Abstract
Ligand functional groups can modulate the contributions of one another to the ligand-protein binding thermodynamics, producing either positive or negative cooperativity. Data presented for four thermolysin phosphonamidate inhibitors demonstrate that the differential binding free energy and enthalpy caused by replacement of a H with a Me group, which binds in the well-hydrated S2′ pocket, are more favorable in presence of a ligand carboxylate. The differential entropy is however less favorable. Dissection of these differential thermodynamic parameters, X-ray crystallography, and density-functional theory calculations suggest that these cooperativities are caused by variations in the thermodynamics of the complex hydration shell changes accompanying the H→Me replacement. Specifically, the COO− reduces both the enthalpic penalty and the entropic advantage of displacing water molecules from the S2′ pocket, and causes a subsequent acquisition of a more enthalpically, less entropically, favorable water network. This study contributes to understanding the important role water plays in ligand-protein binding.
Keywords: Water-mediated Cooperativity, Double mutant cycle, Differential binding energy, Polarization-enhanced hydrogen bonds, Enthalpy-entropy compensation, Density-functional theory calculation, Structure-thermodynamics-relationship
Introduction
Interpreting the changes in the binding free energy across a series of congeneric ligands (aka the structure-activity-relationship [SAR]) is one of the central pillars in the practice of medicinal chemistry. These free energy changes are the net result of enthalpy and entropy changes that often oppose each other (enthalpy-entropy compensation). A deeper understanding of the changes in the binding thermodynamics across congener series1–2 (structure-thermodynamics-relationship) is consequently fundamental to fully comprehend SAR relationships. Binding thermodynamics are determined by the thermodynamic status of the solvated ligand-protein complex relative to that of the solvated uncomplexed ligand and protein. Thermodynamic changes can therefore be produced when the solvated ligand-protein complex and/or the solvated uncomplexed ligand systems are perturbed by ligand structural modifications. One of the most versatile elements of these systems, which can be easily perturbed, is water. Water is a major player in ligand-protein binding whose role in the binding process is not yet fully understood3. In recent years, there has been growing awareness of many of the previously unexplored aspects of water involvement in ligand-protein binding4–8. For example, a very recent study published by Snyder et al provided new insight into the hydrophobic effect through analyzing the thermodynamics of water molecules in carbonic anhydrase active site9. An intriguing characteristic of water molecules in active sites is their ability to participate in extended H-bond networks10 which can have favorable enthalpies (due to multiple H-bonds) and unfavorable entropies (due to reduction in mobility). Perturbations of such networks by incremental structural changes in the ligand can have their own enthalpy-entropy compensation effects that are layered upon those of the bound complexes and the unbound ligands. Investigating the thermodynamic consequences of these perturbations is therefore crucial for understanding the relative enthalpy, entropy, and free energy of binding across series of congeneric ligands.
Understanding structure-activity and structure-thermodynamics relationships is also contingent upon unraveling the details of another important aspect in ligand-protein binding: cooperativity among ligand functional groups. Changes in the binding thermodynamics across a series of ligand congeners can be influenced by the presence of other functional groups in the ligand. For example, the improvement in the binding free energy across a series of congeneric thrombin inhibitors, in which the hydrophobic side chain binding the S3 pocket was systematically varied, was shown to be more pronounced in the presence of a ligand NH2 group capable of forming a H-bond with the protein11. The NH2 group and the hydrophobic side chain in this series of thrombin inhibitors are therefore synergistic; in other words they demonstrate positive cooperativity. Two ligand functional groups are said to be cooperative when their simultaneous existence in the ligand structure contributes more, or less, favorably to the binding free energy than the sum of the contributions of these groups when present individually. Cooperativity between ligand groups X and Y, for instance, can be evaluated using double mutant cycles12–15 by comparing the binding free energy change occurring when both groups exist in the ligand (ΔΔG(H,H→X,Y)) with the sum of the binding free energy changes occurring when each group exists individually (ΔΔG(H,H→X,H)+ ΔΔG(H,H→H,Y)). When ΔΔG(H,H→X,Y) has a more negative value than ΔΔG(H,H→X,H)+ ΔΔG(H,H→H,Y), groups X and Y are synergistic (i.e. ΔΔG(H,H→X,Y)< ΔΔG(H,H→X,H)+ ΔΔG(H,H→H,Y): positive cooperativity). On the contrary, when ΔΔG(H,H→X,Y) has less negative value than ΔΔG(H,H→X,H)+ ΔΔG(H,H→H,Y), groups X and Y are antagonistic (i.e. ΔΔG(H,H→X,Y)> ΔΔG(H,H→X,H)+ ΔΔG(H,H→H,Y): negative cooperativity). Another way to estimate the amount of cooperativity is through comparing either the differential binding energy16 caused by the replacement of the ligand H with group X in the presence “ΔΔG(H,Y→X,Y)” and absence of group Y “ΔΔG(H,H,→X,H)”, or the differential binding energy caused by the H→Y replacement in the presence “ΔΔG(X,H→X,Y)” and absence of group X “ΔΔG(H,H→H,Y)”. If, for example, ΔΔG(H,Y→X,Y) has more negative value than ΔΔG(H,H→X,H), groups X and Y show positive cooperativity and vice versa.
Cooperativity can be produced if the strength of direct ligand-protein interactions is modified. For example, mutual reinforcement of direct ligand-protein interactions17, which might be associated with reducing the residual mobility of the ligand, can cause positive cooperativity (e.g. thrombin inhibitors)11. Cooperativity can be also mediated by other factors which influence the binding process. For example, it may arise from introducing conformational bias in one of the ligand side chains by another, or from modifying the ligand binding mode12. One of the potential sources of cooperativity which, to the best of our knowledge, has not been explored is the influence of a ligand functional group on the structural changes caused by another group in the hydration layers of the uncomplexed ligand and/or the ligand-protein complex (water-mediated cooperativity). For instance, water displacement, reorganization, and other structural changes in the hydration shells caused by a ligand group X can be either facilitated or hindered by another ligand group Y. This could occur when group Y directly interacts with the water molecules being displaced, reorganized, etc. by group X; or indirectly perturbs the extended H-bond networks that these water molecules might participate in. The influence of group Y on the hydration shell changes produced by group X might consequently cause the differential binding energy of the H→X replacement to be more, or less, favorable (ΔΔG(H,Y→X,Y)< ΔΔG(H,H→X,H): positive cooperativity, ΔΔG(H,Y→X,Y)> ΔΔG(H,H→X,H): negative cooperativity).
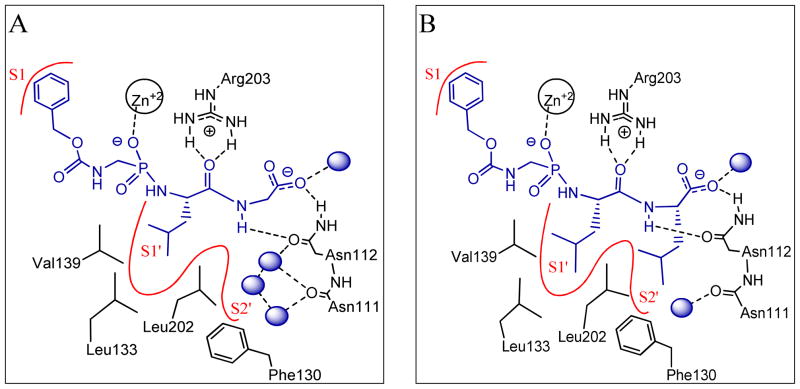
In this study, we tested the hypothesis that cooperativity can be mediated by changes in the hydration shells. Thermolysin (TLN), a Zn-endopeptidase18–20 obtained from Bacillus thermoproteolyticus, was selected as a biological model system to test this hypothesis. One reason for selecting this system was that, in the course of investigating hydrophobic binding in the thermolysin S1′ and S2′ pockets, the binding of ZGPLG16 to thermolysin was found to leave the S2′ pocket well hydrated with a number of water molecules hydrogen-bonded to the protein and to each other (Fig. 1a; details are provided later). Following the hypothesis, it appeared that some of these water molecules can be displaced, and/or rearranged, by the introduction of a hydrophobic side chain capable of binding the S2′ pocket. For example, the crystal structure of the previously reported thermolysin inhibitor, ZGPLL21–22, shows that an isobutyl side chain displaces some water molecules from the S2′ pocket (Fig. 1b). Since these water molecules form H-bonds with Asn112, which is in turn H-bonded to the ligand COO−, it was postulated that the waters that are displaced or rearranged as a consequence of side chain binding to the S2′ pocket could be influenced by the presence of the COO−, and their thermodynamics could be modulated by this group giving rise to variations in the side chain contribution to the binding thermodynamics (i.e. cooperativity).
Fig 1.
Schematic representation of the interactions between the phosphonamidate inhibitors (blue) and thermolysin; H-bonds are shown as dotted lines, water molecules are shown as blue balls, and the enzyme hydrophobic pockets are represented as red curves A) ZGPLG: three crystallographic water molecules are shown in the S2′ pocket; others are not shown; diagram is based on one of the crystal structures discussed later in this paper. B) ZGPLL: two of the three water molecules shown in the S2′ pocket of ZGPLG are displaced by the isobutyl side chain; diagram is based on the crystal structure of ZGPLL (3FWD).
The approach used to investigate the potential cooperativity between side chains that bind in the S2′ pocket and the ligand COO− group was to compare the changes in binding affinities and thermodynamic profiles associated with the introduction of these S2′ side chains in presence, and absence, of the COO− group. The side chain that was selected for this initial investigation was a Me group. The phosphonamidate ligands 8a: ZGPLG and 8b: ZGPLA, both of which have the terminal COO− group, as well as their analogues 8c and 8d, which lack the COO−, were therefore synthesized and analyzed by kinetic assay, isothermal titration calorimetry, and X-ray crystallography in order to answer the following questions: 1) Is there cooperativity between the ligand methyl group which binds in the thermolysin S2′ pocket and the ligand terminal COO− group? 2) Is this cooperativity positive or negative? 3) Is this cooperativity enthalpically, or entropically, driven? 4) Does the aqueous environment of the S2′ pocket play a role in this cooperativity? In order to better reveal the origin of cooperativity, a detailed theoretical approach, along with an initial computational analysis, was used to penetrate further into the differential thermodynamic parameters caused by the H→Me group replacement. Additional inhibitors, with various larger alkyl side chains binding the S2′ pocket, are currently under investigation and will be reported in subsequent publications.
Results and discussion
Chemistry
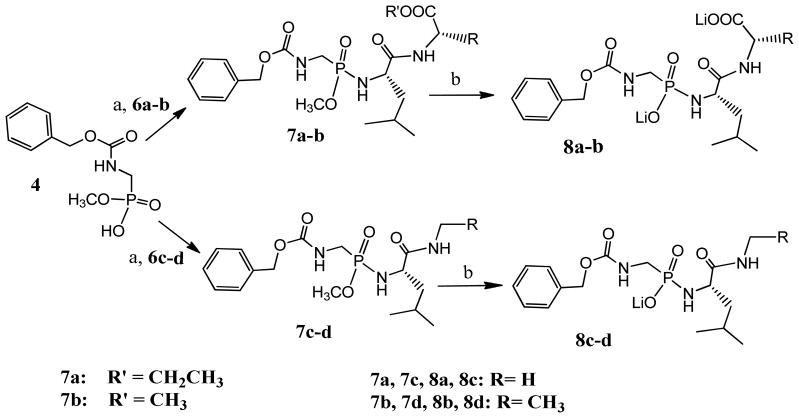
The synthesis of the intermediate 4 is illustrated in Scheme 1. Commercially available benzylcarbamate was heated in an aqueous basic formaldehyde solution to give benzyl N-(hydroxymethyl) carbamate 1 23. The terminal hydroxyl group of intermediate 1 was acetylated using acetic anhydride to give benzyl N-(acetoxymethyl) carbamate 2. Intermediate 2 was converted to dimethyl N-benzyloxycarbonyl aminomethylphosphonate 3 by refluxing with trimethyphosphite. Intermediate 4 was obtained by the hydrolysis of one of the two methyl phosphonate esters in 3 using 10% NaOH solution.
