chromatin Immunoprecipitation
Chromatin Immunoprecipitation (ChIP) was performed using a ChIP kit (Millipore), with the modifications previously described (19). The antibodies used were: MMSET (described in reference (19)), H3K36me2 (Upstate, 07-369) and rabbit IgG (Abcam, ab37415). qPCR was performed as above (primers in Table S3). Enrichment was calculated as percentage of total input DNA precipitated.
Statistical Analysis
Data obtained from MTT, soft agar, migration, invasion, apoptosis, cell cycle, real time PCR and ChIP assays were analyzed using the Student’s T test or Mann-Whitney U test. The Pearson’s correlation test was used to evaluate the association between MMSET and TWIST1 expression. Statistical analysis was done using the SPSS software package, version 15.0 (IBM).
Supplementary Material
Acknowledgments
We acknowledge Associazione Italiana Ricerca sul Cancro (AIRC).
This work was supported by a Fundacion Alfonso Martin Escudero fellowship (T.E.), a Ruth Kirschstein National Research Service Award F32HL099177 (R.P.), R01GM067193 (N.L.K.), RO1CA123204 (J.D.L.) a Leukemia and Lymphoma Society Specialized Center of Research Award (J.D.L.) and Physical Sciences Oncology Center grant U54CA143869 (J.D.L. and N.L.K.).
Footnotes
Conflict of interest
The authors declare no competing conflicts of interest.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Jeronimo C, Bastian PJ, Bjartell A, et al. Epigenetics in prostate cancer: biologic and clinical relevance. Eur Urol. 60:753–66. doi: 10.1016/j.eururo.2011.06.035. [DOI] [PubMed] [Google Scholar]
- 3.Perry AS, Watson RW, Lawler M, Hollywood D. The epigenome as a therapeutic target in prostate cancer. Nat Rev Urol. 7:668–80. doi: 10.1038/nrurol.2010.185. [DOI] [PubMed] [Google Scholar]
- 4.Schulz WA, Hatina J. Epigenetics of prostate cancer: beyond DNA methylation. J Cell Mol Med. 2006;10:100–25. doi: 10.1111/j.1582-4934.2006.tb00293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yegnasubramanian S, Kowalski J, Gonzalgo ML, et al. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004;64:1975–86. doi: 10.1158/0008-5472.can-03-3972. [DOI] [PubMed] [Google Scholar]
- 6.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 7.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Lennartsson A, Ekwall K. Histone modification patterns and epigenetic codes. Biochim Biophys Acta. 2009;1790:863–8. doi: 10.1016/j.bbagen.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Mellor J. The dynamics of chromatin remodeling at promoters. Mol Cell. 2005;19:147–57. doi: 10.1016/j.molcel.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 10.Halkidou K, Gaughan L, Cook S, Leung HY, Neal DE, Robson CN. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate. 2004;59:177–89. doi: 10.1002/pros.20022. [DOI] [PubMed] [Google Scholar]
- 11.Kahl P, Gullotti L, Heukamp LC, et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 2006;66:11341–7. doi: 10.1158/0008-5472.CAN-06-1570. [DOI] [PubMed] [Google Scholar]
- 12.Metzger E, Wissmann M, Yin N, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–9. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 13.Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 14.Bianco-Miotto T, Chiam K, Buchanan G, et al. Global levels of specific histone modifications and an epigenetic gene signature predict prostate cancer progression and development. Cancer Epidemiol Biomarkers Prev. 19:2611–22. doi: 10.1158/1055-9965.EPI-10-0555. [DOI] [PubMed] [Google Scholar]
- 15.Seligson DB, Horvath S, Shi T, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–6. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 16.Stec I, Wright TJ, van Ommen GJ, et al. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet. 1998;7:1071–82. doi: 10.1093/hmg/7.7.1071. [DOI] [PubMed] [Google Scholar]
- 17.Volkel P, Angrand PO. The control of histone lysine methylation in epigenetic regulation. Biochimie. 2007;89:1–20. doi: 10.1016/j.biochi.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Kim JY, Kee HJ, Choe NW, et al. Multiple-myeloma-related WHSC1/MMSET isoform RE-IIBP is a histone methyltransferase with transcriptional repression activity. Mol Cell Biol. 2008;28:2023–34. doi: 10.1128/MCB.02130-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Garcia E, Popovic R, Min DJ, et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood. 117:211–20. doi: 10.1182/blood-2010-07-298349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Trojer P, Xu CF, et al. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem. 2009;284:34283–95. doi: 10.1074/jbc.M109.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92:3025–34. [PubMed] [Google Scholar]
- 22.Garlisi CG, Uss AS, Xiao H, et al. A unique mRNA initiated within a middle intron of WHSC1/MMSET encodes a DNA binding protein that suppresses human IL-5 transcription. Am J Respir Cell Mol Biol. 2001;24:90–8. doi: 10.1165/ajrcmb.24.1.4224. [DOI] [PubMed] [Google Scholar]
