Abstract
Lacking from the rapidly evolving field of chromatin regulation is a discrete model of chromatin states. We propose that each state in such a model should meet two conditions: a structural component and a quantifiable effect on transcription. The practical benefits to the field of a model with greater than two states (including one with six states, as described herein) would be to improve interpretation of data from disparate organ systems, to reflect temporal and developmental dynamics and to integrate the, at present, conceptually and experimentally disparate analyses of individual genetic loci (in vitro or using single gene approaches) and genome-wide features (including ChIP-seq, chromosomal capture and mRNA expression via microarrays/sequencing).
Keywords: Chromatin structure, transcription, genome complexity
Introduction
Two established principles of chromatin biology present an obvious paradox for gene regulation: eukaryotic chromosomes are long, contiguous molecules that must be packaged in an ordered manner to fit into the nucleus and global changes in gene expression occur with specificity, reproducibility and speed, across the genome. The first principle has been known for decades; the second has emerged as a result of rigorous molecular dissection of individual transcriptional events in isolation and, in the past decade or so, through the application of global measures of gene expression (microarrays and RNA sequencing). The structure of the nucleosome, the octameric protein complex which binds ~146-147 base pairs of DNA and is the building block of chromatin, is known with atomic level resolution.[1] Likewise, the characteristic appearance of mitotic chromosomes has been known for nearly a century[2]. How the genome is organized in three dimensions in other phases of the cell cycle, or in non-dividing cells, is only beginning to be understood. Moreover, much less is known about the principles of chromatin packing that convert the beads on a string nucleosomal DNA into an ordered three dimensional unit, and how this structure is subsequently reorganized in a non-random way to enable gene expression. Lastly, the intermediate structural states the genome can assume in the non-mitotic somatic nucleus are unknown, although emerging evidence supports discrete intermediate fiber dimensions of 10 or 30 nm (based mostly on reconstitution studies[3] and some microscopic evidence in situ[4]) as well as reproducible inter- and intra-chromosomal interactions between endogenous genetic elements (based largely on chromosomal capture techniques[5-7]).
We propose that the rapidly evolving field of chromatin regulation is inherently limited by a lack of understanding of the structural basis for packaging of the genome. In addition to work addressing this problem experimentally through techniques such as chromosomal conformation capture,[5] we believe the field can benefit from a state model of chromatin. Such a model could serve as a framework to bring together experimental data from reductionist transcription assays in cell culture and in vitro, along with global measures of protein occupancy across the genome and direct measurements of genomic structure. A chromatin state model should be able to evolve as new experiments reveal additional states (or disprove existing hypothesized ones) and importantly, provides a basis for mathematical representation of local and global transitions in chromatin structure.
Considerations for Models of Chromatin Structure
Transcription Classified in a Binary Manner
For the purpose of this thesis we will define transcriptionally on regions of the genome as those areas directly being transcribed to RNA, including genes (exons and introns), validated non-protein- coding RNAs, and characterized proximal promoters. Transcriptionally off regions include distal enhancers (with some exceptions[8]), intergenic DNA and portions of the genome currently unrecognized to code for RNA. Off regions also include genes and other coding regions that can be transcribed but that are not actively being transcribed in the given cell or cellular environment. Sequencing studies estimate that at any given time, ~70% of the mRNA encoding genome is transcribed,[9] keeping in mind that this amounts to a little greater than one-third of the total genome, which is only ~3% exon and ~35% intron in humans and mice (the rest is intergenic DNA; source: NCBI). It is tempting to immediately classify this group of genes (i.e. those that can be activated/transcribed, but that are not, given the overriding cellular condition) as a third group of transcriptional activity. This is avoided in the current analysis based on the following rationale: differentiating off permanently (that is, areas that can never be transcribed) from off temporarily (the type described in the previous sentence) requires omniscient knowledge of all transcriptional states, for all RNAs, in all cells. A more conservative analysis that avoids a false negative assignment of one region as permanently inactive due to a lack of evidence leaves two transcriptional states—on and off— with genetic elements moving between these transcriptional states when polymerases use them as a template for RNA synthesis.