Scheme 1. The synthesis of intermediate 4.
a) 0.5 eq. Na2CO3, 1.5 eq. 37%HCHO, H2O, heat till dissolve, stir at r.t., overnight, 74% b) Excess Ac2O, 6eq pyridine, THF, r.t, 2hr, 67% c) 3–4eq P(OCH3)3, reflux, 3hr, 98% d) 10% NaOH 6eq, r.t, 2hr, 77%
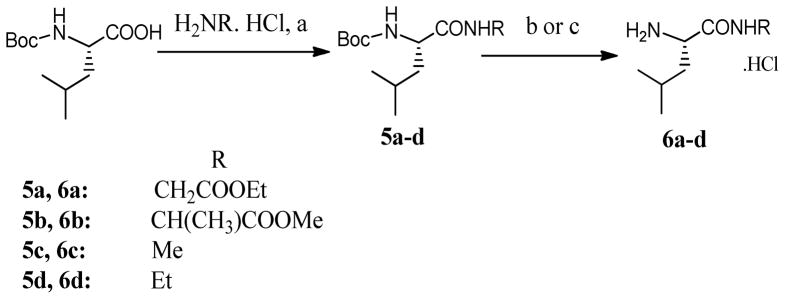
The synthesis of intermediates 6a-d is illustrated in Scheme 2. Boc protected L-leucine was coupled to the hydrochloride salts of glycine ethyl ester and L-alanine methyl ester to give 5a and 5b respectively; and coupled to methylamine hydrochloride and ethylamine hydrochloride to give 5c and 5d respectively. Either EDCI/HOBT or PyBop in anhydrous DMF were used effectively to achieve the coupling in presence of diisopropylethylamine. Compounds 6a-d were obtained as hydrochloride salts upon the removal of the Boc groups from intermediates 5a-d. In order to remove the Boc groups, either 3M HCl/MeOH solution was used (5b-5d), or HCl gas was bubbled into an ethyl acetate solution of the Boc protected intermediate (5a to avoid transesterification with MeOH).
Scheme 2. The synthesis of intermediates 6a–d.
a) 1.2eq PyBop, 1.5eq aminoacid/amine HCl, 4eq DIEA, anhydrous DMF, r.t., 2–5hr 72–80% b) 3M HCl, MeOH, r.t, 1.5–2hr, 99% c) HCl/AcOEt, r.t, 1.5–2hr, 99%
Intermediate 4 was coupled to each of the intermediates 6a-d in anhydrous dichloromethane using PyBop as the coupling reagent to give 7a-d (Scheme 3). Compounds 7a-d were then hydrolyzed using lithium hydroxide to give the final compounds 8a-d either as dilithium salts (when the carboxylate group is present: 8a and 8b) or monolithium salts (when the carboxylate group is absent: 8c and 8d). All the final compounds (8a-d) were purified by reverse-phase HPLC to at least 95% purity.
Scheme 3. The synthesis of ligands 8a-d.
a) 1.2eq PyBop, 1.5eq 6a-d, 4eq DIEA, Anhydrous DCM, r.t. 6hr-overnight 45–75% b) LiOH 2–4eq, H2O/MeCN, r.t. 2hr-overnight, 60–95%
Binding affinity
The inhibition constants (Ki) for compounds 8a-d were determined in a standard thermolysin biochemical assay using 2-furanacryloyl-Gly-Leu-NH2 as a substrate24. The assay was carried out in high salt concentration which improves the enzyme activity as well as the substrate binding to the enzyme. More details about the assay condition are given in the experimental section. The Ki and the corresponding free energy of binding values for the thermolysin inhibitors 8a-d are provided in Table 1.
Table 1.
The Ki and the corresponding ΔG values of the thermolysin inhibitors 8a-d
| Compound | Ki (nM) | ±sd | ΔGKi (kJ/mol) | ±sd |
|---|---|---|---|---|
|
| ||||
| 8a | 472 | 36.0 | −36.1 | 0.2 |
| 8b | 19.1 | 0.55 | −44.0 | 0.1 |
| 8c | 902 | 197 | −34.5 | 0.4 |
| 8d | 289 | 35.1 | −37.3 | 0.3 |
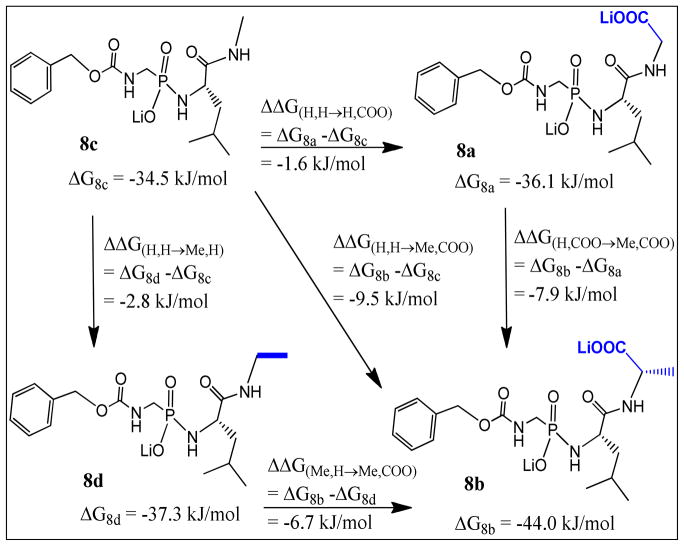
Data in Table 1 were analyzed by the double mutant cycle shown in Fig 2. This cycle reveals that there is positive cooperativity between the Me and the COO− groups which account for 5.1 kJ/mol improvement in binding caused by combining the Me and the COO− groups in 8b (ΔΔG(H,H→Me,COO)− (ΔΔG(H,H→Me,H)+ ΔΔG(H,H→H,COO))= −5.1 kJ/mol). Cooperativity were also estimated by comparing the differential binding energy of H→Me in presence and absence of the COO− group (ΔΔG(H,COO→Me,COO) vs. ΔΔG(H,H→Me,H)). This comparison showed that the Me group is more favorable in presence of the COO− group by 5.1 kJ/mol representing the amount of cooperativity between these two groups (i.e. ΔΔG(H,COO→Me,COO)− ΔΔG(H,H→Me,H)= −5.1 kJ/mol).
Fig 2.
Double mutant cycle showing positive cooperativity of −5.1 kJ/mol between the COO− group and the Me side chain. Cooperativity= ΔΔG(H,H→Me,COO)− (ΔΔG(H,H→Me,H)+ ΔΔG(H,H→H,COO))= −5.1 kJ/mol. Also cooperativity= ΔΔG(H,COO→Me,COO)− ΔΔG(H,H→Me,H)= −7.9- (−2.8) = −5.1 kJ/mol. ΔΔG(H,COO→Me,COO): the differential binding free energy of the H→Me replacement in presence of the COO− group. ΔΔG(H,H→Me,H): The differential binding free energy of the H→Me replacement in absence of the COO− group.
ITC data25
Isothermal Titration Calorimetry (ITC) was used to dissect the changes in binding affinities caused by the replacement of H with Me, both in the presence and the absence of the ligand COO− group, into their enthalpic and entropic components. Binding enthalpies, entropies and free energies of 8b relative to 8a (differential enthalpy, entropy and binding free energy in presence of the COO− group) and of 8d relative to 8c (differential enthalpy, entropy and binding free energy in absence of the COO− group) are given in Table 2. Absolute ITC values of the four inhibitors under investigation are not given because these values reflect not only the thermodynamics of the binding process, and any associated molecular changes, but also the thermodynamic signal of the dipeptide Val-Lys displacement from the enzyme active site by the inhibitors22. This dipeptide is produced by autoproteolysis of thermolysin in the enzyme concentrated solutions used in ITC experiments. The Val-Lys displacement contributions to the observed ITC values are assumed to be the same for all of the inhibitors; consequently they are cancelled out in the relative values given in Table 2.
Table 2.
The relative binding thermodynamic values of 8b relative to 8a, and 8d relative to 8c as determined by ITC
| ΔΔGKd (kJ/mol) | ΔΔH (kJ/mol) | −TΔΔS (kJ/mol) | |
|---|---|---|---|
| 8b–8a | −5.6 | −13.3 | 7.7 |
| 8d–8c | −2.2 | 2.5 | −4.7 |
The experimentally determined values of enthalpy can be influenced by the buffer ionization capacity if the ligand and/or the protein undergo protonation/deprotonation upon binding26. The buffer ionization capacity is however anticipated to equally affect the observed enthalpies of ligands which pick up/lose the same number of protons upon binding. Since the thermolysin inhibitors 8b and 8a have the same ionizable groups, the binding of either 8b or 8a, to the enzyme presumably causes the ligand-protein system to pick up/lose the same number of protons. The ionization contributions to the observed enthalpies of 8b and 8a are therefore equal and cancelled out in their relative value. The same applies to 8d and 8c relative to each other as they also have the same ionizable groups. Changes in the protonation state that these ligands may undergo upon binding are currently under investigation.
Table 2 shows that the replacement of H with Me group in absence of the COO− group causes an entropy-driven enhancement of binding free energy (favorable differential entropy dominates unfavorable differential enthalpy: −TΔΔS8d-8c (−TΔΔS(H,H→Me,H))= −4.7 kJ/mol; ΔΔH8d-8c (ΔΔH(H,H→Me,H))= 2.5 kJ/mol). The replacement of H with Me group in presence of the COO− group, however, causes an enthalpy-driven enhancement of binding free energy (favorable differential enthalpy dominates unfavorable differential entropy: ΔΔH8b-8a (ΔΔH(H,COO→Me,COO))= −13.3 kJ/mol; −TΔΔS8b-8a (−TΔΔS(H,COO→Me,COO))= 7.7 kJ/mol). The presence of the COO− group, therefore, improves the enthalpic contribution of the Me group by 15.8 kJ/mol (enthalpic positive cooperativity; ΔΔH(H,COO→Me,COO)− ΔΔH(H,H→Me,H)= −13.3–2.5= −15.8 kJ/mol). This large enthalpic positive cooperativity is substantially compensated by an entropic negative cooperativity of 12.4 kJ/mol [−TΔΔS(H,COO→Me,COO)− (−TΔΔS(H,H→Me,H))= 7.7- (−4.7)= 12.4 kJ/mol]. The data of the binding free energies calculated from the dissociation constants Kd’s, that were determined in ITC experiments, shows positive cooperativity between the Me and the COO− groups of −3.4 kJ/mol [ΔΔG(H,COO→Me,COO)− ΔΔG(H,H→Me,H)= −5.6–(−2.2)= −3.4 kJ/mol]. The magnitudes of the ITC-determined free energy cooperativity and the kinetically determined cooperativity are reasonably comparable (−3.4 vs. −5.1 kJ/mol). Free energy cooperativity can be also obtained when both the enthalpic and the entropic cooperativities are added together as shown in equation 1.
