Summary
The bacteria in the fruitfly Drosophila melanogaster of different life stages was quantified by 454 pyrosequencing of 16S rRNA gene amplicons. The sequence reads were dominated by 5 operational taxonomic units (OTUs) at ≤ 97% sequence identity that could be assigned to Acetobacter pomorum, A. tropicalis, Lactobacillus brevis, L. fructivorans and L. plantarum. The saturated rarefaction curves and species richness indices indicated that the sampling (85 000–159 000 reads per sample) was comprehensive. Parallel diagnostic PCR assays revealed only minor variation in the complement of the five bacterial species across individual insects and three D. melanogaster strains. Other gut-associated bacteria included 6 OTUs with low %ID to previously reported sequences, raising the possibility that they represent novel taxa within the genera Acetobacter and Lactobacillus. A developmental change in the most abundant species, from L. fructivorans in young adults to A. pomorum in aged adults was identified; changes in gut oxygen tension or immune system function might account for this effect. Host immune responses and disturbance may also contribute to the low bacterial diversity in the Drosophila gut habitat.
Introduction
Healthy animals are a habitat for microorganisms, most of which are benign or beneficial (Wilson, 2005; Douglas, 2010). Mammals and other vertebrates appear to support many more microbial species than most invertebrates. For example, the gut microbiota in an individual mammal comprises > 1000 taxa, most of which are unique to each host individual (Dethlefsen et al., 2007; Ley et al., 2008; Costello et al., 2009; Qin et al., 2010). The diversity of the gut microbiota in most invertebrates that have been studied is apparently one to two orders of magnitude lower than in the mammals (Dillon and Dillon, 2004; Dunn and Stabb, 2005; Behar et al., 2008; Lehman et al., 2009; Morales-Jimenez et al., 2009; Grunwald et al., 2010; Robinson et al., 2010). Nevertheless, vertebrates, especially mammals, have been the subject of far greater sampling effort than invertebrates, raising the possibility that this difference between vertebrates and invertebrates may be partly artefactual. Two further issues affect the interpretation of data on the diversity of the microbiota in animal guts. First, the composition of the gut microbiota can vary with diet, and developmental age and physiological condition of the animal host (e.g. Dethlefsen et al., 2007; Lehman et al., 2009; Sharon et al., 2010). Second, microorganisms recovered from the gut comprise two ecologically distinct groups: the autochthonous (resident) taxa and the allochthonous (non-resident) forms that are ingested with, and pass through, the gut with the food. The allochthonous microbes can artificially inflate both the reported microbial diversity in an individual host, and among-host variation in microbial diversity, especially where the animals sampled utilize different sources of food.
The purpose of this study was to determine the diversity of the gut bacteria of the fruitfly Drosophila melanogaster using 454 pyrosequencing of PCR-generated amplicons from the 16S rRNA gene. We used Drosophila raised on an axenic diet of fixed composition, to minimize the diversity of allochthonous taxa, and sampled the animals across the full life cycle, to establish the total diversity and how it varies with life stage. Our analysis builds on previous research, which has identified various taxa, including Lactobacillus, Enterococcus and Acetobacter associated with Drosophila [Corby-Harris et al., 2007; Cox and Gilmore, 2007; Ren et al., 2007; Ryu et al., 2008; also see the review (Crotti et al., 2010) of Acetobacter as insect symbionts]. Our study overcomes three key limitations of previous studies: all may have failed to detect low-abundance taxa through shallow sampling using limited Sanger sequencing of cloned 16S rRNA gene sequences; most were conducted on the whole insect, making it impossible to identify the bacteria specifically associated with the gut; and several studies did not attempt to limit the incidence of allochthonous taxa.
Results
Pyrosequencing data
The 454 pyrosequencing analysis of 16S rRNA gene amplicons from the dissected guts of D. melanogaster strain Canton S produced 923 109 reads, with an average length of 361 nucleotides (including the multiplex identifier ‘MID’ and primer sequences), after quality filtering and removal of chimaeric sequences. The reads could be assigned to 720 operational taxonomic units (OTUs) at 93% sequence identity threshold, 894 and 1135 OTUs at 95% and 97% threshold, and 8935 OTUs at 99% threshold. A substantial number of the OTUs identified were represented by just one to several reads in both the experimental samples and the reagent-only control. These were interpreted as contaminants and they were discarded, leaving 808 483 reads that were distributed among the samples as follows: D. melanogaster eggs (0.2%), early-instar larvae (10.6%), pupae (13.9%) and guts from third-instar larvae (13.4%), 3- to 7-day-old males (14.0%) and females (19.7%), 3- to 5-week-old males (10.5%) and females (17.5%). Altogether, the reads yielded 122 OTUs at the 97% identity threshold recommended for accurate diversity estimation (Kunin et al., 2010).
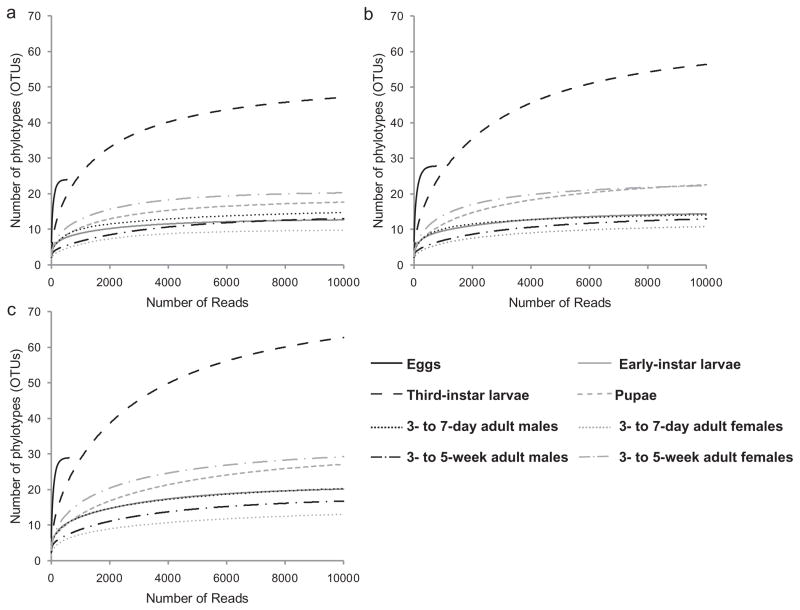
For each sample, the rarefaction curves tended towards saturation at similar numbers of clusters at 97%, 95% and 93% pairwise ID thresholds (Fig. 1). Subsequent analysis was, therefore, conducted at 97% ID. All values of richness indices (Chao1, ACE and Jackknife) equalled the number of OTUs (Table 1), confirming the conclusion from rarefaction analysis that sampling of each life stage had reached saturation. The third-instar larvae bore the most species-rich bacterial community, comprising 71 OTUs. The egg surface had the most diverse bacterial community by both Simpson’s and Shannon indices (Table 1), including 19/28 (68%) unique clusters. In all other samples, five OTUs (clusters 1, 2, 5, 6 and 7) accounted for > 80% of all reads (Table S1), and OTUs unique to one life stage were rare (early instars, 3- to 5-week-old adults) or absent (3- to 7-day-old adults). Exceptionally, 12/30 (40%) of OTUs in pupae and 48/71 (68%) OTUs in third-instar larvae were unique.