Non-coding Regions and Other Structural Elements Contribute to Chromatin States
Again, we define on as a region of the genome being read as a template to synthesize RNA in a given population of cells. It is possible to argue, however, that the physically contiguous nature of the genome makes it difficult or impossible to functionally separate two regions of DNA adjacent to each other on the physical strand—regardless of whether these regions are undergoing active transcription. Unlike bacteria which lack introns and large regions of intergenic DNA, it is well established that in eukaryotes, coding areas are widely interspersed with regions of non-protein coding, intergenic DNA. In humans, the size of intergenic DNA varies widely from several bases to several megabases. The implication, therefore, is that it is virtually impossible to separate regions involved in transcription from those that are not solely on the basis of whether RNA is actively produced. Indeed these large regions of untranscribed DNA are clearly essential for the genome to assume the convoluted three dimensional structure[2,5] in the interphase nucleus that enables specific gene expression: without intergenic DNA, long range connections between disparate loci required for establishment of chromosomal territories could not be formed. Furthermore, the large amounts of intergenic DNA also likely play a role in genome evolution, acting as a substrate for gene duplication, modification and functional selection. Lastly, these non-coding regions of the genome have been shown to host non-nucleosomal chromatin structural proteins[10] and extensive DNA methylation,[11] both of which are dynamically regulated over the life of a cell, highlighting the role of DNA-binding proteins and DNA modification of intergenic regions in transcriptional regulation (be it indirectly, from a physical standpoint).
The three dimensional configuration of the genome, which is limited (but not encoded) by the primary sequence and the distribution of coding and non-coding regions, then, determines the phenotype of the cell by limiting the range of possible transcriptional states—gene expression states—at any given time. Converting between these states requires changes in the structure of the genome; since the DNA component is unchanged (over transcriptional time scales), this change must be endowed by the non-DNA component of the chromatin backbone: transcription factors, histones, non-histone chromatin structural proteins, post-translational modifications and non-coding RNAs. This relationship also establishes the means for the three dimensional structure of the genome—like the three dimensional structure of a protein—to be a substrate of selection and thereby evolution. In such a scheme, rearrangements of the linear DNA in three dimensions that enabled favorable transcriptional states to arise in the third dimension (that is, in the context of the in vivo packaging of the genome in the interphase nucleus) could undergo positive selection. Negative selection in this same manner is also possible. This logic can explain how transcriptional modules or gene expression profiles that contain genes distributed throughout the genome can be selected for, and thereby conserved, en toto.
The two premises we have to this point are: firstly, from the perspective of chromatin, there are (only) transcriptionally on and off states (how many chromatin states there are will be addressed shortly); secondly, intergenic DNA along with DNA-bound chromatin structural proteins are involved in and probably necessary for the immense variety of cell types in individual eukaryotic organisms, for the countless transcriptional programs they can exhibit, and for their evolution. These two premises present an obvious problem, in that the second seems to contradict the digital logic of the first. Our requirements for a chromatin state model follow from these premises and attempt to resolve this conflict: the model must have a quantifiable transcriptional readout and it must have a detectable structural impact on the genome. An ancillary goal is to provide a rhetoric for understanding the time dimension in chromatin biology, but structuring results in a state model whereby transitions between, and dwell times within, individual states can be captured and compared across different cell types. While pioneering work has suggested that chromatin can directly do things other than endow transcriptional states—such as functioning as a lens in the rod photoreceptor of the nocturnal eye[12]—these actions are at present esoteric and not codified (and certainly not known to be present in all nuclei). For this discussion, we restrict our remarks to actions of chromatin as they relate to transcription, although it is certainly possible that more bizarre behaviors of chromatin remain to be discovered and may be proven evolutionarily important.