| (1) |
Dissecting the differential thermodynamic parameters associated with the H→Me replacement
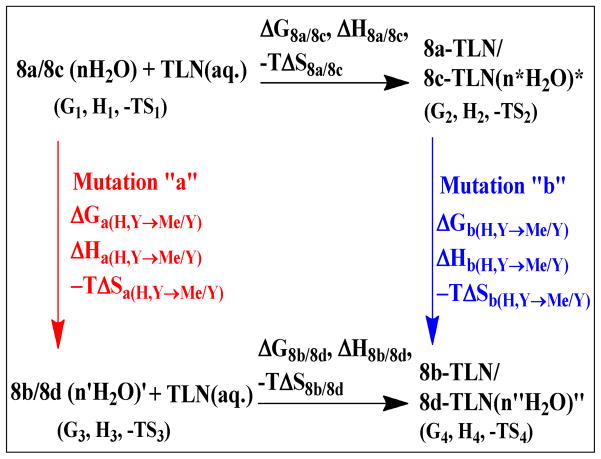
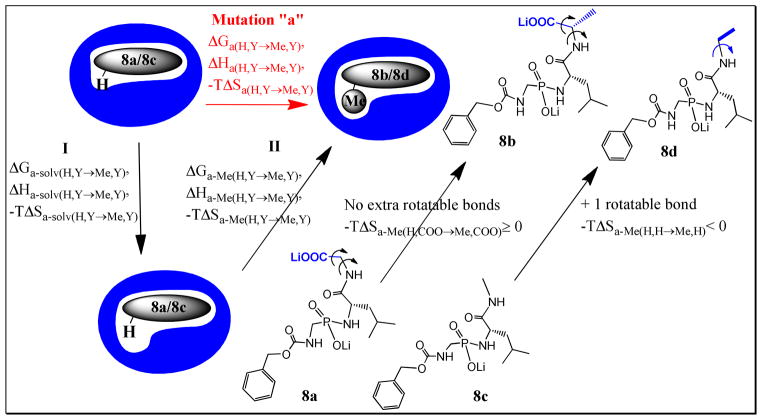
The differential thermodynamic parameters caused by the structural modification H→Me were examined using the thermodynamic cycle shown in Fig 327. This thermodynamic cycle includes four systems: (1) the uncomplexed solvated ligand 8a, or 8c, together with the uncomplexed solvated TLN, (2) the solvated ligand-protein complex 8a-TLN, or 8c-TLN, (3) the uncomplexed solvated ligand 8b, or 8d, together with the uncomplexed solvated TLN, and (4) the solvated ligand-protein complex 8b-TLN, or 8d-TLN. Both (1)→ (2), and (3)→ (4) represent the binding of 8a/8c, and 8b/8d to TLN respectively; while (1)→ (3), and (2)→ (4) represent the mutation of the uncomplexed 8a/8c→8b/8d (mutation a), and the mutation of the 8a/8c-TLN→8b/8d-TLN complexes (mutation b). As illustrated in Fig 3, mutations “a” and “b” can be accompanied with significant changes in the hydration states of the uncomplexed ligand and the ligand-protein complex.
Fig 3.
Theoretical thermodynamic cycle showing the relative binding of ligands 8a and 8b, or 8c and 8d, to thermolysin (TLN). It also shows the mutations 8a→8b, or 8c→8d, in both the free (mutation a) and the enzyme-bound (mutation b) states (Y= H in the ligand pair 8c and 8d, and =COO− in the ligand pair 8a and 8b). The hydration state of each species is illustrated as a number (n, n′, n″, or n*) of H2O molecules and are marked by *, ‘, or “ to indicate that the properties of the hydration water molecules might be different from one species to another. The thermodynamic parameters of each system (e.g. G1, H1, −TS1), the binding thermodynamic parameters (e.g. ΔG8b/8d, ΔH8b/8d, −TΔS8b/8d), as well as the thermodynamic parameters of mutations “a” and “b” are shown.
The thermodynamic cycle in Fig 3 shows that a differential binding parameter such as ΔΔG(H,Y→Me,Y) (Y=H/COO−), which is by definition equal to the difference between the binding free energies of the Me and the H analogues (ΔG8b/8d- ΔG8a/8c), is equal to [G4- G3- (G2- G1)]. Rearranging [G4- G3- (G2- G1)] to [(G4- G2- (G3- G1)] which is equal to ΔGb(H,Y→Me,Y)− ΔGa(H,Y→Me,Y), we can equate ΔΔG(H,Y→Me,Y) with ΔGb(H,Y→Me,Y)− ΔGa(H,Y→Me,Y) as well (equation 2A; ΔGb(H,Y→Me,Y): the free energy change caused by mutation “b”, ΔGa(H,Y→Me,Y): the free energy change caused by mutation “a”). Similar equations can be written for both ΔΔH(H,Y→Me,Y) and −TΔΔS(H,Y→Me,Y) (equations 2B and 2C). The thermodynamic cycle shown in Fig 3, therefore, enables one to express the differential binding thermodynamics of two closely related analogues in terms of the thermodynamics of the mutation of the complex of one of these analogues with the protein, to the complex of the other, relative to the thermodynamics of the mutation of the uncomplexed first analogue to the other. The thermodynamic parameters of mutations “b” and “a” can be partitioned, according to the structural changes taking place in these mutations, into more basic terms28 as described in the next sections. In these sections, changes in these basic terms caused by the presence of the COO− group, and how these changes affect the differential thermodynamic parameters, will be investigated in order to find out the term(s) that might be responsible for the experimentally observed enthalpy, entropy and free energy cooperativities between the Me and the COO− groups.
| (2A) |
| (2B) |
| (2C) |
Where Y= H (8c, 8d) or COO− (8a, 8b)
1) Dissecting the thermodynamic parameters of mutation “b”
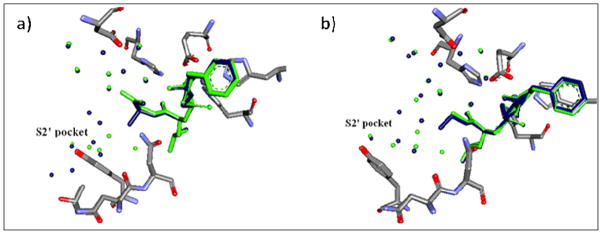
The fact that the free energy, enthalpy and entropy are state functions (dependant on the initial and final states, but not on the path taken to reach the final state) can be utilized to dissect the thermodynamic parameters of mutations “a” and “b”. In mutation “b”, going from the 8c-/8a-TLN to the 8d-/8b-TLN complexes can be achieved through different paths each of which comprises a number of hypothetical steps and artificial intermediate states. In each step, one or more of the structural changes associated with the mutation is accomplished. High-resolution crystal structures for the complexes of the four ligands with thermolysin were taken so that the structural differences between 8c-TLN and 8d-TLN complexes, and between 8a-TLN and 8b-TLN complexes could be determined25. Superimposing the crystal structures of each ligand pair (Fig. 4) reveals the following: 1) No change in the binding mode occurs due to the replacement of H with Me in either pairs of ligands. 2) No significant conformational change in the enzyme occurs due to the H→Me replacement in either pairs of ligands. 3) Significant changes in the hydration layer of the S2′ pocket and the nearby regions take place as the H is replaced by Me in both 8c-TLN and 8a-TLN complexes. It should be noted that an accurate comparison of the hydration shell of a ligand-protein complex, with the hydration shell of another, requires that the two complexes have comparably high resolutions because the detection of crystallographic water molecules is highly dependent on the structure resolution29. This requirement was reasonably fulfilled in the crystal structures obtained for the four ligands in question (resolution ranges from 1.6–1.28Å)25.
Fig 4.
a) Overlay of the crystal structures 8a-TLN and 8b-TLN. Ligand 8a and water molecules that belong to its complex are shown in green. Ligand 8b and water molecules that belong to its complex are shown in blue. Protein atoms of both complexes are shown in the following colors: C (grey); O (red) and N (blue). Some water molecules and protein residues are not shown for clarity. No conformational or binding mode changes, but significant changes in hydration layer, are observed. b) Overlay of the crystal structures 8c-TLN and 8d-TLN. Ligand 8c and water molecules that belong to its complex are shown in green. Ligand 8d and water molecules that belong to its complex are shown in blue. No conformational or binding mode changes, but significant changes in hydration layer, are observed.
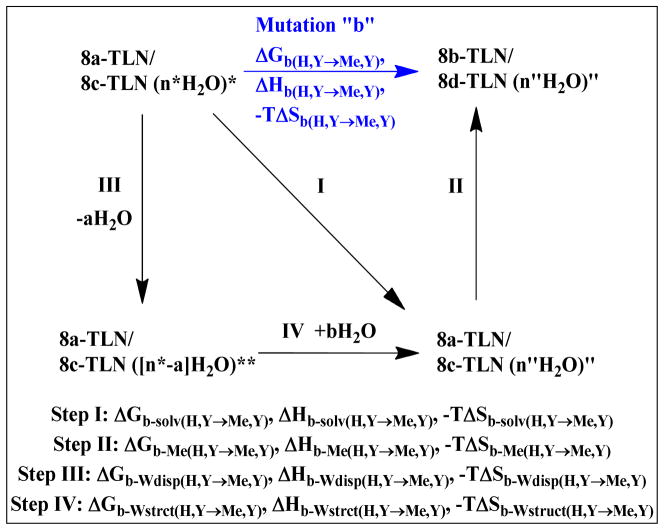
Since no conformational or binding mode changes were observed in the crystal structures, mutation “b” can be accomplished in two steps; the first involves the changes taking place in the complex hydration shell (step I, Fig 5), and the second involves the replacement of H with Me (step II, Fig 5). Changes in the hydration shell that can be detected by X-ray crystallography involve the disappearance of some crystallographic water molecules which exist in the initial, but not in the final complex, and the appearance of others which exist in the final, but not the initial complex. The disappearance of crystallographic water molecules can be attributed to the transfer of these water molecules to the bulk solvent (i.e. water displacement), or to increase in their motions, while the appearance of new water molecules can be attributed to the acquisition of these water molecules from the bulk solvent, or to decrease in their motions.
Fig 5.
Thermodynamic cycle showing that the mutation of 8a-TLN→8b-TLN, or 8c-TLN→8d-TLN, can be achieved via either two (I and II) or three (III, IV and II) hypothetical steps. Step I: The change in the complex hydration state from (n*H2O)* to (n″H2O)″. Step II: The replacement of H with Me group. Step III: Removal of some water molecules from the complex hydration shell. Step IV: The appearance of a number of new water molecules, which are either captured from the bulk solvent or experience reduction in their mobility, in the hydration shell. Thermodynamic parameters of each step are shown.
As a relevant background for the discussion that follows, it is helpful to consider that regions in protein active sites can be roughly categorized according to their hydration status to: (1) Regions hydrated with ordered water molecules which are visible by X-ray crystallography. These regions often have polar groups which can form strong H-bonds with water. (2) Regions potentially hydrated with disordered water molecules which are not visible by X-ray crystallography30–33 (3) void regions. Regions that belong to both the second and the third categories are typically apolar pockets which do not provide sufficient H-bonding groups to make strong interactions with water. Whether these apolar pockets are actually empty, or have disordered waters, is controversial and difficult to determine34. Fortunately, we will not have to deal with this case in the current study because crystallographic data shows that the S2′ pocket is hydrated with ordered waters. Since almost all of the water molecules in the first hydration layer of the area we are interested in could be detected by X-ray crystallography, our discussions will be focused only on these waters.
Considering the thermodynamic principle that nature adopts the status that maximizes the free energy benefit, the fact that the S2′ pocket is hydrated with ordered water molecules indicates that this hydration status is more favorable than either the hydration of this pocket with disordered waters, or its being void. We can therefore conclude that perturbing this hydration status, for example through the transfer of crystallographic waters from the S2′ pocket to the bulk solvent (to create a void area), is unfavorable in terms of free energy (ΔG> 0), and vice versa. The free energy of transferring water molecules to protein cavities hydrated with crystallographic water molecules was previously studied and shown to be negative35, 36. Ordered waters in the active site adopt an entropically unfavorable status because of their restricted motions which allow their crystallographic detection. In order for these waters to remain in the active site, their free energy must be favorable. This can be accomplished only if these waters can form stronger interactions, and consequently have more favorable enthalpy, in the active site relative to the bulk solvent. The transfer of ordered water molecules to the bulk is therefore entropically favorable, due to a relative increase in the motions of these waters in the bulk solvent, and enthalpically unfavorable due to the weaker binding they experience there‡ (binding opposes motions). The unfavorable free energy anticipated for the transfer of crystallographic water molecules to the bulk water can therefore be attributed to the enthalpic penalty which overcompensates the favorable entropy of displacing these waters from the active site (the magnitude of the unfavorable enthalpy is larger than that of the favorable entropy; |ΔH|> |−TΔS|).