Fig. 1.
Rarefaction curves of OTUs clustered at different %ID across life stages of D. melanogaster Canton-S. (A) 93%, (B) 95%, (C) 97%.
Table 1.
Richness and diversity estimation of the 16S rRNA gene libraries from the pyrosequencing analysis.
| Sample | Number of reads | Number of OTUsa | Species richness indices
|
Species diversity indices
|
|||
|---|---|---|---|---|---|---|---|
| Chao1 | ACE | Jackknife | Shannon | Simpson | |||
| Eggs | 1 798 | 28 | 28 | 28 | 28 | 2.62 | 0.88 |
| Early-instar larvae | 86 038 | 21 | 21 | 21 | 21 | 0.87 | 0.37 |
| Third-instar larvae | 112 382 | 71 | 71 | 71 | 71 | 1.40 | 0.58 |
| Pupae | 108 609 | 30 | 30 | 30 | 30 | 1.47 | 0.70 |
| 3- to 7-day-old adult males | 113 614 | 19 | 19 | 19 | 19 | 1.26 | 0.59 |
| 3- to 7-day-old adult females | 159 309 | 15 | 15 | 15 | 15 | 0.35 | 0.15 |
| 3- to 5-week-old adult males | 85 095 | 17 | 17 | 17 | 17 | 0.72 | 0.32 |
| 3- to 5-week-old adult females | 141 761 | 31 | 31 | 31 | 31 | 1.02 | 0.43 |
The operational taxonomic units (OTUs) were defined with pairwise 97% ID.
Taxonomic composition of bacteria identified by pyrosequencing
At the phylum level, Firmicutes and Proteobacteria accounted for the vast majority of reads (> 97%) in the larvae, pupae and adults, and 66% of the reads for the eggs. Actinobacteria, Bacteroides and Cyanobacteria were also detected (Table 2a).
Table 2.
Abundance of 16S rRNA gene amplicons in D. melanogaster samples, expressed as % of total in each life stage.
| (a) Bacterial phyla
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Phylum | % of total sequence reads in each life stage
|
|||||||
| Eggs | Early-instar larvae | Third-instar larvae | Pupae | 3- to 7-day-old males | 3- to 7-day-old females | 3- to 5-week-old males | 3- to 5-week-old females | |
| Actinobacteria | 23.28 | 0 | 0.10 | 0 | 0 | 0 | 0.12 | 0 |
| Bacteroidetes | 5.91 | 0.03 | 2.19 | 0 | 0 | 0 | 0 | 0.03 |
| Cyanobacteria | 3.88 | 0 | 0.14 | 0 | 0 | 0 | 0 | 0 |
| Firmicutes | 35.32 | 85.97 | 87.83 | 43.67 | 80.45 | 93.60 | 15.52 | 20.29 |
| Proteobacteria | 30.34 | 14.01 | 9.66 | 56.31 | 19.55 | 6.40 | 84.37 | 79.67 |
| Other | 1.27 | 0 | 0.09 | 0.02 | 0 | 0 | 0 | 0 |
| (b) Bacterial species
| |||||||
|---|---|---|---|---|---|---|---|
| Species | % of total sequence reads in each life stage
|
||||||
| Early-instar larvae | Third-instar larvae | Pupae | 3- to 7-day-old males | 3- to 7-day-old females | 3- to 5-week-old males | 3- to 5-week-old females | |
| Acetobacter pomorum | 8.97 | 1.75 | 8.98 | 5.79 | 5.81 | 81.65 | 74.43 |
| Acetobacter tropicalis | 5.03 | 3.56 | 47.31 | 13.74 | 0.60 | 2.72 | 4.18 |
| Lactobacillus brevis | 1.94 | 22.42 | 3.11 | 15.13 | 1.03 | 7.05 | 2.42 |
| Lactobacillus fructivorans | 80.30 | 4.30 | 3.28 | 61.01 | 92.50 | 7.60 | 10.22 |
| Lactobacillus plantarum | 3.73 | 60.90 | 21.44 | 4.31 | 0.07 | 0.82 | 7.65 |
| Staphylococcus sp. | 0 | 0 | 15.73 | 0 | 0 | 0 | 0 |
| Other | 0.04 | 7.07 | 0.15 | 0.02 | 0 | 0.16 | 1.09 |
All data refer to guts isolated from the insects, apart from early instars and pupae.
The five OTUs dominating most samples (see above) corresponded to Acetobacter and Lactobacillus species: A. pomorum, A. tropicalis, L. brevis, L. fructivorans and L. plantarum (Table 2b, Table S1). The relative abundance of these taxa varied with developmental age (Table 2b). Lactobacillus fructivorans accounted for > 60% of the reads in early-instar larvae and 3- to 7-day-old adults (both sexes); L. plantarum dominated the gut bacteria of third-instar larvae; and A. tropicalis and A. pomorum were strongly represented in pupae and 3- to 5-week-old adults respectively. These species were detected in eggs at varying abundance (Table S1): L. fructivorans (21%), A. pomorum (14%), A. tropicalis (2%), L. brevis (1.5%) and L. plantarum (four reads, which was below the cut-off for contaminants). The sequences of the five OTUs were submitted to NCBI GenBank (accession HQ173707–HQ173711).