Chromatin Model with Two States
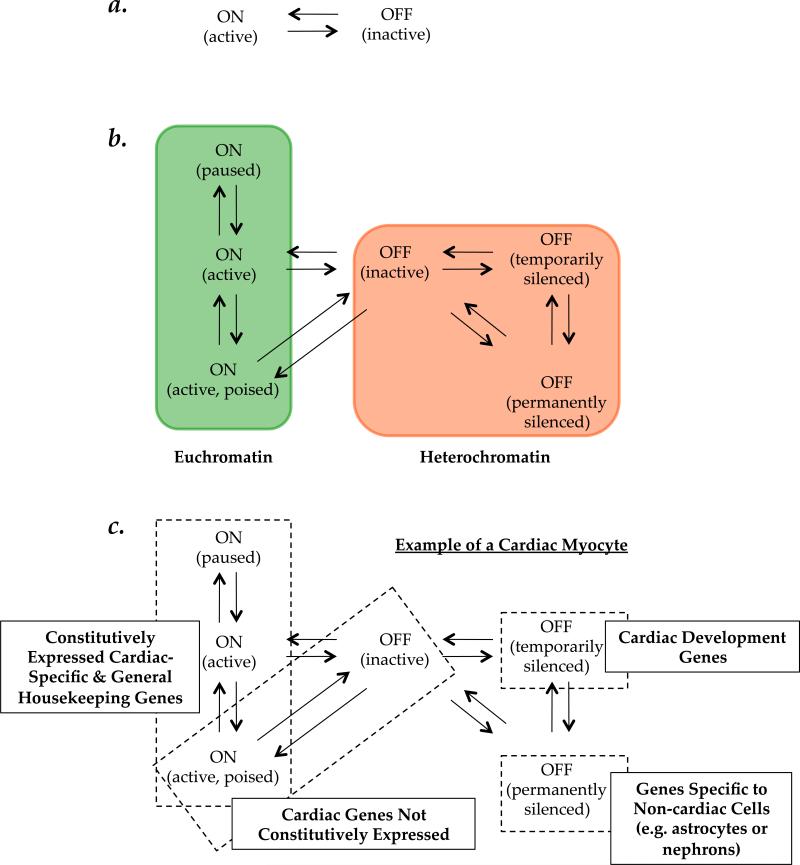
A two state model is appealing because it is for this model alone that unequivocal evidence exists for a transcriptional phenotype: a segment of DNA is either being transcribed, or it is not. A binary model does not require a more nuanced transcriptional or structural readout. Accordingly, in this model the functional chromatin states are either on or off, open or closed, euchromatin or heterochromatin (Panel A in Figure 1). One should not conclude that a two state model is equivalent to a simple one; the everyday example of the enormous complexity that can be generated with 1's and 0's in computer programming is an intuitive example of how two states, applied across a continuum and with differential reading frame, can create complexity (this has also been shown in more relevant examples of biological networks[13]).
Figure 1. Chromatin state models.
A, Although unequivocal data may exist for only a two state model of chromatin with transcription as a readout, with on and off states corresponding to DNA being transcribed or not, this model does not facilitate hypothesis generation for unraveling new properties of chromatin. B, We propose a model with 6 states based on the following criteria: each state is defined by chromatin structural features and a transcriptional readout; major chromatin behaviors meeting the previous requirement are incorporated in one of the states; an open format is used in which more states can be added based on the determination of global features meeting the structure/transcription criteria; and the model incorporates high-throughput ChIP-seq and related data and is constrained by the known physiological properties of chromatin (as opposed to being solely data-driven). The arrows indicate hypothesized routes of transition between states and the shaded areas indicate how we envision these distinct states fitting into a broader concept of chromatin as either hetero- or euchromatic. C, Different types of genes in various states are given for the example of a cardiac myocyte to illustrate the hypothesized relationship between chromatin states and cell-type specific gene expression and phenotype.
Superimposition of signal transduction preceding transcription, transcript/protein abundance and lifetime in the cell, and the role of interaction networks (physical and functional) onto the two state model can give rise to all the variation in transcriptional activities observed experimentally. In other words, the chromatin model itself need have no more than two states to account for the known behaviors of genomes. A cogent argument can be made, in fact, that unequivocal evidence exists for no more than two states of chromatin across model systems, genomes and importantly, individual loci (in contrast to detailed analyses of varied transcriptional behavior observed at isolated loci in vitro or in heterologous cell systems).
While perhaps appealing from an analytical standpoint, there are several practical problems with a two state chromatin model, perhaps foremost that it is hard to disprove and thus a weak vehicle for hypothesis generation. There is also a wealth of knowledge about global chromatin regulation that is not incorporated into two states, including: distal enhancers/repressors, chromosomal territories, three dimensional genomic structure, nucleosome positioning and altered histone variant deposition, distinct activities of RNA polymerase, different rates of transcription, DNA methylation and histone post-translational modifications, to name the major categories that have been shown to affect the two properties—transcriptional readout and structural rearrangement of chromatin—we propose as necessary components of a strong model. While we would argue that these factors have not been shown to universally control gene expression in a given manner, research in these areas is certainly not equivocal and an ideal chromatin state model should incorporate as much of our knowledge on transcription and chromatin structure as possible—even if some of it will ultimately be disproven—to stimulate further experimentation. Thus, there is a need for a chromatin model with greater than two states. Note that in such a model, our definition of two states of transcription still remain; it is a chromatin model that we are positing has a greater number of states.