Conversely, the aforementioned appearance of new crystallographic water molecules in the complex, which is caused by either the transfer of water molecules to the complex, or the restriction of their motions, is anticipated to have an opposite thermodynamic profile (enthalpically favorable and entropically unfavorable). Since some of the changes in the hydration layer are entropic in nature and others are enthalpic in nature, it is appropriate to partition the first step of mutation “b” into two steps; one involves the entropically favorable, enthalpically unfavorable changes (e.g. crystallographic water displacement; step III, Fig 5), and the other involves the enthalpically favorable, entropically unfavorable changes (e.g. crystallographic water acquisition from the bulk solvent; step IV, Fig 5). The mutation of 8c-/8a-to 8d-/8b-thermolysin complexes, along with its potential steps, is described by the thermodynamic cycle shown in Fig 5. This thermodynamic cycle was used to write equation set 3.
| (3A) |
| (3B) |
| (3C) |
Details of the Mutations of 8c-TLN→ 8d-TLN, and 8a-TLN→ 8b-TLN complexes
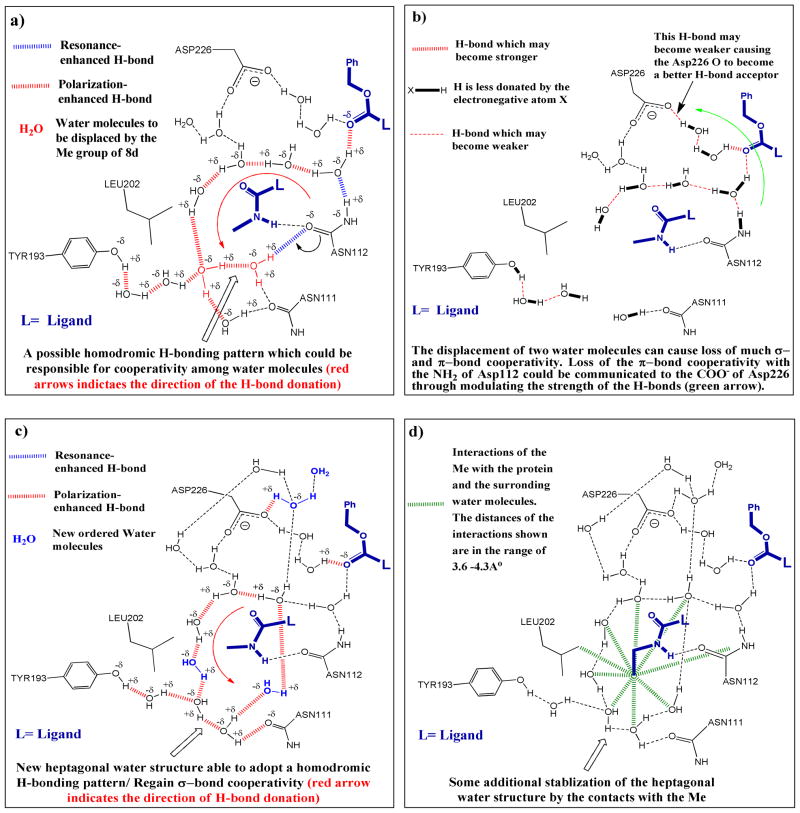
As previously noted in Fig. 5, mutation “b” can be carried out in three steps (steps III, IV, and II). First, water molecules which do not exist in the 8d-TLN (8b-TLN) complex (the final state of the mutation) are removed from the active site of the 8c-TLN (8a-TLN) complex (the initial state). Second, water molecules which appear in the 8d-TLN (8b-TLN) complex, but not in the 8c-TLN (8a-TLN) complex, are incorporated into the complex hydration shell. Third, the H in 8c (8a) is replaced by Me to give 8d (8b). A schematic representation of the S2′ pocket and its adjacent regions in 8c-TLN, 8d-TLN, as well as the two artificial intermediates produced by the first two steps of this mutation, are shown in Fig 6. In this figure, Fig 6a→ Fig 6b corresponds to step III/Fig 5, Fig 6b→ Fig 6c corresponds to step IV/Fig 5, and Fig 6c→ Fig 6d corresponds to step II/Fig 5. An analogous representation for the 8a-TLN→ 8b-TLN three-step mutation is shown in Fig 7. As illustrated in these figures, the region of the active site being investigated is characterized by the presence of many crystallographic water molecules which are, together with the ligand and the protein polar groups, involved in a web-like H-bonding network. Many of these water molecules form chains or polygonal structures37 that are presumably stabilized by the mutual reinforcement (cooperativity) of the H-bonds formed among their water molecules. Mutual reinforcement of H-bonds in these structures can be attributed to resonance-enhancement (π-bond cooperativity)38–41 and/or polarization-enhancement (σ-bond cooperativity) of these H-bonds41–44. Π-bond cooperativity is exemplified in the 8c-TLN complex by the mutual reinforcement of the H-bonds formed by the C=O and the NH2 of the Asn112 amide (Fig. 6a). Σ-bond cooperativity is observed in water arrangements which can adopt homodromic patterns45, 46 (Fig 6a, c). The reinforcement of H-bonds by the π- and the σ-bond cooperativities can be responsible for a great deal of the favorable free energy, favorable enthalpy and unfavorable entropy which crystallographic waters presumably have in ligand-protein complexes.
Fig 6.
Schematic representation of the complex active site showing the most important water molecules and the H-bond networks among them; a) The crystallographic complex 8c-TLN b) An artificial state produced by the removal of two water molecules from the S2′ pocket of the 8c-TLN crystallographic complex. c) An artificial state produced by the incorporation of new crystallographic water molecules in the active site (the ones detected in the 8d crystallographic complex, but not in the 8c complex). d) The 8d-TLN crystallographic complex produced by the replacement of the H of ligand 8c in the artificial state c with Me group. Hs of the water molecules and polar groups were intuitively placed where they can form H-bonds, or where the H-bonds they form can maximize the energetic benefit. a→b corresponds to step III in Fig 5; b→c corresponds to step IV in Fig 5; and c→d corresponds to step II in Fig 5.
Fig 7.
Schematic representation of the complex active site showing the most important water molecules and the H-bond networks among them; a) The crystallographic complex 8a-TLN b) An artificial state produced by the removal of two water molecules from the S2′ pocket of the 8a-TLN crystallographic complex. c) An artificial state produced by the incorporation of new crystallographic water molecules in the active site (the ones detected in the 8b complex, but not in the 8a complex). d) The 8b-TLN crystallographic complex produced by the replacement of the H of ligand 8a in the artificial state c with Me group. Hs of water molecules and polar groups were intuitively placed where they can form H-bonds, or where the H-bonds they form can maximize the energetic benefit. a→b corresponds to step III in Fig 5; b→c corresponds to step IV in Fig 5; and c→d corresponds to step II in Fig 5.
A) Water Displacement from the S2′ pocket
The first step in the mutation 8c-TLN→8d-TLN involves the removal of two water molecules from the S2′ pocket of the 8c-TLN complex (Fig 6a) to produce the artificial state shown in Fig 6b (corresponds to step III/Fig. 5). This step creates the cavity required to accommodate the Me group when it replaces the H of 8c. Similarly, two water molecules are removed from the S2′ pocket of the 8a-TLN complex to create a cavity for the Me group (Fig 7a). The removal, or displacement, of ordered water molecules from the S2′ pocket (i.e. their transfer to the bulk solvent) involves breaking the H-bonds they form with other water molecules and with the protein (e.g. with the Asn111 and the Asn112 residues), as well as reducing the σ- and π-bond cooperativities in the complex H-bond network. This loss of H-bonding correlates with an enthalpic penalty. It is not likely that this enthalpic penalty is fully compensated when the displaced waters form new H-bonds in the bulk solvent because, as it was previously mentioned, water molecules are anticipated to experience weaker H-bonding in the bulk water than when ordered in the complex. Consequently ΔHb-Wdisp(H,Y→Me,Y)> 0. With regard to the entropy and the free energy of this water displacement step, it was pointed out earlier that the displacement of crystallographic water molecules, such as those in the S2′ pocket, from the ligand-protein complex is favorable in terms of entropy and unfavorable in terms of free energy (−TΔSb-Wdisp(H,Y→Me,Y)< 0, ΔGb-Wdisp(H,Y→Me,Y)> 0).
In order to confirm that the removal of water molecules from the S2′ pocket reduces the σ- and π-bond cooperativities in the active site H-bond network, and as an initial effort to investigate the role these cooperativities play in stabilizing this network, the polarization of water molecules in the active site was studied using QM calculations. Model systems were constructed based on the crystal structures of 8c-TLN and 8a-TLN. These model systems included parts of the Tyr193, Leu202, Asn111 and Asn112 residues, the terminal part of the ligand, and many of the water molecules shown in Fig 6a and Fig 7a. The model systems were optimized using the DFT/B3LYP method47–49 with 3-21G basis, and the partial charges of their atoms were calculated using B3LYP functional and triple-zeta cc-pVTZ basis50. The two S2′ pocket water molecules were then removed from each model, and the new partial charges of the remaining atoms were then calculated. The results show that, after water removal, a significant global decrease in the polarization of the remaining water molecules’ atoms takes place. For example, In the model representing 8c-TLN, the average partial negative charge on the O atoms decreases from −0.685 to −0.478, and the average partial positive charge on the H atoms decreases from +0.353 to +0.258 (Fig 8a). Also, in the model representing 8a-TLN, the average partial negative charge on the O atoms decreases from −0.70 to −0.52, and the average partial positive charge on the H atoms decreases from +0.33 to +0.25 (Fig 8b). This indicates an underlying decrease in the cooperativity among the H-bonds formed by the water molecules left after the removal of the two S2′ waters (i.e. a less polarized O or H atom is less capable of participating in H-bonds)51.
Fig 8.
Schematic representation of parts of the model systems which were constructed based on 8c-TLN and 8a-TLN active sites and used for calculations; all of the water molecules included in a model, along with the H-bond networks among them, are shown a) The model system of 8c-TLN before and after the removal of the two red-colored water molecules from the S2′ pocket. Partial charges of the water molecules atoms are shown in blue, each close to its corresponding atom. b) The model system of 8a-TLN before and after the removal of the two red-colored water molecules from the S2′ pocket. Partial charges of the water molecules atoms are shown in blue, each close to its corresponding atom. In both a and b, the numerical values of the partial charges of all of the water molecules’ atoms decrease when the two water molecules are removed from the S2′ pocket.
Influence of the COO− group on the thermodynamics of water displacement
Comparing the structures of the two crystallographic complexes, 8a-TLN and 8c-TLN, reveals that one of the two water molecules whose displacement from the S2′ pocket is discussed herein, in presence of the COO− group, forms one less H-bond than in absence of this group (Fig 7a vs. Fig 6a). The presence of the COO− in the 8a-TLN complex also causes the H-bond network of this complex to be less interconnected than that of the 8c-TLN complex (e.g. in Fig 7a, the southern H-bond network is isolated from the northern one). This indicates a potential reduction in the σ- and π-bond cooperativities in the 8a-TLN complex (relative to 8c-TLN) which can be translated into a decrease in the binding (and an increase in the mobility) of the water molecules in the active site, including those that are displaced from the S2′ pocket. Both the formation of one less H-bond by one of the S2′ waters, and the decrease in the σ- and π-bond cooperativity, can cause the displacement of water from the S2′ pocket to have a smaller enthalpic penalty, and a less favorable entropic change in presence, vs. absence, of the COO− group (ΔHb-Wdisp(H,COO→Me,COO)< ΔHb-Wdisp(H,H→Me,H); −TΔSb-Wdisp(H,COO→Me,COO)> −TΔSb-Wdisp(H,H→Me,H)).