Pupae bore appreciable numbers of Staphylococcus, accounting for 16% of the reads, of which > 99% were assigned to Staphylococcus sp. K6-17B (Table S1D), while Staphylococcus represented < 0.1% of reads in all other life stages.
Candidate novel bacterial taxa
The %ID between some 454 reads and the BLAST top hits was less than 97% (Table S1 and Table 3). Two approaches were adopted to assess whether these low %IDs were likely a consequence of sequencing error. First, the polymorphisms were confirmed not to be in homopolymeric regions, which are common sites of 454 sequencing error. Second, the Bonferroni-corrected Poisson probabilities were calculated for each biological sample. At %IDs of 96% or less, the probability of the polymorphism arising by sequencing error was ≤ 0.0002 (Table S2). These data suggest that the low %ID of the clusters in Table 3 are not the result of sequencing error.
Table 3.
Phylotype clusters with low % sequence identity (≤ 97%) to the top hit sequences in the NCBI database.
| Phylotype clustera | Accession | BLAST top hit | Scoreb | E valueb | %ID | Candidate novel taxa
|
|
|---|---|---|---|---|---|---|---|
| Minimum V2 %ID | 97% full 16S rRNA gene sequence cut-off | ||||||
| a-Cluster16042 | EU096229.1 | Acetobacter pomorum strain EW816 | 433 | 3.00E-118 | 95.9 | + | |
| b-Cluster932 | EU096229.1 | Acetobacter pomorum strain EW816 | 411 | 1.00E-111 | 94.1 | + | + |
| c-Cluster5070 | FJ915625.1 | Acetobacter tropicalis strain IMAU30060 | 444 | 1.00E-121 | 96.3 | ||
| d-Cluster7664 | FJ915625.1 | Acetobacter tropicalis strain IMAU30060 | 281 | 1.00E-72 | 92.2 | + | + |
| e-Cluster3222 | X76330.1 | Lactobacillus fructivorans strain DSM 20203 T | 436 | 2.00E-119 | 96.6 | ||
| f-Cluster668 | X76330.1 | Lactobacillus fructivorans strain DSM 20203 T | 381 | 1.00E-102 | 92.8 | + | + |
| g-Cluster467 | X76330.1 | Lactobacillus fructivorans strain DSM 20203 T | 385 | 8.00E-104 | 91.9 | + | + |
| h-Cluster8879 | X76330.1 | Lactobacillus fructivorans strain DSM 20203 T | 438 | 6.00E-120 | 95.9 | + | |
| i-Cluster94 | X76330.1 | Lactobacillus fructivorans strain DSM 20203 T | 390 | 2.00E-105 | 92.5 | + | + |
| j-Cluster1982 | AB289116.1 | Lactobacillus fructivorans strain JCM 1198 | 320 | 2.00E-84 | 93.6 | + | + |
| k-Cluster4458 | GU415690.1 | Lactobacillus brevis clone CX018 | 455 | 6.00E-125 | 97.0 | ||
| l-Cluster115 | GU415690.1 | Lactobacillus brevis clone CX018 | 375 | 5.00E-101 | 92.3 | + | |
| m-Cluster1522 | AB025971.1 | Lactobacillus brevis | 331 | 1.00E-87 | 94.4 | + | |
| n-Cluster1494 | GU430842.1 | Lactobacillus plantarum clone OCR057 | 442 | 5.00E-121 | 96.3 | ||
| o-Cluster77 | FJ532361.1 | Lactobacillus plantarum strain 14W | 405 | 6.00E-110 | 93.8 | + | |
| p-Cluster4922 | AB362740.1 | Lactobacillus plantarum strain NRIC 1749 | 326 | 5.00E-86 | 93.5 | + | |
| q-Cluster6807 | AB362740.1 | Lactobacillus plantarum strain NRIC 1749 | 357 | 2.00E-95 | 90.5 | + | |
| r-Cluster1601 | GU125508.1 | Lactobacillus plantarum strain IMAU80086 | 394 | 1.00E-106 | 93.5 | + | |
Letter (a–r) abbreviations of cluster numbers are indicated in Fig. 2E and F.
The derivation and meaning of Score and E value are provided at http://www.ncbi.nlm.nih.gov/BLAST/tutorial/Altschul-1.html.
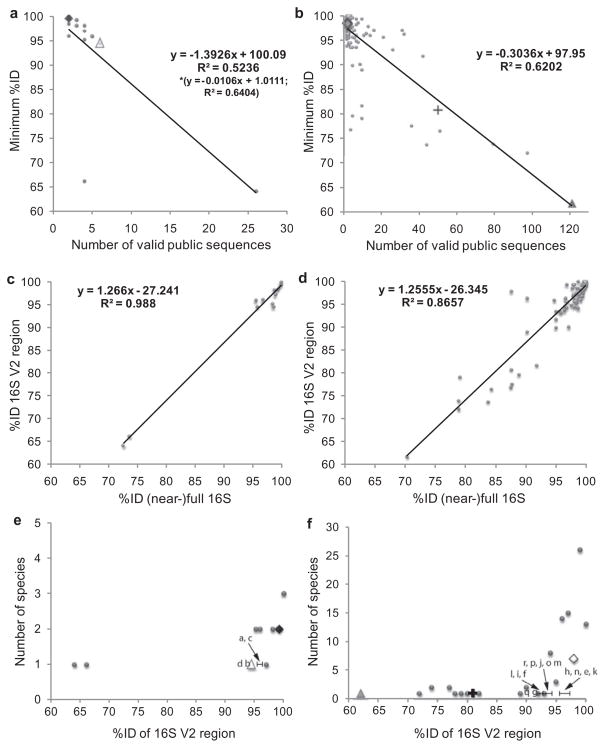
To investigate the possibility that the sequences might represent novel taxa, the %ID between each cluster and its top BLAST hit (Table 3) was compared with pairwise %ID comparisons among publicly available sequences representing the same bacterial species in the 16S rRNA databases (Text S1). The minimum values of pairwise %ID of the V2 region among publicly available sequences of A. pomorum and A. tropicalis are 99.6% and 94.5% respectively; equivalent values for L. brevis, L. fructivorans and L. plantarum are 80.8%, 98.5% and 61.8% (Fig. 2A and B). The variation in minimum %ID could be explained by its significant negative regression on the number of publicly available sequences (Fig. 2A and B), which can be attributed to inadequate sampling at high %ID and possible mis-identifications, especially at low %ID.