Multi-State Chromatin Models
The recent deluge of chromatin immunoprecipitation studies coupled with either microarrays or next generation DNA sequencing has fueled the urge to define transcriptional states on the basis of protein binding profiles.[14] While these studies have led to unprecedented insights into genome-wide protein occupancy, a chromatin state model that can be tested and refined across laboratories is unlikely to result solely from this approach based on the following pieces of evidence: global studies are rarely all done in the same cell type, making universality difficult to conclude; the chromatin proteome is known to contain hundreds of proteins, as confirmed from multiple cell types;[15,16] and histone proteins, themselves occurring in numerous isoforms and variants, can undergo scores of modifications in the same cell type.[17] Just considering the last two, a state model incorporating only the 4 core histone proteins and only 10 post-translational modifications could theoretically specify 2^40 (or ~10^12) states, although the actual number would likely be less as some modifications are mutually exclusive. In our view, this is not a meaningful synthesis and, as has been recognized in other areas of cell biology, identifying modular features of biological systems can make their representation tractable[18-21]. Therefore, it is not experimentally (or conceptually) helpful to define states in a model of chromatin based solely on combinatorial patterns of protein occupancy—states in a manageable model should exhibit a transcriptional readout and a chromatin structure phenotype, rather than just reproducible patterns of protein occupancy.
There are numerous histone variants[22] and post-translational modifications[23,24] for which transcriptional readouts, and/or chromatin features, have been described. This is true for states ostensibly “on” or “off” in terms of transcription. For some modifications, effects on higher order chromatin structure have been observed. The recent development of genome-wide ChIP-seq studies has in turn provided extensive information about localization of these modifications in distinct cell types.[25-27] In a similar vein, it is apparent that nucleosome positioning is non-random, dynamic and plays a fundamental role in gene expression.[28-30] What is lacking is a universal dogma for how these features operate in concert to specify global gene expression and thereby phenotype. An emerging theme is that no modification acts in isolation and that, to the extent such modifications specify chromatin states, global changes in gene expression are achieved through combinatorial complexity. A classic example of this phenomenon is bivalent chromatin marks,[31] that is, genes labeled with histone post-translational modifications that alone specify opposing chromatin accessibility and/or transcriptional activity, but combined create decision-making nodes in gene regulation networks.
Permanent inactivation of genes, a well-known example being inactivation of the X chromosome in the female, leads to a distinct type of off state. This process is mediated in large part by histone post-translational modifications, long non-coding RNAs and DNA methylation.[32] Genes in this region of DNA are permanently silenced in all cell types in the organism (“off permanently silenced”, panel B in Figure 1), although which copy of the X chromosome is inactivated varies between cells.
Another functionally obvious chromatin state is characterized by genes not expressed in a given cell type due to silencing associated with normal differentiation.[33] Such genes clearly exist, but a universal dogma to explain their regulation in the context of chromatin states—why each cell has a different gene/protein expression profile—is lacking. Nevertheless, these genes clearly occupy a different off state, in that they can be activated in other cells but never will be in the normal life of the given cell (“off temporarily silenced”, panel B in Figure 1). We propose this chromatin state to be plastic as evinced by dedifferentiation in heart disease,[34] cancer[35] and induced pluripotency.[36]
The last species of functionally distinct off state we seek to define is that containing genes that can be expressed in the normal life of a differentiated cell but that are off at a given point in time (“off inactive”, panel B in Figure 1). This includes genes activated by stress, mitogens, injury, environmental cues and so forth. One can conceptually envision a “poised off” state that is distinct from those discussed so far and which may exist, for example when RNA polymerase disengages from DNA before nucleosomes reassemble, however we know of no functional readout for such a state in terms of transcription and/or chromatin structure.
The most basic “on” state is that in which transcription is actively occurring (“on, active”, panel B in Figure 1) and is identical to the “on” state in the two state model. There is ample evidence that an additional on state exists when RNA polymerase “pauses” on the template, temporarily delaying elongation.[37,38] Another distinct on state is one in which chromatin is “poised” for transcription,[39,40] at which sites transcription is imminent but not active. Both of these states imply directionality and whether they are biologically (rather than just semantically) distinct, remains unknown.
It is well established that RNA polymerase exhibits different firing rates—that it, different rates at which the RNA is generated on a given template. In the present definition of a chromatin state model, we avoid delineation of these different rates as different states due to the absence of evidence for coordinate variable chromatin structure; it is very possible that such structural differences exist and have yet to be detected (or, perhaps these are subspecies of the “on, active” state).