In order to qualitatively support this, the interaction energy of these water molecules with the complex was calculated in each of the two model systems shown in Fig 8. The enthalpic penalty of removing a water molecule from the ligand-protein complex is the sum of the penalty of losing the interactions of this water in the complex and the smaller-in-magnitude advantage of forming new interactions in the bulk solvent. Since the interaction energy of water in the complex is the only component that varies when the COO− group exists (i.e. interactions with the bulk solvent is the same), the difference between the calculated interaction energies of the two water molecules of interest in the model that has the COO− group, and the model that does not have it, can be qualitatively correlated with −(ΔHb-Wdisp(H,COO→Me,COO)− ΔHb-Wdisp(H,H→Me,H)). Dispersion-corrected DFT energy calculations show that the interaction energy of the two S2′ water molecules in the model system that has the COO− group is 9.3 kcal/mol less favorable than their interaction energy in the model system that lacks this group (Eint,coo- Eint,No-coo= 9.3 kcal/mol; supporting information: Table 1). This indicates that, as was predicted based upon structural data, the enthalpic penalty of the removal of the indicated two water molecules from the complex in the absence of the COO− group is larger than the enthalpic penalty of their removal in the presence of this group (−(ΔHb-Wdisp(H,COO→Me,COO)− ΔHb-Wdisp(H,H→Me,H)) correlates with the 9.3 kcal/mol difference in interaction energy→ ΔHb-Wdisp(H,COO→Me,COO)< ΔHb-Wdisp(H,H→Me,H)).
B) Crystallographic water gain
The removal of two water molecules from the S2′ pocket, which produces the intermediates shown in Fig 6b and Fig 7b, most likely causes the configurations of the remaining water molecules in these intermediates to become unstable. This is because, for the rest of the crystallographic water molecules, to remain fixed at their particular positions without the level of enthalpic support provided by the σ- and π-bond cooperativity in the original complexes, would be costly in terms of both entropy and free energy. New water molecules are therefore captured from the bulk solvent, or become more ordered so as to be detectable by X-ray crystallography, allowing the water configurations of Fig 6b and Fig 7b to regain stability (Fig 6c, Fig 7c). Two of the new water molecules appear close to where the two water molecules that were previously removed had existed. These water molecules, together with others left in the S2′ pocket, form a heptagonal (Fig 6c), or a semicircular (Fig 7c), structure which can hydrate the Me group when it replaces the ligand H. It is not uncommon for hydrophobic side chains in protein systems to be hydrated with similar polygonal water structures (e.g. the methyl side chain of Ala92 of the human lysozyme52, the Val3 side chain in the human insulin53, etc), particularly when these water structures are anchored on the protein by one or more polar groups (e.g. Asn111 C=O group). These water structures can restore a great deal of the σ-bond cooperativity, which was lost upon the removal of the previously described water molecules from the S2′ pocket (Fig 6c).
Other new water molecules appear in the intermediates shown in Fig 6c and 7c. For example, additional new waters could be found near the COO− group of Asp226 in Fig 6c. In Fig 7c, the additional new waters appear in two regions: one is bordered by Asn111 and Thr129, and the other lies between the Asp226 COO− and the ligand COO− groups. These new waters in Fig 7c, together with others, form an intricate water network which bridges many of the protein polar groups, and the ligand COO− together. It is possible that the appearance of these additional new waters in the crystal structures is caused by reducing their motions due to stronger binding they might experience following the removal of the two waters from the S2′ pocket: Water displacement from the S2′ pocket could be communicated to other regions of the active site through H-bond networks, bringing about the appearance of new waters in these regions (e.g. the network extending from the Asn112 NH2 to the Asp226 COO−; Fig 6b). The H-bonds formed by the new water molecules can be mutually cooperative. They can also reinforce several existing H-bonds in the active site through σ-bond cooperativity (Fig 7c). As noted earlier, this water gain step is anticipated to be enthalpically favorable due to the formation and the reinforcement of the H-bonds in the complex active site, and entropically unfavorable due to restricting the motions of the water molecules after they join the crystallographic water inventory (ΔHb-Wstrct(H,Y→Me,Y)< 0, −TΔSb-Wstrct(H,Y→Me,Y)> 0).
Influence of the COO− group on the gain of crystallographic waters
The water gain step in both the 8a-TLN→8b-TLN and the 8c-TLN→8d-TLN mutations (i.e. Fig 7b→7c vs. Fig 6b→6c) reveals that the intermediate shown in Fig 7b, which has the COO− group, gains more crystallographic water molecules than the intermediate shown in Fig 6b which lacks this group (e.g. two more waters per the complex region shown in Fig 6 & 7). This step, in presence of the COO− group, produces a more developed H-bond network as well. The number of water chains bridging the ligand and/or the protein polar atoms can be taken as a measure of how developed the active site H-bond network is. Counting the bridging water chains gained in this step in the complex region shown in Fig 6 and 7, both in absence and presence of the COO− group, shows that 12 water bridges§ are gained in presence of the COO−, while only 6 are gained in its absence (Supporting information: Fig 1). Correlating these structural features with the thermodynamic profile of this step, we can conclude that the COO− group causes the water gain step to be more enthalpically favorable, and more entropically unfavorable (ΔHb-Wstrct(H,COO→Me,COO)< ΔHb-Wstrct(H,H→Me,H); −TΔSb-Wstrct(H,COO→Me,COO)> −TΔSb-Wstrct(H,H→Me,H)).
It is important to note that when a particular ligand-protein complex mutation is compared with another, with respect to the number of crystallographic water molecules gained or lost, experimental variations that could lead to artifacts should be taken into account. For example, even slight variations in resolution among ligand-protein complexes could lead to variations in the number of the detected crystallographic water molecules, and may consequently cause an apparent gain or loss of water. It is therefore more appropriate that, for each complex involved in a mutation, the percentage of ordered water molecules observed in the region of interest, relative to the entire complex, be determined and used to calculate a “water gain/loss index” for this mutation. This water gain/loss index is a ratio of the percentage determined for the final complex of the mutation to the percentage determined for the initial complex (eq. 4: the water gain/loss index for the 8c-TLN→8d-TLN mutation). The use of percentages, rather than numbers, of water molecules eliminates the experimental variability among different crystallographic complexes (normalization). If the water gain/loss index for a particular mutation is larger than 1, it can be concluded that this mutation causes gain of crystallographic water, and vice versa.
| (4) |
The water gain/loss indexes for both the 8c-TLN→8d-TLN and the 8a-TLN→8b-TLN mutations in a sphere of 10Ǻ-radius around the terminal Me group of 8c were calculated, and found to be 0.98 and 1.14 respectively (supporting information: Table 2). The water gain/loss index of the 8c-TLN→8d-TLN mutation indicates that the net gain of water observed in this mutation might be an artifact caused by a global enhancement of the detection of water molecules in the 8d-TLN complex. The water gain/loss index of the 8a-TLN→8b-TLN mutation, on the contrary, suggests that the water gain in this mutation is genuine, and reflect an improvement in the organization of water molecules in the S2′ pocket and the nearby region. The water gain/loss index might be correlated with the thermodynamic profile of the overall hydration shell changes (i.e. ΔHb-solv(H,Y→Me,Y) and −TΔSb-solv(H,Y→Me,Y); Step I/Fig 5: net water gain→ favorable enthalpic change and unfavorable entropic change; and vice versa). This correlation, together with the fact that the water gain/loss index of the 8a-TLN→8b-TLN mutation is larger than that of the 8c-TLN→8d-TLN mutation, leads to conclude that the presence of the COO− group causes the thermodynamics of the overall hydration shell changes to be enthalpically favorable and entropically unfavorable relative to when this group is absent (ΔHb-solv(H,COO→Me,COO)< ΔHb-solv(H,H→Me,H); −TΔSb-solv(H,COO→Me,COO)> −TΔSb-solv(H,H→Me,H)). Since the hydration shell changes include both water displacement and water gain, the decrease in ΔHb-solv(H,Y→Me,Y) caused by Y=COO− compared to Y=H comprises the decreases in both ΔHb-Wdisp (H,Y→Me,Y) and ΔHb-Wstrct(H,Y→Me,Y), and the increase in −TΔSb-solv(H,Y→Me,Y) comprises the increases in both −TΔSb-Wdisp (H,Y→Me,Y)) and −TΔSb-Wstrct (H,Y→Me,Y).
Are changes in the hydration shell responsible for the observed cooperativity?
Investigating whether or not the decrease in ΔHb-solv(H,Y→Me,Y) and the increase in −TΔSb-solv(H,Y→Me,Y) caused by the COO− group are responsible for the experimentally observed enthalpic and entropic cooperativities requires substituting the thermodynamic parameters of mutation “b” in equation set 2 with their fundamental components given by equation set 3. Doing so, we can write equation set 5. Equation 5B shows that the decrease in ΔHb-solv(H,Y→Me,Y) caused by the COO− group decreases ΔΔH(H,Y→Me,Y). Since this is in agreement with the experimental data (ΔΔH(H,COO→Me,COO)− ΔΔH(H,H→Me,H)= −15.8 kJ/mol), we conclude that changes in the hydration shell of the complex can be responsible for the enthalpic positive cooperativity between the Me and the COO− groups. Similarly, the increase in −TΔSb-solv(H,Y→Me,Y) caused by the COO− group increases −TΔΔS(H,Y→Me,Y) (eq. 5C). Since this is also in agreement with the experimental data (−TΔΔS(H,COO→Me,COO)− (−TΔΔS(H,H→Me,H))= 12.4 kJ/mol), we can conclude that changes in the hydration shell of the complex can cause the entropic antagonism (entropic negative cooperativity) between the Me and the COO− groups.
| (5A) |
| (5B) |
| (5C) |
What could cause the COO− to produce this effect?
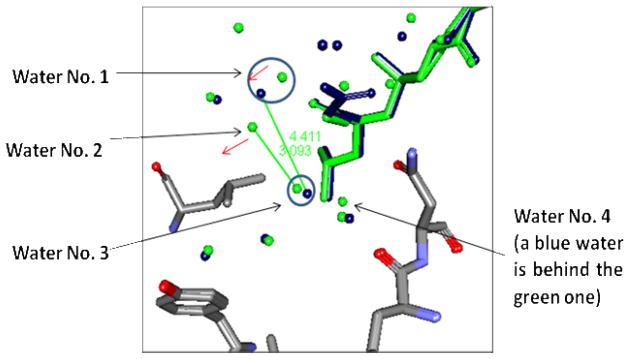
In order to understand why the COO− group is responsible for influencing the hydration shell of the ligand-protein complex in such a way that could produce cooperativity, we need to investigate how the COO− group in the 8a-TLN complex, but not the water molecules which occupy the same region in the 8c-TLN complex, could cause: (1) One of the water molecules being displaced in the first step to have an incomplete set of H-bonds (one H-bond less). (2) The gain of more water molecules and the formation of a more developed H-bond network in the second step of this mutation (i.e. mutation b). First, the effect of the presence of the COO− group on the H-bonding status of the two water molecules being displaced from the S2′ pocket can be illustrated by superimposing the crystal structures of the 8a-TLN complex and the 8c-TLN complex (the initial states of the mutations 8a-TLN→8b-TLN and 8c-TLN→8d-TLN). Fig 9 shows that the COO− group pushes water molecule No. 1 towards water No. 2 which cannot stay in its position anymore because, if it did, it would be in steric clash with water No 1. As a consequence, water No. 3 has one H-bond less in the 8a-TLN complex than in the 8c-TLN complex, and the H-bond network that water No. 3 used to participate in (i.e. in the 8c-TLN) is disrupted (water No. 3 and water No. 4 are the ones being removed in the water displacement step of the mutations 8c-TLN→8d-TLN and 8a-TLN→8b-TLN).
Fig 9.