Fig. 2. Identification of candidate novel taxa from pairwise comparisons of %ID among 16S rRNA gene sequences.
A and B. Minimum pairwise %ID of the V2 region of the 16S rRNA genes for publicly available sequences of each species of Acetobacter and Lactobacillus respectively. The values for the species detected in this study are highlighted as ◆ A. pomorum, △ A. tropicalis in (A), and + L. brevis, ◇ L. fructivorans and ▲ L. plantarum in (B). The equations and r2 values for regression of minimum %ID (y) on number of valid sequences (x) are shown. [For (A), the asterisk refers to the regression equation and r2 excluding outlier species (minimum %ID < 70%).]
C and D. Relationship between minimum pairwise %ID of the V2 region of the 16S rRNA genes and corresponding (near-)full 16S rRNA gene sequences for the publicly available Acetobacter and Lactobacillus species used in (A) and (B) respectively. Regression equations for %ID V2 region of the 16S rRNA gene (y) on %ID near-full 16S rRNA gene (x) are shown.
E and F. Comparison of the %ID of sequences in Table 3 of this study with known species of Acetobacter and Lactobacillus respectively. Species detected in this study are highlighted as in (A) and (B), and clusters are indicated by letter notations (a–r) used in Table 3. For clarity, only clusters with %ID < minimum %ID for the species represented by the top hit are shown.
We adopted two criteria to investigate whether the sequences in Table 3 might be candidate novel taxa. The first criterion applied the minimum %ID obtained for publicly available sequences of the target bacterial species as the cut-off value (Fig. 2A and B). When applied to the 18 clusters listed in Table 3, this criterion yielded eight clusters, three of Acetobacter and five of Lactobacillus (Fig. 2C and D, summarized in Table 3).
The second criterion was based on the widely used 97% ID of the full 16S rRNA gene as a cut-off threshold to define bacterial species (Drancourt et al., 2004; Drancourt and Raoult, 2005; Janda and Abbott, 2007). A 97% ID of the (near-)full 16S rRNA gene is equivalent to 95.6% ID of the V2 region for Acetobacter (96.0% after excluding the outlier species with very low %ID) and 95.4% ID for the V2 region of Lactobacillus (whether or not outliers are included) (Fig. 2E and F). To be conservative, we rounded down these values to 95% ID cut-off for both species. The 14 clusters in Table 3 with < 95% ID to the top hit overlapped with the eight clusters identified by the first criterion [Fig. 2C and D, yielding six sequences as representative of candidate novel taxa (Table 3): one each related to A. pomorum (cluster932, NCBI GenBank accession HQ168004) and A. tropicalis (cluster7664, HQ168006), and four clusters related to L. fructivorans (cluster94, HQ168011; cluster467, HQ168009; cluster668, HQ168008; cluster1982, HQ168012)]. The candidate novel species accounted for 0.1–0.8% of the total reads in a sample (calculated from data in Table S1).
QRT-PCR and diagnostic PCR analyses
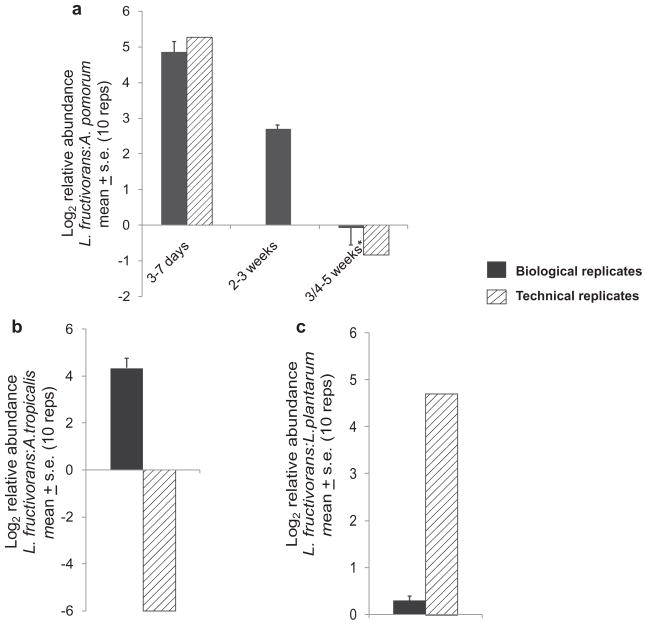
QRT-PCR conducted on adult flies of different ages in June 2010 confirmed the change in relative number of 16S rRNA gene copies of L. fructivorans and A. pomorum, from dominance by L. fructivorans sequences in young adults to A. pomorum sequences in old flies, identified by 454 analysis in November 2009 (Fig. 3A). The dominance of 16S rRNA gene copies of L. plantarum and A. tropicalis in the third-instar larvae and pupae, respectively, were not observed in June 2010, suggesting those life stage-specific effects are not consistent (Fig. 3B and C).
Fig. 3. QRT-PCR of relative abundance of 16S rRNA gene sequence in technical replicates of the pyrosequencing experiment (November 2009) and independent biological samples (June 2010).
A. Lactobacillus fructivorans: A. pomorum in adults of different ages (*3–5 weeks for November 2009 samples; 4–5 weeks for June 2010 samples); 2- to 3-week-old flies were not obtained for the pyrosequencing experiment and its technical replicates in November 2009.
B. Lactobacillus fructivorans: L. plantarum in third-instar larvae.
C. Lactobacillus fructivorans: A. tropicalis in pupae. Ten replicates per sample.
Each DNA sample used for 454 sequencing comprised many insects of strain Canton-S. To assess the prevalence of 16S rRNA gene copies of the various bacterial taxa in individual insects, the guts from five adult males and females were tested for the five dominant bacteria by diagnostic end-point PCR. All Canton S flies were positive for every bacterium, apart from one female which yielded a negative result for L. fructivorans (Fig. S1A). Gut samples from D. melanogaster strains Oregon-R and Ithaca-83 also bore A. pomorum, A. tropicalis, L. fructivorans and L. plantarum, but were negative for L. brevis (Fig. S1B).