Based on currently available experimental data and the preceding structure-function considerations, we favor a six state chromatin model (Panel B in Figure 1). Of course these states are defined not for entire genomes but for regions of the genome within a single nucleus—different regions of the same genome can simultaneously exist in multiple states. Constitutive heterochromatin is thought to be the same between cell types (although there are exceptions to this, for example in the regulation by centromeric satellite DNA[41]) whereas facultative heterochromatin will vary between cell types and within a cell given the state of development. In the present synthesis, these forms of chromatin would be considered “off permanently silenced” and “off temporarily silenced”, respectively (Panel B, Figure 1). In Panel C of Figure 1, we consider the types of genes that would reside in individual states in the example of a fully differentiated cell like a cardiac myocyte; while we can predict functional classes of genes for on active, off temporarily silenced, off permanently silenced, it is a priori possible to distinguish neither between on active poised and off inactive, nor between on paused, on active and on poised inactive. These distinctions can only be made experimentally. Our goal for this model is to create a framework that incorporates as much of the current data (and ongoing technique development) as possible while at the same time restricting the number of states based on the field's experimental knowledge of chromatin and transcription. An example of how we envision specific transcriptional phenotypes, along with to-be-determined structural features, contributing to chromatin states, is represented in Figure 2. Undoubtedly the result of this approach is an over- simplification of the endogenous behavior of chromatin and the actions of various proteins. To the latter point, non-nucleosomal chromatin structural proteins, such as CCCTC binding factor (CTCF)[42,43] and high mobility group proteins (HMG),[10,15] have been shown to control gene expression and phenotype, while their roles in chromatin packing appear to include formation of intermediate chromatin domains and global endogenous genomic structure. Incorporating the actions of these types of proteins into a chromatin state model with transcription as a readout requires conceptualizing their actions in the context of the endogenous genome: unlike a conventional transcription factor model, these proteins do not have their effects only in and around the transcription start site.
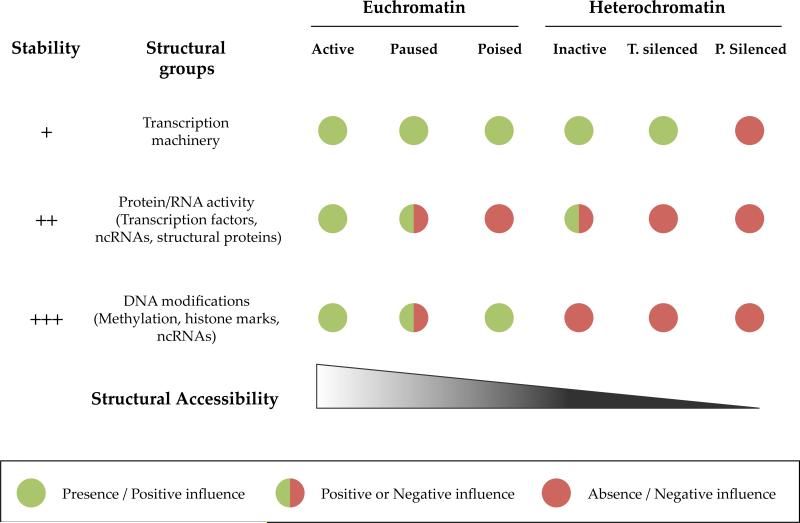
Figure 2. Hypothesized structural differences between chromatin states.
We hypothesize that three key structural groups can, in conjunction with endogenous 3D genomic structure, distinguish the different proposed states at the level of the gene, namely the accessibility of the gene to RNA polymerase II and transcriptional machinery binding (top row), the presence of the appropriate transcription factor in the nucleus and the localization of specific chromatin structural proteins/non- coding RNAs to the gene (middle row) and the presence of conducive, and absence of inhibitory, DNA and histone modifications at the gene (bottom row). Together these help define the structural accessibility of the gene (bottom triangle, with decreasing accessibility as one moves from left to right through the states in as displayed). For each state we propose which class of structural elements are conducive to transcription (green) and which are limiting (red). We further propose that for two states in particular, paused and inactive, more experimental data is needed to determine which class of structural features cause the different transcriptional read-outs (half red/half green demarks possibilities which must be confirmed experimentally). Finally, for each class of structural groups we propose their relative stability and thus the energy required to interconvert between states when these levels of structural regulation are modified.
Implications and Future Experiments
The objective of this thesis is to lay the groundwork for a chromatin model that incorporates three-dimensional structural changes with a transcriptional readout. Such a model is an essential analytical tool, we reason, to resolve the issue of how genome-wide regulation of gene expression is accomplished in eukaryotes in vivo. We present two models that fulfill the criteria of having structural changes in chromatin and transcriptional readouts: a two state and greater than two state model. For reasons of completeness with existing data and ability to generate hypotheses, we favor a model with greater than two states and suggest that a six state model is best representative of current knowledge from both isolated genetic elements and genome-wide studies. While other models of chromatin have been proposed,[14,27,44,45] there are none to our knowledge with these criteria linking structure to transcription as described herein. Of course a model of chromatin states must be linked inextricably with data—our point herein is that incorporating knowledge of the biological function of chromatin can result in a more tractable model. The next experimental step is to directly measure structural features that define individual states.