Overlay of the crystal structures 8a-TLN and 8c-TLN. Ligand 8a and the water molecules that belong to its complex are shown in blue. Ligand 8c and the water molecules that belong to its complex are shown in green. Protein atoms of both complexes are shown in the following colors: C (grey); O (red) and N (blue). Some water molecules and protein residues are not shown for clarity. In ligand 8a complex, water No. 1 is pushed by the COO− group towards water No. 2 as indicated by the red arrow. Water No. 2 ceases to appear in the crystal structure (probably displaced). Water No. 3 loses its 3.1Å H-bond with water No. 2, and cannot form a H-bond with water No. 1 (O--O distance is 4.4Å). Water No. 3 and water No. 4 are the ones being displaced by the Me group in both the mutations 8c-TLN→8d-TLN and 8a-TLN→8b-TLN.
Second, the gain in ordered water molecules and the other associated changes can be considered as compensatory changes that occur secondary to the removal of the two water molecules from the S2′ pocket (e.g. to partially neutralize the unfavorable free energy of water removal from the S2′ pocket). These compensatory changes could be mediated by the charge redistribution that was shown by calculations to occur in the system after the removal of the water molecules from the S2′ pocket (Fig 8). Both the COO− group in the 8a-TLN complex and the water molecules which occupy the same region in the 8c-TLN complex are involved in transmitting this charge redistribution effect through the H-bond network they participate in. Since the COO− group in the 8a-TLN complex, due to being ionized and due to having π-electrons, could be more responsive to charge redistributions than the water molecules in the 8c-TLN complex, and in turn more efficient in relying this effect to other parts of the system, the subsequent compensatory changes in the system might be more prominent when the COO− is present than when it is absent. More detailed computational studies are being carried out in order to investigate this effect, and their results will be reported in due course.
C) The replacement of H with Me
The final step in this mutation involves the replacement of the Hs of 8c and 8a with Me groups to give the 8d-TLN and 8b-TLN complexes respectively (Fig 6d and Fig 7d; corresponds to step II/Fig 5). These Me groups interact favorably with surrounding water molecules, and to a lesser extent with the protein, through dispersion forces. The interactions of the Me groups with their hydrating waters can provide additional stabilization to the heptagonal and the semi-circular water structures as depicted in figures 6d and 7d. The Me interactions with both water and the protein can give rise to some favorable enthalpy (ΔHb-Me(H,Y→Me,Y)< 0). On the other hand, the H→Me replacement is not anticipated to significantly influence the entropy (−TΔSb-Me(H,Y→Me,Y) ≈ 0). The conformational entropy, for instance, is not likely affected by this functional group replacement, even when the ligand acquires an additional rotatable bond in the 8c-TLN→ 8d-TLN mutation (i.e. NH=CH2CH3). This additional rotatable bond is already restricted as the ligand Me group is enclosed in the heptagonal water structure shown in Fig 6d, and the H-bond network among the water molecules of this heptagonal structure would be greatly disrupted if this bond were to rotate freely.
It was previously mentioned that cooperativity can be caused by the mutual reinforcement of direct ligand-protein interactions10. Reinforcement of the Me interactions with the protein by the COO− group, however, does not seem to be the origin of the cooperativity observed in this study. There are two reasons for this. First, the Me group has only few interactions with the protein (i.e. 2–3 contacts within 3.6–4.3Å, Fig 6d, 7d), and reinforcement of such few interactions would not be anticipated to produce large cooperativity terms like those observed here. Second, unlike the cooperativity studied in Ref. 10 which is proportional to the ligand-protein hydrophobic contact, analogues for ligands 8a-d with larger side chains that make more contacts with the protein (e.g. sec-butyl) do not show this proportionality (some even show diminished cooperativity)54. It can therefore be concluded that the cooperativity observed here, and that observed in Ref. 10 have different molecular origins, and that the presence of the COO− group does not significantly change ΔHb-Me(H,Y→Me,Y) (i.e. ΔHb-Me(H,COO→Me,COO)− ΔHb-Me(H,H→Me,H)≈ 0).
2) Dissecting the thermodynamic parameters of mutation “a”
Even though we could attribute the cooperativity observed in this study to changes in the ligand-protein complex hydration waters, the unbound ligand thermodynamics still need to be analyzed because other terms arising from such analysis could also be implicated in the cooperative behavior being investigated here. In this section, we dissect the thermodynamic parameters of mutation “a” (Fig 3) into their basic components. Mutation “a” can be achieved in two steps as shown in Fig 10. First, water molecules reorganize and form a small cavity capable of accommodating the Me group that is to replace the ligand H. Water molecules at the cavity-water interface are not capable of using their full H-bonding capacity if some of their Hs/lone pairs of electrons face the empty cavity and later the hydrophobic surface of the Me group. They, consequently, tend to restrict their motions and assume orientations that maximally preserve their H-bonds (e.g. H-bonded to adjacent water molecules or to ligand H-bonding group). The increase in the order of the interfacial water molecules correlates with an unfavorable entropic change (−TΔSa-solv(H,Y→Me,Y)> 0), and represents the basis for the classical entropy-driven hydrophobic effect. The efficiency in preserving the H-bonds by increasing the order of water molecules around a nonpolar solute largely depends on the size and the shape of the solute. With small side chains like the Me, H-bonds among the interfacial waters are most likely well preserved55. The formation of a cavity in the bulk water for the incoming Me group is therefore anticipated to be accompanied with little change in enthalpy (ΔHa-solv(H,Y→Me,Y)≈ 0). Given that −TΔSa-solv(H,Y→Me,Y)> 0, and ΔHa-solv(H,Y→Me,Y)≈ 0, we can conclude that ΔGa-solv(H,Y→Me,Y)> 0. Both the entropy and the free energy of the formation of cavities in water were extensively studied by Graziano, and were shown to be unfavorable.56–58
Fig 10.
Thermodynamic cycle describing the mutation of the uncomplexed ligand 8a→8b, or 8c→8d via two steps (I and II). Step I: Reorganization of the hydration waters (blue) in order to create a cavity for the Me group. Step II: The replacement of H with Me to produce the final state of the mutation. The changes in the number of rotatable bonds as the H in 8a is replaced by Me to give 8b, and as the H in 8c is replaced by Me to give 8d are shown. The thermodynamic parameters of each step are shown on the arrow representing this particular step of the mutation.
The second step in Fig 10 involves the replacement of H with Me. This replacement produces some interactions between the Me and the aqueous medium (e.g. dispersion forces). This step might therefore have some negative enthalpy change (ΔHa-Me(H,Y→Me,Y)< 0). In order to investigate the entropic term associated with this step, we need to consider the influence of the replacement of H with Me on the ligand conformational entropy. When H is replaced by Me in absence of the COO− group (8c→8d), a larger conformational space becomes accessible to the ligand as it acquires an additional rotatable bond. This causes favorable entropic change (−TΔSa-Me(H,H→Me,H)< 0). On the other hand, the replacement of H with Me in presence of the COO− group (8a→8b) would limit the ligand conformational space because (1) Some of the ligand conformers become more sterically demanding when two functional groups simultaneously exist at the ligand terminal end (i.e. the COO− and the Me groups). (2) No additional rotatable bonds are acquired in presence of the COO−. Reducing the ligand conformational space in presence of the COO− correlates with an unfavorable entropic change (−TΔSa-Me(H,COO→Me,COO)> 0), and leads us to conclude that the COO− group increases −TΔSa-Me(H,Y→Me,Y) (−TΔSa-Me(H,COO→Me,COO)− (−TΔSa-Me(H,H→Me,H))> 0). Whether or not this increase in −TΔSa-Me(H,Y→Me,Y) can be one of the factors responsible for the observed entropic cooperativity depends on how this variation affects the differential entropy. In order to evaluate this, the thermodynamic parameters of mutation “a” in equation set 2 were substituted with their basic components obtained from the dissection of this mutation [e.g. ΔGa(H,Y→Me,Y) is equal to, and can, in turn, be substituted with (ΔGa-solv(H,Y→Me,Y)+ ΔGa-Me(H,Y→Me,Y)], and equation set 6 was subsequently written.
| (6A) |
| (6B) |
| (6C) |
Equation 6C shows that the potential increase in −TΔSa-Me(H,Y→Me,Y) caused by the COO− group decreases −TΔΔS(H,Y→Me,Y). Since ITC data demonstrates that −TΔΔS(H,Y→Me,Y) increases in presence of the COO− group [−TΔΔS(H,COO→Me,COO)− (−TΔΔS(H,H→Me,H))= 12.4kJ/mol], variation in −TΔSa-Me(H,Y→Me,Y) caused by the COO− group cannot be responsible for the observed entropic negative cooperativity.
The presence of the COO− might also influence water reorganization which occurs in the first step of this mutation. For example, the COO− group, being polar and ionized, might form strong charge-assisted H-bonds with the reorganized water molecules, as well as help these water molecules to form stronger H-bonds among themselves. This decreases ΔHa-solv(H,Y→Me,Y) (ΔHa-solv(H,COO→Me,COO)< ΔHa-solv(H,H→Me,H)). The stronger binding experienced by these water molecules in presence of the COO− might be accompanied by additional restriction in their motions which is reflected as an increase in −TΔSa-solv(H,Y→Me,Y) (−TΔSa-solv(H,COO→Me,COO)> −TΔSa-solv(H,H→Me,H)). The experimentally determined enthalpic positive cooperativity (=−15.8 kJ/mol) cannot be attributed to the COO− influence on ΔHa-solv(H,Y→Me,Y), nor can the entropic negative cooperativity (= 12.4 kJ/mol) be attributed to the COO− influence on −TΔSa-solv(H,Y→Me,Y). This is because, as we can conclude from equations 6B and 6C, the potential decrease in ΔHa-solv(H,Y→Me,Y) and the increase in −TΔSa-solv(H,Y→Me,Y) caused by the COO− should increase ΔΔH(H,Y→Me,Y) and decrease −TΔΔS(H,Y→Me,Y) respectively, not decrease ΔΔH(H,Y→Me,Y) and increase −TΔΔS(H,Y→Me,Y) as we observe in the ITC data.
Table 3 summarizes the enthalpic and the entropic terms that constitute both the differential enthalpy and the differential entropy respectively. It also shows the influence of the COO− group on each term and the kind of change that, if a term underwent in presence of the COO− group, could produce the experimentally observed cooperativity. As shown in the table, the only enthalpic term which is influenced by the COO− group in a manner clearly correlating with the experimental enthalpic cooperativity, is the enthalpy of the modifications in the complex hydration shell “ΔHb-solv(H,Y→Me,Y)”. The existence of another enthalpic term (i.e. ΔHa-solv(H,Y→Me,Y)), influenced by the COO− in a manner opposite to what would be contributive to the observed experimental cooperativity, indicates that the influence of the COO− group on ΔHb-solv(H,Y→Me,Y) is large enough to overcome this term and still produces cooperativity (ΔHb-solv(H,COO→Me,COO)− ΔHb-solv(H,H→Me,H)≤ −15.8 kJ/mol). The same applies to the entropy of the complex hydration shell modifications “−TΔSb-solv (H,Y→Me,Y)” which is shown to be the sole term whose influence by the COO− group can rationalize the experimental entropic antagonism. The presence of other terms opposing the experimental cooperativity indicates that the influence of the COO− group on −TΔSb-solv(H,Y→Me,Y) is also large enough to overcome these opposing terms and still produces the observed 12.4 kJ/mol negative cooperativity [−TΔSb-solv(H,COO→Me,COO)− (−TΔSb-solv(H,H→Me,H))]≥ 12.4 kJ/mol).