Discussion
Previous research on the microbiota of D. melanogaster (Ryu et al., 2006; Corby-Harris et al., 2007; Cox and Gilmore, 2007; Ren et al., 2007) employed relatively shallow sampling strategies that would not have detected low-abundance bacteria, and, apart from Ryu et al. (2008), sampled whole insects. These sampling limitations are overcome in this study by the pyrosequencing of dissected guts. The saturation of the rarefaction curves and species richness indices for all samples of larval, pupal and adult flies (Fig. 1, Table 1) suggests that the entire gut microbiota had been sampled effectively. Nevertheless, it is formally possible that the microbial diversity was underestimated because either the general primers used in this study failed to amplify sequences from certain bacteria, or the amplification of very low-abundance sequences in the template was consistently inadequate for detection. Despite these caveats, which are common to any study founded on PCR, the data indicate that the bacterial community of the Drosophila studied here is, indeed, small, with 17–71 OTUs at 97% ID detected, and dominated by just five species in the two genera, Acetobacter and Lactobacillus. The Shannon index of diversity, at 0.35–1.47 (Table 1), is lower than values for the microbiota in many habitats, including soils (2.4–3.7) (Fierer and Jackson, 2006), coral-associated assemblages (1.54–3.33) (Garren et al., 2009) and vertebrate gut communities [e.g. 4.29 in ostrich caecum (Matsui et al., 2010)], and it overlaps with values (0.8–1.7) obtained for the gut microbiota in the butterfly, Pieris rapae (Robinson et al., 2010).
The diversity obtained from our inventory of the bacteria in the gut habitat of Drosophila is orders of magnitude lower than in the mammalian gut habitat, validating the general pattern in the literature (see Introduction). Low-diversity communities are generated in habitats with extreme disturbance regimes or inhospitable conditions in which few organisms can grow (Grime, 1977; Connell, 1978). The D. melanogaster gut is a transient and disturbed environment at multiple spatiotemporal scales, and arguably more so than in mammals. The larval gut persists for about 4 days before dissolution at metamorphosis, followed by the development of the adult gut and its colonization by bacteria; and the lifespan of the adult gut is 4–5 weeks. Additional sources of disturbance include the passage of food, the elimination of the cuticle lining the foregut and hindgut at each larval moult, and sloughing of gut epithelial cells by a process that is accelerated by the presence of microorganisms (Buchon et al., 2009). Features of animal guts that render them inhospitable to many microorganisms include active enzymes (proteases, lysozyme, etc.) and unfavourable oxygen tensions or pH. The oxygen tension in the D. melanogaster gut has not been studied directly, but its colonization by Acetobacter, which require molecular oxygen, and Lactobacillus, which is intolerant of fully oxic conditions (Yamada and Yukphan, 2008; Ljungh and Wadstrom, 2009), suggests that the conditions in the D. melanogaster gut are either microaerobic or spatially variable with respect to oxygen tension. The composition of the gut microbiota may also be influenced by the composition of the food ingested by the insect host (see Introduction). Of particular relevance to the data obtained here, the Drosophila used in this analysis had been reared on a nutritionally complex diet of yeast extract, fortified with glucose and supplemented with organic acid preservatives for many generations. This regime is predicted to have exerted a strong and consistent selection pressure, for example against taxa intolerant of the organic acids, and favouring taxa at a competitive advantage in high-glucose environments. Further research is needed to understand the detail of interactions between diet and composition of the gut microbiota for Drosophila and other animals.
The low bacterial diversity in the Drosophila gut habitat is evident at the within-species level as well as higher taxonomic levels, such that the same OTU at 97% ID is the most abundant representative for each of the five dominant species in every host life stage from early-instar larvae to aged adults. The additional OTUs of each species (Table S1) may represent low-abundance taxa present in many or all individual hosts, or taxa that dominate a few hosts but are absent from most individuals. Low-abundance ‘cryptic’ taxa have been reported in a various symbiotic systems, including rhizobia in legume root nodules (Denison and Kiers, 2004) and dinoflagellate Symbiodinium in corals (Baker et al., 2004). They may be competitively inferior to the dominant OTU under the prevailing conditions, but become dominant under different circumstances, as reported, for example in coral hosts (Venn et al., 2008). Such shuffling of microbial symbionts can be advantageous to the host, offering insurance against failure of the previous dominant to tolerate or deliver services under different environmental conditions (Douglas, 2010). Alternatively, the minor OTUs may be deleterious to the host, acting as opportunistic pathogens when controls over their growth and division are relaxed. For example, Gluconobacter morbifer is generally occurs at low abundance in D. melanogaster guts, but it proliferates rapidly in immunocompromised flies to become the dominant gut inhabitant with deleterious consequences for the insect (Roh et al., 2008; Ryu et al., 2008). (This species was not detected in our study.)
Central to the design of this study was the variation in the gut bacteria with developmental age and stage of D. melanogaster. The pyrosequencing and QRT-PCR analyses concur that the bacterial composition changed with increasing adult age from dominance of 16S rRNA gene sequences of L. fructivorans to A. pomorum sequences (Table 2b and Fig. 3). Acetobacter, unlike Lactobacillus, grows rapidly under fully aerobic conditions, raising the possibility that the conditions in the D. melanogaster gut become more oxic in ageing insects. Immunological dysfunction associated with ageing can also affect the composition of the gut microbiota, as illustrated by elevated Bacteroides populations in elderly people with persistent activation of the NF-κB transcription factor that plays a central role in innate immunity (Claesson et al., 2011). In this study, two further developmental changes in relative abundance of 16S rRNA gene sequences were identified by pyrosequencing: to high levels of L. plantarum sequences in third-instar larvae and A. tropicalis sequences in pupae (Table 2b). Although confirmed by QRT-PCR of technical replicates, these results were not reproduced in separate biological samples (Fig. 3). In the absence of any overt variation in culture conditions, these data point to potentially important sources of environmental variation that remain to be identified.