Use of a Chromatin State Model
Implicit in a transcriptional model of chromatin states are the structural changes at intermediate levels of organization, that is, below the level of the chromosome and above the level of the individual nucleosome. Recent work[30,46] with DNase hypersensitivity, MNase-sequencing analyses and computational approaches has revolutionized the way we think about how nucleosomes associate with different regions of the genome and the logic for the repositioning of these protein complexes commensurate with transcription factor binding. For instance, we now know that upstream of transcription start sites, nucleosomes exhibit very regular spacing,[29] likely contributing to one of the “on” states in this or other chromatin state models. On a more global level, nucleosomes are enriched in exons and depleted in introns,[47] further supporting the role nucleosome density plays in chromatin structure. One limitation of these nuclease-sequencing studies is that the information, although now acquired with single base resolution, is projected onto the linear representation of the genome; in the process, three-dimensional information is lost. Indeed these intermediate areas of chromatin structure represent, in our view, a key frontier in the study of chromatin and chromatin states: new techniques that can directly measure intermediate chromatin states (e.g. by imaging[12,48] and/or chromosomal capture techniques[5,6])—between the level of the nucleosome and the whole genome—and link these states with transcription, will advance our understanding of genome packing and chromatin biology. These approaches can enable representations of chromatin that include both three-dimensional structure and gene expression networks, such that features of cell type-specific chromatin structure (and thereby, transcriptome generation) can be modeled,[7] and ultimately, compared.
The worth of this model is that it provides testable hypotheses. Converting between chromatin states requires something other than the DNA substrate itself; two obvious and extensively studied candidates are RNA and protein. Notwithstanding DNA sequence preferences for remodelers and transcription factors, how inter-state conversion is coordinated on a genome-wide scale is unknown. To test this, one would need to experimentally characterize transitions to establish directionality of movement among states. To the extent possible, this would include recapitulation of large-scale chromatin structures in vitro and/or use of mathematical modeling combined with experimental data to define distinct three-dimensional structural features, should they exist, that are hallmarks of different states. Once structural hallmarks can be established, dwell times in each state can be measured (similar to what has been done genome-wide for individual nucleosomes[49]) and this quantitative information added to the model. A major challenge with this type of experimentation is that it at present is conducted on populations of (usually millions) cells or on tissues. The heterogeneity present in such populations presents a major technical hurdle for rigorous delineation of any discrete subcellular event, chromatin remodeling included, and new methods to measure single-cell level changes[50] will revolutionize the way we examine subcellular processes.
Importantly, should a model such as the one proposed herein be tested experimentally, analyses of genome occupancy (ChIP-seq) must be coupled with expression (microarrays/sequencing) and chromosomal conformation capture analysis, to define whether nuanced structural states accompany the various patterns that emerge from ChIP-seq and gene expression studies. Any of these three global methods alone is insufficient to define chromatin states. If one accepts the premise of chromatin states defined by structural features and transcriptional states, then any investigation of chromatin measuring these outputs directly or indirectly could be interpreted in the context of this model—that is, assigning individual loci measured in a given system to one chromatin state. In more comprehensive studies, this would allow determination of what percentage of the coding genome resides in any given state, and if carried out before and after stimuli (or perhaps in healthy and diseased cells), would shed light on how physiological processes result from shifting portions of the genome between different chromatin states. Such comprehensive studies, which are increasingly common, would allow the model to be disproven by showing that additional chromatin states exist based on significant portions of the genome exhibiting structural and transcriptional features not defined in the existing states, or to demonstrate the absence of structural features distinguishing two states posited to exist, implying only transcriptional states, not those of chromatin, determine a physiological response. Regardless of the system, key data inputs for such a model include loci-specific transcriptional data (microarrays and RNA seq—the higher the resolution, the better) and chromatin structural readouts (principally techniques like MNase-seq, DNase hypersensitivity analysis and ChIP-seq, but increasingly super resolution imaging and chromosomal capture techniques like 3C/HiC will play a role). We speculate that regions of the genome near each other in three-dimensions based on 3C/HiC data would occupy the same chromatin state. Insights into cellular phenotype will certainly aid in the use of these models of chromatin states but in principle are not required for their generation. We envision these models being probabilistic to begin with, due to the large amounts of data and distinct number of loci to be considered, although when possible it will be appealing to apply numerical and potentially analytical methods (employing differential equations) to encapsulate the behaviors of chromatin.