Table 3.
a. The enthalpic and entropic terms constituting the differential enthalpy and the differential entropy, the anticipated influence of the COO− on each term, and the influence required to consider such term a cause for the experimentally observed cooperativity
| Enthalpic Term | Entropic Term | Influence of COO− on Enthalpic Term | Influence of COO− on Entropic Term | Influence of COO− required for Enthalpic term to cause cooperativity | Influence of COO− required for Entropic term to cause cooperativity |
|---|---|---|---|---|---|
| ΔHa-solv (H,Y→Me,Y) | (−) | (+) | |||
| −TΔSa-solv (H,Y→Me,Y) | (+) | (−) | |||
| ΔHa-Me (H,Y→Me,Y) | Insignificant | (+) | |||
| −TΔSa-Me (H,Y→Me,Y) | (+) | (−) | |||
| ΔHb-solv (H,Y→Me,Y) | (−) | (−) | |||
| −TΔSb-solv (H,Y→Me,Y) | (+) | (+) | |||
| ΔHb-Me (H,Y→Me,Y) | Insignificant | (−) | |||
| −TΔSb-Me (H,Y→Me,Y) | Insignificant | (+) |
The signs shown in the table are the algebraic signs of the difference between the enthalpic, or the entropic terms, in presence and absence of the COO− (e.g. (ΔHb-solv(H,COO→Me,COO)− ΔHb-solv(H,H→Me,H)). (+): increase in the thermodynamic term; (−): decrease in the thermodynamic term.
It should be noted that the free energy cooperativity is the net result of the two underlying enthalpic and entropic cooperatvities (eq. 1). Even though the magnitude of each of these cooperativities is large, the free energy cooperatvity is comparatively smaller because the enthalpic and the entropic cooperativities in part compensate each other. Since the magnitude of the enthalpic positive cooperativity is larger than that of the entropic negative cooperativity, the free energy shows positive cooperativity. It is not necessary, however, for the enthalpic cooperativity to be the favorable cooperativity term or to be greater than the entropic cooperativity. The kind, as well as the magnitude, of cooperativity is highly dependent on the structural details of the system, and on how this system responds to the structural perturbations being studied. This indicates that in other ligand-protein binding cases the free energy cooperativity could be positive, negative or nonexistent (i.e. additivity) depending upon the details.
Another aspect which greatly adds to the intricacy of the free energy cooperativity evaluation is that this cooperativity is produced by multiple enthalpy-entropy compensation effects. Each of these effects yields a free energy term that contributes to the overall free energy cooperativity. To illustrate this, the influence of the COO− group in question on the differential binding energy components is assumed to be negligible except for ΔGb-solv(H,Y→Me,Y) which is composed of ΔGb-Wdisp(H,Y→Me,Y) and ΔGb-Wstruct(H,Y→Me,Y). Based upon this assumption, and using equations 2A and 3A, equation 7, which shows the free energy terms contributing to cooperativity, can be derived (for detailed derivation, see supporting information). Since each of the four free energy terms in equation 7 represents the net effect of an enthalpy-entropy compensation relationship (e.g. ΔGb-Wdisp(H,H→Me,H)= ΔHb-Wdisp(H,H→Me,H)− TΔSb-Wdisp(H,H→Me,H); ΔHb-Wdisp(H,H→Me,H) is unfavorable but partially compensated by the favorable −TΔSb-Wdisp(H,COO→Me,COO)), free energy cooperativity can be viewed as an outcome of all the enthalpy-entropy compensation effects. As these enthalpy-entropy compensations largely depend on the structural details of the system, the type and the magnitude of the free energy cooperativity appears to be highly context-dependent and therefore requires an in-depth experimental and theoretical analysis of the individual system under consideration in order to be well understood.
| (7) |
Conclusions
In this study, we have demonstrated that the contribution of the Me side chain, which binds in the thermolysin S2′ pocket, to the ligand binding affinity can be enhanced by about an order of magnitude in the presence of a ligand COO− group (positive cooperativity). This positive cooperativity is the net result of a large favorable enthalpic cooperativity term (−15.8 kJ/mol) and a smaller unfavorable entropic cooperativity term (12.4 kJ/mol). In order to determine the origin of these individual cooperativities, the differential thermodynamic parameters of the Me group, in the absence and presence of the COO− group, were dissected into fundamental components, and the influence of the COO− on each of these components was investigated. An approach based on investigating the mutations of 8a and 8c to 8b and 8d respectively, in both the enzyme-bound and the unbound states, was used to achieve this.
The crystallographic data of the 8a-, 8b-, 8c- and 8d-thermolysin complexes were used to compare the structural changes occurring in the complex when the Me group is introduced into the ligand in absence of the COO− group, with the changes occurring when this group is introduced into the ligand in presence of the COO−. This comparison revealed that the changes occurring in the hydration shell of the complex (e.g. displacement of two waters from the S2′ pocket, and acquisition of new waters) in presence of the COO− group are more enthalpically, and less entropically, favorable than those occurring in absence of this group. For example, with regard to the enthalpically favorable, entropically unfavorable acquisition of new crystallographic waters by the complex, the presence of the COO− causes the complex to acquire more of these waters, and to form a more extensive H-bond network characterized by enhanced σ-bond cooperativity. This correlates with the improved differential enthalpy, and the worsened differential entropy observed in the ITC data when the COO− is present.
In addition, a smaller enthalpic penalty, and a reduced entropic advantage, for the displacement of two water molecules from the S2′ pocket, in presence of the COO− group, were indicated to contribute to the positive enthalpic, and the negative entropic cooperativities respectively. Both structural data and calculation results were found to support this smaller enthalpic penalty which is accompanied by the smaller entropic advantage. For example, crystal structures reveal that the COO− group causes one of the two displaced waters to form one less H-bond than what it forms in absence of this group. The COO− of 8a also interferes with the interconnectedness of the active site H-bond network causing an enthalpic destabilization of the S2′ water molecules, as well as the other waters in the active site. The thermodynamic implications of these data are consistent with the results of the QM calculations of the interaction energies of the two displaced waters in model systems representing the 8a-TLN, and 8c-TLN complexes. Such calculations showed that these waters have weaker interactions in presence of the COO−. Enthalpically less-stabilized waters like those in 8a-TLN are likely to have a higher degree of mobility, which is reflected as a decrease in the entropic advantage of water displacement from the 8c-TLN S2′ pocket.
It was also revealed in the current study that cooperativity among ligand functional groups can be more intricate than might have been appreciated in the past. This is because the binding free energy is determined by a complex interplay among a number of enthalpy-entropy compensation effects. Consequently, ligand functional group cooperativity can be highly dependent on the structural details of the system being investigated and on how this system responds to structural perturbations in the ligand. Cooperativity might therefore be positive, negative or nonexistent.
To the best of our knowledge, this is the first time water molecules within binding cavities have been shown to be involved in cooperativity among ligand functional groups. Snyder et al, though, suggested that changes in binding affinity among closely related analogues can be significantly influenced by changes in the number and organization of water molecules localized in and beyond the active site in the bound complex9. Studies like the one presented herein can set the stage, and provide direction, for more in-depth experimental and theoretical studies which may further analyze and better quantify the contributions of the individual thermodynamic factors to the relative binding, and the SARs in ligand congener series. Some of the general considerations suggested by this study are: (1) High-resolution crystal structures for the complexes of closely-related analogues (e.g. congeneric ligand series) can reveal important changes in the complex hydration shell that might occur when the ligand structure is modified. (2) These hydration shell changes can have significant effect on the thermodynamic parameters of binding as determined by ITC. (3) Changes in the hydration water of the ligand-protein complex can produce cooperativity among ligand side chains. (4) The consideration of the changes of the water structures, in response to ligand structural modification, can assist SARs analyses and contribute to a better understanding of the underlying science of ligand-protein binding. This enhanced fundamental understanding may be useful for improving the accuracy of scoring functions in predicting binding affinity.
Experimental Section
Chemistry
General methods
Reagents were obtained from commercial suppliers and used without further purification. Anhydrous solvents were purchased as sealed bottles from either Fisher-Acros (AcroSeal™) or Aldrich (Sure-seal™) and were maintained under an argon atmosphere. All amino acids used are L unless otherwise noted. Proton, phosphorus and carbon nuclear magnetic resonance was performed in deuterated solvents purchased from Cambridge Isotope Laboratories, Inc (Andover, MD) on one of the following instruments: Varian Gemini 300MHz, Varian Inova 400MHz, or Varian Inova 500MHz. 1HNMR data is reported in the following format: chemical shift (ppm values in relation to TMS or appropriate solvent peak), multiplicity (s= singlet, d= doublet, t= triplet, q= quartet, dd= doublet of doublets, d of t= doublet of triplets, complex= combinations of peaks resulting from different molecular conformations, m= multiplet, br= broad peak) coupling constant(s), and integration. Low resolution ESI mass spectrometry was performed on a Thermo Finnigan LCQ Advantage instrument using 60% methanol in water with 1% acetic acid or 60% acetonitrile in water with 0.1% trifluoroacetic acid as the mobile phase. Preparative and semi-preparative HPLC instrumentation included a Milton Roy gm4000 gradient programmer, Milton Roy Constametric I and III pumps, a Rheodyne 7125 injector with a 5.00mL sample loop, and a Knauer Variable Wavelength Detector set at 218nm with a preparative flow cell. All final compounds tested were at least 95% pure by HPLC analysis.
Synthesis of benzyl N-(hydroxymethyl) carbamate (1)23
Benzyl carbamate (6g, 40mmol) was added to a solution of 37% formalin (4.4 g, 56 mmol) and sodium carbonate (2.2 g, 20 mmol) in 65 ml water. The mixture was heated until all the solids were dissolved, then cooled to room temperature and stirred overnight. The precipitated solid was then filtered, dried, and redissolved in dichloromethane. The solution was dried using anhydrous magnesium sulfate and the solvent was removed under vacuum to give the product as a white solid which was used in the next step without further purification (5.40g, 74.6%) 1H-NMR (CDCl3-d) δ 4.10 (s, 1H) 4.71 (d, J= 6.5Hz, 2H) 5.13 (s, 1H) 6.07 (s, 1H) 7.36 (s, 5H), m/z (LCMS, ESI): found 181.2 [M+H]+, C9H11NO3 requires 180.1
Synthesis of benzyl N-(acetoxymethyl) carbamate (2)
Compound 1 (3.62g, 20mmol) was dissolved in 25ml anhydrous THF and was added slowly to an ice-cooled stirred solution of 23ml acetic anhydride and 6.5ml anhydrous pyridine under Argon. The mixture was stirred at r.t. for 2hrs, then the solution was diluted with 150ml ethyl acetate and washed with 1N HCl (3× 150ml) and brine (2× 150ml). The organic layer was dried with anhydrous magnesium sulfate and the volatile materials were removed under vacuum to give an oily residue which was purified with flash chromatography (2.95g, 66.1%) of the pure product. 1H-NMR (CDCl3-d1) δ 2.08 (s, 3H) 5.16 (s, 2H) 5.23 (d, J= 7.5Hz, 2H) 6.08 (s, 1H) 7.38 (s, 5H). m/z (LCMS, ESI): found 246.0 [M + Na]+, C11H13NO4 requires 223.1
Synthesis of dimethyl N-benzyloxycarbonyl-aminomethyl-phosphonate (3)
A mixture of compound 2 (2.90g, 13mmol) and trimethylphosphite (4.6ml, 39mmol) was refluxed for 3hrs. The volatile materials were removed by distillation at 60 °C under reduced pressure to give the product as an oily residue which was used in the next step without further purification (3.44g, 97.5%) 1H-NMR (CDCl3-d1) δ 3.62 (dd, J= 6.5Hz, JH-P= 11Hz, 2H) 3.72 (d, JH-P= 11Hz, 6H) 5.09 (s, 2H) 5.83 (s, 1H) 7.32 (m, 5H). 31P-NMR (CDCl3-d1) δ 25.32, m/z (LCMS, ESI): found 296.1 [M + Na]+, C11H16NO5P requires 273.1
Synthesis of methyl N-benzyloxycarbonyl-aminomethyl-phosphonate (4)
Compound 3 (3.28g, 12mmol) was shook vigorously with 10% NaOH (14.5ml, 3eq) until it was completely dissolved. The mixture was stirred at room temperature for 2 hrs then diluted with water, extracted with ethyl acetate (2× 30ml), and acidified to pH 1 with 2N HCl. The aqueous solution was extracted with dichloromethane (2× 100ml) and ethyl acetate (2× 50ml). The dichloromethane layers were combined, washed with brine (2× 50ml), and dried using anhydrous magnesium sulfate. The ethyl acetate layers were also combined, washed with brine (2× 25ml), and dried with anhydrous magnesium sulfate. The organic layers were then combined and the volatile solvents were removed under high vacuum to give the product as a pure white solid (2.26g, 72.7%) 1H-NMR (CDCl3-d1) δ 3.64 (d, JH-P= 11Hz, 2H) 3.71 (d, JH-P= 11Hz, 3H) 5.12 (s, 2H) 5.7 (br s, 1H) 7.35 (m, 5H) 11.8 (br s, 1H). 31P-NMR (CDCl3-d1) δ 24.12, m/z (LCMS, ESI): found 282.1 [M + Na]+, C10H14NO5P requires 259.1
General procedure for amide coupling- Compounds 5a-d
To a cooled solution of the carboxylic acid (1eq), the amine/aminoacid HCl (1.5eq) and PyBop (1.2 eq) in anhydrous DMF was added diisopropylethylamine (4 eq) gradually. The reaction mixture was stirred at r.t. for 2–5hrs, diluted with ethyl acetate (50ml for every 5ml DMF), then extracted with 1N HCl (3×), saturated sodium bicarbonate (3×), and brine (2×). The organic layer was then dried with anhydrous sodium sulfate and the solvent was evaporated under vacuum to give the products which were purified by flash chromatography.