This study is based exclusively on 16S rRNA gene sequence data. It should be interpreted with caution in that information on the complement and expression of genes mediating bacterial colonization and proliferation in the gut environment is entirely lacking. This limitation is potentially significant because functionally distinct bacteria with identical or near-identical 16S sequence are known (Scanlan et al., 2009), and differences in gene sequence or expression can have far-reaching phenotypic consequences. For example, gene expression levels are important determinants of the abundance of Leptospirillum bacteria in natural biofilms in acid mine drainage (Denef et al., 2010), and the host range of symbiotic Vibrio is determined by a single regulatory gene (Mandel et al., 2009). These considerations raise the possibility that both the divergent representatives of Acetobacter and Lactobacillus species in D. melanogaster (Table 3) and the bacteria that can confidently be allocated to known species by 16S criteria may be genetically distinct from free-living conspecifics in the content, sequence or regulation of protein-coding genes.
In conclusion, this comprehensive analysis of 16S rRNA gene diversity indicates that the D. melanogaster gut bears a low-diversity bacterial community. Further research focusing on the functional traits of the bacteria is critically important to establish the scale of evolutionary change and diversification of protein-coding genes associated with life in an animal gut.
Experimental procedures
The experimental material
Drosophila melanogaster was reared at 25°C with a 12 h:12 h light–dark cycle on autoclaved yeast-glucose medium [Y-G diet, comprising Brewer’s yeast and glucose (both at 83 g l−1, from MP Biomedicals), agar (10 g l−1, from Frutarom) and preservatives (0.04% phosphoric acid, 0.42% propionic acid, from Sigma)], and transferred to fresh medium weekly. Outbred populations of strains Canton-S and Oregon-R had been maintained on Y-G diet for at least 18 years. Strain Ithaca-83 is an isofemale line established from a single female collected at Littletree Orchard, New-field, New York in 2004, and maintained on Y-G diet since collection.
The experimental samples comprised: guts (from proventriculus to rectum, excluding Malpighian tubules) dissected from third-instar larvae and adults; whole first- to second-(‘early’) instar larvae (< 48 h after hatching: these insects were too small for gut dissections); pupae (which lack a gut); and eggs (< 20 h after deposition). All samples except the eggs were surface-sterilized in 10% sodium hypochlorite solution, followed by three rinses in sterile distilled water. Gut dissections were conducted in sterile Ringer’s solution on clean glass slides with sterilized forceps, using a dissecting microscope at × 7 magnification. This sampling design followed preliminary experiments that confirmed the presence of bacteria in all surface-sterilized samples except eggs (data not shown), consistent with published evidence that bacteria are borne within larvae, pupae and adults, but not internal to the eggshell (Bakula, 1969). All experiments used reagent-only controls comprising a drop of Ringer’s solution treated as for dissections (including swirling the dissection instruments in the solution), but without D. melanogaster materials.
DNA isolation
For pyrosequencing, total genomic DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, California, USA) following a protocol modified from the manufacturer’s instructions for Gram-positive bacteria. Briefly, samples were hand-homogenized in 20 mM Tris-HCl (pH 8.0), 2 mM sodium EDTA, 1.2% Triton® X-100 containing 20 mg lysozyme ml−1. The homogenates were incubated at 37°C for 1.5 h with a 5 min bead-beating in a Disruptor Genie® using 0.1 mm glass beads (Scientific Industries) at 45 min. Pilot experiments confirmed that this treatment disrupted Gram-positive bacteria including Bacillus and Lactobacillus, and achieved 10–50% greater yield than lysozyme digestion without bead-beating (data not shown). All DNA samples were quantified by Nanodrop 1000 (Thermo Scientific) and the PCR products for pyrosequencing were analysed by Agilent 2100 Bioanalyser.
Multiplex 454 pyrosequencing of 16S rRNA gene sequences
Each DNA sample comprised three biological replicates of D. melanogaster strain Canton S: 100 eggs, 50 early-instar larvae, guts from 50 third-instar larvae, 30 pupae, and guts from 50 each of male and female adults at 3–7 days and 3–5 weeks post eclosion. The variable region 2 (V2) of the bacterial 16S rRNA gene was amplified with the general 16S rRNA gene primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 338R (5′-TGCTGCCTCCCGTAGGAGT-3′), with the sample-specific 27F primer bearing a multiplex identifier (MID) sequences and all 27F and 338R primers modified with 5′-Adaptor A and 5′-Adaptor B sequences, respectively, for pyrosequencing (Roche) (Table S3A). PCRs for the biological samples and reagent control were conducted in triplicate with 0.6 U Platinum® Taq DNA Polymerase (Invitrogen) in 1 × PCR buffer, 2 mM MgCl2, 8 pmol each primer, 0.24 mM dNTP and c. 100 ng of DNA sample in 25 μl final volume, at 94°C for 10 min followed by 25 cycles of 94°C for 1 min, 58°C for 1 min and 72°C for 1 min. DNA from an aliquot of each PCR reaction was purified using the Agencourt Ampure® SPRI kit and quantified using the Quant-iT™ PicoGreen® Kit. Each reaction product was diluted to 1 × 109 molecules μl−1, except MID-1 (egg DNA) and MID-9 (reagent-only control), which were diluted to 1 × 108 molecules μl−1. Equal volumes of the three reaction products per sample were mixed together and diluted to 1 × 107 (samples 2–8) or 1 × 106 (MID-1 and MID-9) molecules μl−1 for emulsion PCR at one copy per bead using only ‘A’ beads for unidirectional sequencing. Beads were subjected to sequencing on one full plate of the 454 GS-FLX pyrosequencing instrument using standard Titanium chemistry.
Pyrosequencing flowgrams were converted to sequence reads using 454 Life Science software (http://www.454.com). Reads with ambiguous nucleotides (N) and < 270 nucleotides after the forward primer, and mismatches with the 16S rRNA gene primers were excluded in the initial filtering. To ensure accurate determination of microbial diversity, the data were processed with Perl scripts (Kunin and Hugenholtz, 2010) (http://pyrotagger.jgi-psf.org/release) modified to remove reads with 0.2% per-base error probability (≥ 3% of bases with Phred scores < 27). The remaining sequences were trimmed to 270 nucleotides, dereplicated and clustered into OTUs with 93%, 95% and 97% sequence identity (ID) thresholds. The most abundant unique sequence of each OTU cluster was selected as representative, aligned by P-CLUSTALW at BioHPC (http://cbsuapps.tc.cornell.edu/clustalw.aspx) and subjected to chimera check by the Mallard algorithm (Ashelford et al., 2006). Taxonomy of the non-chimaeric sequences was assigned by NCBI StandAlone BLAST (megaBLAST program) using the nucleotide (nt) database (13 June 2010) with default settings. Identified reads were counted and distributed to their respective MID samples. Phylotypes with < 10 reads or fewer reads than in the reagent-only control were interpreted as contaminants, and removed. The richness [Chao1, abundance-based coverage estimators (ACE) and Jackknife] and diversity (Simpson’s and Shannon) indices for each biological sample were calculated using R. Rarefaction curves were generated using Analytic Rarefaction v1.3 (http://www.uga.edu/~strata/software/index.html).