- 23.Keats JJ, Maxwell CA, Taylor BJ, et al. Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients. Blood. 2005;105:4060–9. doi: 10.1182/blood-2004-09-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brito JL, Walker B, Jenner M, et al. MMSET deregulation affects cell cycle progression and adhesion regulons in t(4;14) myeloma plasma cells. Haematologica. 2009;94:78–86. doi: 10.3324/haematol.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lauring J, Abukhdeir AM, Konishi H, et al. The multiple myeloma associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity. Blood. 2008;111:856–64. doi: 10.1182/blood-2007-05-088674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marango J, Shimoyama M, Nishio H, et al. The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor. Blood. 2008;111:3145–54. doi: 10.1182/blood-2007-06-092122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hudlebusch HR, Santoni-Rugiu E, Simon R, et al. The histone methyltransferase and putative oncoprotein MMSET is overexpressed in a large variety of human tumors. Clin Cancer Res. 17:2919–33. doi: 10.1158/1078-0432.CCR-10-1302. [DOI] [PubMed] [Google Scholar]
- 28.Hudlebusch HR, Skotte J, Santoni-Rugiu E, et al. MMSET is highly expressed and associated with aggressiveness in neuroblastoma. Cancer Res. 71:4226–35. doi: 10.1158/0008-5472.CAN-10-3810. [DOI] [PubMed] [Google Scholar]
- 29.Kassambara A, Klein B, Moreaux J. MMSET is overexpressed in cancers: link with tumor aggressiveness. Biochem Biophys Res Commun. 2009;379:840–5. doi: 10.1016/j.bbrc.2008.12.093. [DOI] [PubMed] [Google Scholar]
- 30.Toyokawa G, Cho HS, Masuda K, et al. Histone Lysine Methyltransferase Wolf-Hirschhorn Syndrome Candidate 1 Is Involved in Human Carcinogenesis through Regulation of the Wnt Pathway. Neoplasia. 13:887–98. doi: 10.1593/neo.11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Yin C, Okamoto H, et al. Identification of a novel proliferation-related protein, WHSC1 4a, in human gliomas. Neuro Oncol. 2008;10:45–51. doi: 10.1215/15228517-2007-036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okabe H, Satoh S, Kato T, et al. Genome-wide analysis of gene expression in human hepatocellular carcinomas using cDNA microarray: identification of genes involved in viral carcinogenesis and tumor progression. Cancer Res. 2001;61:2129–37. [PubMed] [Google Scholar]
- 33.Kwok WK, Ling MT, Lee TW, et al. Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res. 2005;65:5153–62. doi: 10.1158/0008-5472.CAN-04-3785. [DOI] [PubMed] [Google Scholar]
- 34.Yang J, Mani SA, Donaher JL, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 36.Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744–9. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 37.Lapointe J, Li C, Higgins JP, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A. 2004;101:811–6. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varambally S, Yu J, Laxman B, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Yu YP, Landsittel D, Jing L, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–9. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 40.Lombardi L, Poretti G, Mattioli M, et al. Molecular characterization of human multiple myeloma cell lines by integrative genomics: insights into the biology of the disease. Genes Chromosomes Cancer. 2007;46:226–38. doi: 10.1002/gcc.20404. [DOI] [PubMed] [Google Scholar]
- 41.Agnelli L, Bicciato S, Mattioli M, et al. Molecular classification of multiple myeloma: a distinct transcriptional profile characterizes patients expressing CCND1 and negative for 14q32 translocations. J Clin Oncol. 2005;23:7296–306. doi: 10.1200/JCO.2005.01.3870. [DOI] [PubMed] [Google Scholar]
- 42.Berger MF, Lawrence MS, Demichelis F, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804–12. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 44.Morishita M, di Luccio E. Cancers and the NSD family of histone lysine methyltransferases. Biochim Biophys Acta. 2011;1816:158–63. doi: 10.1016/j.bbcan.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 45.Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol. 2005;23:6333–8. doi: 10.1200/JCO.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 46.Zhuo WL, Wang Y, Zhuo XL, Zhang YS, Chen ZT. Short interfering RNA directed against TWIST, a novel zinc finger transcription factor, increases A549 cell sensitivity to cisplatin via MAPK/mitochondrial pathway. Biochem Biophys Res Commun. 2008;369:1098–102. doi: 10.1016/j.bbrc.2008.02.143. [DOI] [PubMed] [Google Scholar]
- 47.Yang MH, Hsu DS, Wang HW, et al. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat Cell Biol. 12:982–92. doi: 10.1038/ncb2099. [DOI] [PubMed] [Google Scholar]
- 48.Vedadi M, Barsyte-Lovejoy D, Liu F, et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol. 7:566–74. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kubicek S, O’Sullivan RJ, August EM, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–81. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 50.Daigle SR, Olhava EJ, Therkelsen CA, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia BA, Mollah S, Ueberheide BM, et al. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protoc. 2007;2:933–8. doi: 10.1038/nprot.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacLean B, Tomazela DM, Shulman N, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 26:966–8. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.