Other Contributors to Chromatin States?
While DNA methylation is clearly associated with modulating chromatin function,[51] the discrete structural features it endows make challenging its direct incorporation into a state model. Like histone post-translational modification, context specificity clearly plays a role in determining the readout. Future studies into histone and DNA modifications need to address directly whether there is universal employment of combinations of marks to endow different readouts. Whether there is a code in the strict sense will require omniscient knowledge of all histone modifications (an unrealistic proposition) or a reimagining of what constitutes the digits in such a code. Regardless, if the goal is to understand global remodeling of chromatin, more studies examining histone and DNA modifications need to test readouts of chromatin structure directly, including: DNase sensitivity, MNase digestion, sedimentation analysis in reconstituted chromatin, high resolution microscopy and perhaps most prominently, chromosomal conformation capture techniques. This model posits six states for coding DNA, but only one (“off permanently silenced”) for non-transcribed regions of the genome. A major challenge for structural studies of chromosomes in situ is to determine structures of non-coding regions in a manner that allows them to be linked to phenotype, regardless of whether these non-coding regions directly or indirectly affect transcription.
How does one distinguish whether a structural or transcriptional action is of paramount importance, when evidence for both exists (whereas often when evidence for which comes first, on an atomic scale, does not)? In the case of a transcription factor binding to the promoter of a gene, it is not much of a jump to assume that if the transcription factor induces a structural change important for transcription, then this change should precede transcription. However, for chromatin structural proteins that decorate various coding and non-coding areas of the genome, the simple linear view of local structural changes preceding transcription of the same region quickly becomes inadequate. Thus, when no data on time-course exist (from a straightforward cause-effect point of view), our preference is to consider them as a single event. Novel approaches that can link structural changes to transcriptional outputs when these two events are not connected by the linear DNA strand (i.e. that rely on the endogenous three-dimensional architecture of the genome) will enhance this understanding.
Genomes—the term here referring to the DNA and all the chromatin structural proteins and RNAs that bind it—are self-organizing systems; there is no master regulator mechanism that assembles the three dimensional structure of the genome in vivo. In this property genomes are not unlike proteins themselves,[52] in that structural features arise at secondary, tertiary and quaternary levels to endow functionality not present in the primary protein/DNA sequence. Like protein structures, then, we can examine genome structures in different cells to reveal features endowing cell type specific gene expression profiles. One fundamental difference between these two systems is that proteins are thought to be structurally super-imposable between copies within a cell or between cells; we certainly do not imply this to be the case with the genome, where structural similarities between copies of a genome are more likely global patterns (think: cloud formations). From an evolutionary standpoint, structural features that arose in proteins, untraceable to amino acid sequence, are selected for based on function. So too it may be with genomes, in that the structural features of the genome in three dimensions—and the consequent properties thereby determining how different regions shift their chromatin states— determine phenotype.
Highlights.
Need exists for state models of chromatin to integrate high throughput data
States are defined by both transcriptional and structural features
Transcriptional insights will come from microarrays and RNA sequencing
Structural insights will come from chromosomal conformation capture and imaging
Experimental observations supporting a model with 6 states are discussed
Acknowledgements
We thank members of the Vondriska laboratory and Drs. Zhilin Qu and Siavash Kurdistani (UCLA) for helpful discussions. We also thank three anonymous reviewers and the editor for helping us refine the ideas in this paper. Research in the Vondriska laboratory is supported by the National Institutes of Health and the Laubisch Endowment at UCLA. HC is the recipient of an American Heart Association Pre-doctoral Fellowship; EM is recipient of the Jennifer S. Buchwald Graduate Fellowship in Physiology at UCLA; MP is the recipient of an NIH Ruth Kirschstein Post-doctoral Fellowship; and SF is the recipient of an NIH K99 Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tremethick DJ. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 2007;128:651–4. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Fussner E, Ching RW, Bazett-Jones DP. Living without 30nm chromatin fibers. Trends Biochem Sci. 2011;36:1–6. doi: 10.1016/j.tibs.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 5.van Steensel B, Dekker J. Genomics tools for unraveling chromosome architecture. Nat Biotechnol. 2010;28:1089–1095. doi: 10.1038/nbt.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bau D, Sanyal A, Lajoie BR, Capriotti E, Byron M, Lawrence JB, Dekker J, Marti-Renom MA. The three-dimensional folding of the alpha-globin gene domain reveals formation of chromatin globules. Nat Struct Mol Biol. 2011;18:107–14. doi: 10.1038/nsmb.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramskold D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuddapah S, et al. Genomic profiling of HMGN1 reveals an association with chromatin at regulatory regions. Molecular and cellular biology. 2011;31:700–9. doi: 10.1128/MCB.00740-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo JU, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14:1345–51. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, Guck J, Joffe B. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–68. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 13.Kauffman S. At Home in the Universe. Oxford University Press; New York: 1996. [Google Scholar]