The Bonferroni-corrected Poisson probability of occurrence of 454 reads with %ID ≤ 98% to the BLAST top hits in each biological sample were calculated using R. A pyrose-quencing error rate of 0.3% was used as it was suggested that pyrosequencing errors can be reduced 0.25% (i.e. up to 3 bp per kb) after discarding reads with ambiguous bases (N) (Huse et al., 2007).
All non-chimaeric 454 sequences are deposited in the short read archive at NCBI, Accession No. SRA023605.3.
PCR assays
Taxon-specific 16S rRNA gene primers were designed for A. tropicalis, A. pomorum, L. brevis, L. fructivorans and L. plantarum (Table S3B) using Primer3 software and unique regions identified from alignments of full 16S rRNA gene sequences. Preliminary experiments confirmed that the primers generated no detectable cross-amplification between species (data not shown). PCRs were performed as above with 65°C annealing temperature and 35 cycles. PCR products were separated by gel electrophoresis using 1% agarose gel and visualized with SYBR®Safe (Invitrogen), and their identities were confirmed by Sanger sequencing.
Specific 16S rRNA gene primers were designed for QRT-PCR of the dominant bacteria (Table S3B). The reactions were conducted in triplicate, with a reagent-only negative control, in C1000™ Thermal cycler (Bio-Rad) with 1 × Brilliant III Ultra-Fast QPCR Master Mix (Agilent Technologies), 8 pmol each primer and c. 100 ng in 20 μl volume, under a thermal profile of 95°C for 10 min, then 35 amplification cycles of 95°C for 10 s, 60°C for 30 s and dissociation cycle of 95°C for 10 s, 60°C for 5 s then brought back to 95°C. Fold differences of bacterial genes were calculated by the ΔΔCt method (Livak and Schmittgen, 2001). The dissociation curve confirmed that every reaction yielded a single PCR product with the predicted Tm. QRT-PCR assays were used to check the repeatability of pyrosequencing data for dominant bacterial species in D. melanogaster. Samples comprised DNA samples from the 454 pyrosequencing experiment (November 2009), and Canton-S flies (June 2010: 10 replicate samples of five pupae and five guts from third-instar larvae and adults at 3–7 days, 2–3 weeks and 4–5 weeks post eclosion). Bacterial relative abundances were compared for A. pomorum/L. fructivorans in adults, L. fructivorans/L. plantarum in third-instar larvae and A. tropicalis/L. fructivorans in pupae.
Pairwise comparisons of %ID of 16S rRNA gene sequences
A non-redundant set of (near-)full 16S rRNA gene sequences for 15 species of Acetobacter (79 sequences) and 102 species of Lactobacillus (1082 sequences) was collected from Greengenes (http://greengenes.lbl.gov), Ribosomal Database Project (RDP; http://rdp.cme.msu.edu) and Silva (http://www.arb-silva.de). Species with a single sequence, unidentified species and species without binomial nomenclature were excluded from the analysis. The remaining sequences were trimmed to 1270 bp, and the V2 region was isolated in silico and trimmed to 270 bp from position 48–318. For each species, all possible pairwise alignments were obtained, and %ID between every sequence pair was calculated using algorithm of (Needleman and Wunsch, 1970) for the (near-full) 16S rRNA and V2 sequences. The lowest value of %ID for each species was adopted as a measure of the total sequence variation for that species.
Supplementary Material
Acknowledgments
We thank Dr Rodrigo Vega, Dr Qi Sun and Dr John Yu for advice, and Dr Brian Lazzaro and Dr Mariana Wolfner who provided the Drosophila. The pyrosequencing was conducted at the Cornell University Life Sciences Core Laboratories Center. We are grateful to the Sarkaria Institute of Insect Physiology and Toxicology for financial support.
Footnotes
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Prevalence of the five major bacterial species (A) in the guts of isolated individual Canton-S flies. (B) Across three different D. melanogaster strains: Canton-S, Oregon-R and Ithaca-83 (each sample is pooled guts of three adult males and three adult females). The positive controls were DNA of each species.
Table S1. Complete microbiota profiles (including read counts and BLAST identities) of D. melanogaster eggs and across larval to adult life stages generated by multiplex 454 pyrosequencing.
Table S2. Bonferroni-corrected Poisson probability of 16S rRNA V2 sequences with ≥ 2% dissimilarity to the BLAST top hit accountable for by pyrosequencing errors, based on 0.3% error rate.
Table S3. Primer details
Text S1. The minimum, 5%-quantile, median, 95%-quantile and maximum intra-species pairwise 16S rRNA V2 and (near-)full sequences %ID of Acetobacter and Lactobacillus species.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol. 2006;72:5734–5741. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker AC, Starger CJ, McClanahan TR, Glynn PW. Coral reefs: corals’ adaptive response to climate change. Nature. 2004;430:741. doi: 10.1038/430741a. [DOI] [PubMed] [Google Scholar]
- Bakula M. The persistence of a microbial flora during postembryogenesis of Drosophila melanogaster. J Invertebr Pathol. 1969;14:365–374. doi: 10.1016/0022-2011(69)90163-3. [DOI] [PubMed] [Google Scholar]
- Behar A, Yuval B, Jurkevitch E. Gut bacterial communities in the Mediterranean fruit fly (Ceratitis capitata) and their impact on host longevity. J Insect Physiol. 2008;54:1377–1383. doi: 10.1016/j.jinsphys.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe. 2009;5:200–211. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, et al. Microbes and Health Sackler Colloquium: composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA. 2011;108:4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell JH. Diversity in tropical rain forests and coral reefs. Science. 1978;199:1302–1310. doi: 10.1126/science.199.4335.1302. [DOI] [PubMed] [Google Scholar]
- Corby-Harris V, Pontaroli AC, Shimkets LJ, Bennetzen JL, Habel KE, Promislow DE. Geographical distribution and diversity of bacteria associated with natural populations of Drosophila melanogaster. Appl Environ Microbiol. 2007;73:3470–3479. doi: 10.1128/AEM.02120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CR, Gilmore MS. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect Immun. 2007;75:1565–1576. doi: 10.1128/IAI.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti E, Rizzi A, Chouaia B, Ricci I, Favia G, Alma A, et al. Acetic acid bacteria, newly emerged symbionts of insects. Appl Environ Microbiol. 2010;76:6963–6970. doi: 10.1128/AEM.01336-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denef VJ, Kalnejais LH, Mueller RS, Wilmes P, Baker BJ, Thomas BC, et al. Proteogenomic basis for ecological divergence of closely related bacteria in natural acidophilic microbial communities. Proc Natl Acad Sci USA. 2010;107:2383–2390. doi: 10.1073/pnas.0907041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison RF, Kiers ET. Lifestyle alternatives for rhizobia: mutualism, parasitism, and forgoing symbiosis. FEMS Microbiol Lett. 2004;237:187–193. doi: 10.1016/j.femsle.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon RJ, Dillon VM. The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol. 2004;49:71–92. doi: 10.1146/annurev.ento.49.061802.123416. [DOI] [PubMed] [Google Scholar]
- Douglas AE. The Symbiotic Habit. Princeton, NJ, USA: Princeton University Press; 2010. [Google Scholar]
- Drancourt M, Raoult D. Sequence-based identification of new bacteria: a proposition for creation of an orphan bacterium repository. J Clin Microbiol. 2005;43:4311–4315. doi: 10.1128/JCM.43.9.4311-4315.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drancourt M, Berger P, Raoult D. Systematic 16S rRNA gene sequencing of atypical clinical isolates identified 27 new bacterial species associated with humans. J Clin Microbiol. 2004;42:2197–2202. doi: 10.1128/JCM.42.5.2197-2202.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn AK, Stabb EV. Culture-independent characterization of the microbiota of the ant lion Myrmeleon mobilis (Neuroptera: Myrmeleontidae) Appl Environ Microbiol. 2005;71:8784–8794. doi: 10.1128/AEM.71.12.8784-8794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garren M, Raymundo L, Guest J, Harvell CD, Azam F. Resilience of coral-associated bacterial communities exposed to fish farm effluent. PLoS ONE. 2009;4:e7319. doi: 10.1371/journal.pone.0007319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grime JP. Evidence for the existence of three primary strategies in plants and its relevance to ecological and evolutionary theory. Am Nat. 1977;111:1169–1194. [Google Scholar]
- Grunwald S, Pilhofer M, Holl W. Microbial associations in gut systems of wood- and bark-inhabiting longhorned beetles [Coleoptera: Cerambycidae] Syst Appl Microbiol. 2010;33:25–34. doi: 10.1016/j.syapm.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol. 2007;45:2761–2764. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Hugenholtz P. PyroTagger: a fast, accurate pipeline for analysis of rRNA amplicon pyrosequence data. Open J. 2010;1:1–8. [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Lehman RM, Lundgren JG, Petzke LM. Bacterial communities associated with the digestive tract of the predatory ground beetle, Poecilus chalcites, and their modification by laboratory rearing and antibiotic treatment. Microb Ecol. 2009;57:349–358. doi: 10.1007/s00248-008-9415-6. [DOI] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ljungh A, Wadstrom T, editors. Lactobacillus Molecular Biology: From Genomics to Probiotics. Norfolk, UK: Caister Academic Press; 2009. [Google Scholar]
- Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. A single regulatory gene is sufficient to alter bacterial host range. Nature. 2009;458:215–218. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H, Kato Y, Chikaraishi T, Moritani M, Ban-Tokuda T, Wakita M. Microbial diversity in ostrich ceca as revealed by 16S ribosomal RNA gene clone library and detection of novel Fibrobacter species. Anaerobe. 2010;16:83–93. doi: 10.1016/j.anaerobe.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Morales-Jimenez J, Zuniga G, Villa-Tanaca L, Hernandez-Rodriguez C. Bacterial community and nitrogen fixation in the red turpentine beetle, Dendroctonus valens LeConte (Coleoptera: Curculionidae: Scolytinae) Microb Ecol. 2009;58:879–891. doi: 10.1007/s00248-009-9548-2. [DOI] [PubMed] [Google Scholar]
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Webster P, Finkel SE, Tower J. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 2007;6:144–152. doi: 10.1016/j.cmet.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Robinson CJ, Schloss P, Ramos Y, Raffa K, Handelsman J. Robustness of the bacterial community in the cabbage white butterfly larval midgut. Microb Ecol. 2010;59:199–211. doi: 10.1007/s00248-009-9595-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh SW, Nam YD, Chang HW, Kim KH, Kim MS, Ryu JH, et al. Phylogenetic characterization of two novel commensal bacteria involved with innate immune homeostasis in Drosophila melanogaster. Appl Environ Microbiol. 2008;74:6171–6177. doi: 10.1128/AEM.00301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JH, Ha EM, Oh CT, Seol JH, Brey PT, Jin I, et al. An essential complementary role of NF-κB pathway to microbicidal oxidants in Drosophila gut immunity. EMBO J. 2006;25:3693–3701. doi: 10.1038/sj.emboj.7601233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, et al. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, et al. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev. 2009;73:249–299. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G, Segal D, Fingo JM, Hefetz A, Zilber-Rosenberg I, Rosenberg E. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc Natl Acad Sci USA. 2010;107:20051–20056. doi: 10.1073/pnas.1009906107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venn AA, Loram JE, Trapido-Rosenthal HG, Joyce DA, Douglas AE. Importance of time and place: patterns in abundance of symbiodinium clades a and b in the tropical sea anemone condylactis gigantea. Biol Bull. 2008;215:243–252. doi: 10.2307/25470708. [DOI] [PubMed] [Google Scholar]
- Wilson M. Microbial Inhabitants of Humans. Cambridge, UK: Cambridge University Press; 2005. [Google Scholar]
- Yamada Y, Yukphan P. Genera and species in acetic acid bacteria. Int J Food Microbiol. 2008;125:15–24. doi: 10.1016/j.ijfoodmicro.2007.11.077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.