- 14.Baker M. Making sense of chromatin states. Nat Methods. 2011;8:717–22. doi: 10.1038/nmeth.1673. [DOI] [PubMed] [Google Scholar]
- 15.Franklin S, Chen H, Mitchell-Jordan SA, Ren S, Wang Y, Vondriska TM. Quantitative analysis of chromatin proteome reveals remodeling principles and identifies HMGB2 as a regulator of hypertrophic growth. Mol Cell Proteomics. 2012 doi: 10.1074/mcp.M111.014258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vermeulen M, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–80. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 17.Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA. High throughput characterization of combinatorial histone codes. Mol Cell Proteomics. 2009;8:2266–84. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 19.Vondriska TM, Klein JB, Ping P. Use of functional proteomics to investigate PKC epsilon-mediated cardioprotection: the signaling module hypothesis. Am J Physiol Heart Circ Physiol. 2001;280:H1434–41. doi: 10.1152/ajpheart.2001.280.4.H1434. [DOI] [PubMed] [Google Scholar]
- 20.Franklin S, Vondriska TM. Genomes, proteomes, and the central dogma. Circ Cardiovasc Genet. 2011;4:576. doi: 10.1161/CIRCGENETICS.110.957795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch C. Systems biology. Modular biological complexity. Science. 2012;337:531–2. doi: 10.1126/science.1218616. [DOI] [PubMed] [Google Scholar]
- 22.Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–75. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 23.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 24.Turner BM. Decoding the nucleosome. Cell. 1993;75:5–8. [PubMed] [Google Scholar]
- 25.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2010;470:279–83. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–9. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Filion GJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–24. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–98. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, Wippo CJ, Wal M, Ward E, Korber P, Pugh BF. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science. 2011;332:977–80. doi: 10.1126/science.1200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–6. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 32.Morey C, Avner P. Genetics and epigenetics of the X chromosome. Ann N Y Acad Sci. 2010;1214:E18–33. doi: 10.1111/j.1749-6632.2010.05943.x. [DOI] [PubMed] [Google Scholar]
- 33.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–84. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajabi M, Kassiotis C, Razeghi P, Taegtmeyer H. Return to the fetal gene program protects the stressed heart: a strong hypothesis. Heart Fail Rev. 2007;12:331–43. doi: 10.1007/s10741-007-9034-1. [DOI] [PubMed] [Google Scholar]
- 35.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330:1340–4. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 36.Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–12. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–51. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5' end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 39.Gross D, Garrard WT. Poising chromatin for transcription. Trends Biochem Sci. 1987;12:293–297. [Google Scholar]
- 40.Barski A, Jothi R, Cuddapah S, Cui K, Roh TY, Schones DE, Zhao K. Chromatin poises miRNA- and protein-coding genes for expression. Genome Res. 2009;19:1742–51. doi: 10.1101/gr.090951.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ugarkovic D. Functional elements residing within satellite DNAs. EMBO reports. 2005;6:1035–9. doi: 10.1038/sj.embor.7400558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt D, et al. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell. 2012;148:335–48. doi: 10.1016/j.cell.2011.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–50. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2011;28:817–25. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009;25:335–43. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chodavarapu RK, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–92. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitchell-Jordan SA, Chen H, Franklin S, Stefani E, Bentolila LA, Vondriska TM. Features of endogenous cardiac chromatin revealed by super-resolution STED microscopy. J Mol Cell Cardiol. 2012 doi: 10.1016/j.yjmcc.2012.07.009. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 50.Bendall SC, Nolan GP. From single cells to deep phenotypes in cancer. Nat Biotechnol. 2012;30:639–647. doi: 10.1038/nbt.2283. [DOI] [PubMed] [Google Scholar]
- 51.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 11:204–20. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grosberg A, Rabin Y, Havlin S, Neer A. Crumpled globule model of the three-dimensional structure of DNA. Europhys Lett. 1993;23:373–378. [Google Scholar]