(N-(tert-butoxycarbonyl)-L-leucinyl)-glycine ethyl ester (5a)
Using the above procedure, Boc-L-leucine (2.5mmol, 579mg) was reacted with glycine ethyl ester HCl (3.75mmol, 524mg) to give compound 5a (603mg, 76.3%). 1H-NMR (DMSO-d6) δ 0.86 (d, J= 7Hz, 3H) 0.88 (d, J= 7Hz 3H) 1.19 (t, J= 7Hz, 3H) 1.38 (s, 9H) 1.43 (t, J= 7.5Hz, 2H) 1.58–1.66 (m, 1H) 3.76 (dd, J= 17, 6Hz, 1H) 3.87 (dd, J= 17, 6Hz, 1H) 4.01 (q, J= 7Hz, 1H) 4.08 (q, J= 7Hz, 2H) 6.89 (d, J= 8.5Hz, 1H) 8.21 (t, J= 6Hz, 1H), m/z (LCMS, ESI): found 339.1 [M + Na]+, C15H28N2O5 requires 316.2
(N-(tert-butoxycarbonyl)-L-leucinyl)-alanine methyl ester (5b)
Using the above procedure, Boc-L-leucine (2.5mmol, 579mg) was reacted with alanine methyl ester HCl (3.75mmol, 524mg) to give compound 5b (578mg, 73.1%). 1H-NMR (DMSO-d6) δ 0.86 (d, J= 7Hz, 3H) 0.88 (d, J= 7Hz 3H) 1.28 (d, J= 7Hz, 3H) 1.37 (s, 9H) 1.39 (t, J= 7.5Hz, 2H) 1.58–1.66 (m, 1H) 3.61 (s, 3H) 3.99 (q, J= 7Hz, 1H) 4.26 (m, 1H) 6.82 (d, J= 8.5Hz, 1H) 8.21 (d, J= 7Hz, 1H), m/z (LCMS, ESI): found 339.1 [M + Na]+, C15H28N2O5 requires 316.2
N′-methyl-N-(tert-butoxycarbonyl)-L-leucinamide (5c)
Using the above procedure, Boc-L-leucine (2.5mmol, 579mg) was reacted with methylamine HCl (3.75mmol, 253mg) to give compound 5c (473mg, 77.4%).1H-NMR (DMSO-d6) δ 0.85 (d, J= 6.5Hz, 3H) 0.88 (d, J= 6.5Hz, 3H) 1.39 (s, 9H) 1.30–1.45 (m, 2H) 1.57 (m, 1H) 2.58 (d, J= 4Hz, 3H) 3.92 (m, 1H) 6.82 (d, J= 8.5Hz, 1H) 7.75 (q, J= 3.5Hz, 1H). m/z (LCMS, ESI): found 267.1 [M + Na]+, C12H24N2O3 requires 244.2
N′-ethyl-N-(tert-butoxycarbonyl)-L-leucinamide (5d)
Using the above procedure, Boc-Lleucine (2.5mmol, 579mg) was reacted with ethylamine HCl (3.75mmol, 306mg) to give compound 5d (416mg, 64.5%).1H-NMR (DMSO-d6) δ 0.89 (d, J= 6Hz, 3H) 0.91 (d, J= 6.5Hz, 3H) 1.04 (t, J= 7Hz, 3H) 1.42 (s, 9H) 1.36–1.46 (m, 2H) 1.61 (m, 1H) 3.04–3.15 (m, 3H) 3.93 (m, 1H) 6.82 (d, J= 8.5Hz, 1H) 7.75 (t, J= 3.5Hz, 1H). m/z (LCMS, ESI): found 281.1 [M + Na]+, C13H26N2O3 requires 258.2
General procedure for Boc deprotection- Compounds 6a-d
The Boc-protected compound was dissolved either in 3N HCl/MeOH, or in ethyl acetate. When the compound is dissolved into ethyl acetate, HCl gas was bubbled into the solution. The solution was stirred for 1.5–2 hrs. The volatile materials were removed under vacuum to give the product as hygroscopic solid which was purified with semi-preparative HPLC.
L-leucinyl-glycine ethyl ester hydrochloride (6a)
Compound 5a (554mg, 1.75mmol) was converted to 6a using HCl/ethyl acetate as described in the above procedure (426mg, 99.1%). 1H-NMR (DMSO-d6) δ 0.90 (d, J= 7Hz, 3H) 0.92 (d, J= 7Hz 3H) 1.19 (t, J= 7Hz, 3H) 1.59 (m, 2H) 1.77 (m, 1H) 3.81 (t, J= 7Hz, 1H) 3.83 (dd, J= 17, 5.5Hz, 1H) 3.98 (dd, J= 17, 5.5Hz, 1H) 4.00 (d of q, J= 7, 2Hz, 2H) 8.42 (br, 3H) 9.17 (t, J= 6Hz, 1H). m/z (LCMS, ESI): found 217.0 [M + H]+, C10H20N2O3 requires 216.1
L-leucinyl-alanine methyl ester hydrochloride (6b)
Compound 5b (554mg, 1.75mmol) was converted to 6b using 3N HCl/MeOH as described in the above procedure (408mg, 95.0%). 1H-NMR (DMSO-d6) δ 0.90 (d, J= 6Hz, 3H) 0.93 (d, J= 6.5Hz 3H) 1.33 (d, J= 7Hz, 3H) 1.57 (m, 2H) 1.73 (m, 1H) 3.64 (s, 3H) 3.77 (t, J= 7Hz, 1H) 4.35 (m, 1H) 8.29 (br, 3H) 9.01 (d, J= 6.5Hz, 1H). m/z (LCMS, ESI): found 217.1 [M + H]+, C10H20N2O3 requires 216.2
N′-methyl-L-leucinamide hydrochloride (6c)
Compound 5c (428mg, 1.75mmol) was converted to 6c using 3N HCl/MeOH as described in the above procedure (313mg, 99%), 1H-NMR (DMSO-d6) δ 0.88 (d, J= 6Hz, 3H) 0.90 (d, J= 6Hz, 3H) 1.55 (t, J= 7Hz, 2H) 1.60 (m, 1H) 2.65 (d, J= 5Hz, 3H) 3.70 (t, J= 8Hz, 1H) 8.29 (br, 3H) 8.64 (q, J= 4Hz, 1H). m/z (LCMS, ESI): found 289.0 [2M + H]+, C7H16N2O requires 144.1
N′-ethyl-L-leucinamide hydrochloride (6d)
Compound 5d (400mg, 1.55mmol) was converted to 6d using 3N HCl/MeOH as described in the above procedure (297mg, 98.5%), 1H-NMR (DMSO-d6) δ 0.93 (d, J= 6.5Hz, 3H) 0.95 (d, J= 6.5Hz, 3H) 1.10 (t, J= 7Hz, 3H) 1.59 (t, J= 7Hz, 2H) 1.67 (m, 1H) 3.10–3.25 (m, 2H) 3.72 (t, J= 8Hz, 1H) 8.31 (br, 3H) 8.68 (t, J= 4Hz, 1H). m/z (LCMS, ESI): found 339.0 [2M + Na]+, C8H18N2O requires 158.1
General procedure for the synthesis of compounds 7a-d
To a cooled solution of (4), 6a-d (1.5eq.) and PyBop (1.2eq.) in anhydrous DCM was added diisopropylethylamine (4eq.) gradually. The reaction mixture was stirred at r.t. for 6hr to overnight. The reaction mixture was then diluted up to 25ml DCM, extracted with 5% citric acid (2× 12ml), saturated sodium bicarbonate (2× 12ml), and brine (2× 10ml), and dried over anhydrous sodium sulfate. The solvent was then evaporated under reduced pressure and the residue was purified by semi-preparative HPLC.
Ethyl-N-(N-(N-(benzyloxycarbonyl)-(R/S)-aminomethylphosphonyl)-L-leucinyl)-glycinate methyl ester (7a)
Compound 4 (0.5mmol, 130mg) was reacted with 6a (0.75mmol, 190mg) according to the general procedure described above (PyBop: 0.6mmol, 312mg; DIEA: 2mmol, 0.35ml) to give 7a as 2 diastereomers with an approximate ratio of 3: 2 as determined by 31PNMR (153mg, 66.8%). 1H-NMR (CDCl3) δ 0.90–0.98 (complex, 6H) 1.23 & 1.26 (2 × t, J= 7Hz, 3H) 1.52–1.63 (m, 2H) 1.76 (m, 1H) 3.45–4.05 (complex, 9H) 4.14 & 4.16 (2 × q, J= 7Hz, 2H) 5.10 (complex, 2H) 5.97 & 6.10 (2 × br, 1H) 7.34 (m, 5H) 7.75 & 7.80 (complex, 1H); 31P-NMR (CDCl3) δ 27.69 & 28.50 (approx. 3: 2). m/z (LCMS, ESI): found 480.2 [M + Na]+, C20H32N3O7P requires 457.2
Methyl-N-(N-(N-(benzyloxycarbonyl)-(R/S)-aminomethylphosphonyl)-L-leucinyl)-alaninate methyl ester (7b)
Compound 4 (0.5mmol, 130mg) was reacted with 6b (0.75mmol, 190mg) according to the general procedure described above (PyBop: 0.6mmol, 312mg; DIEA: 2mmol, 0.35ml) to give 7b as 2 diastereomers with an approximate ratio of 3: 2 as determined by 31P-NMR (134mg, 58.5%). 1H-NMR (CDCl3) δ 0.90–0.96 (complex, 6H) 1.36 & 1.38 (2 × d, J= 7Hz, 3H) 1.49–1.60 (m, 2H) 1.75 (m, 1H) 3.40–4.15 (complex, 9H) 4.50 (m, 1H) 5.10 (complex, 2H) 6.01 & 6.15 (2 × br, 1H) 7.33 (m, 5H) 7.62 & 7.67 (2 × d, J= 7Hz, 1H); 31P-NMR (CDCl3) δ 27.61 & 28.04 (approx. 3: 2). m/z (LCMS, ESI): found 480.2 [M + Na]+, C20H32N3O7P requires 457.2
N-(N-(benzyloxycarbonyl)-(R/S)-aminomethylphosphonyl)-N′-methyl-L-leucinamide methyl ester (7c)
Compound 4 (0.5mmol, 130mg) was reacted with 6c (0.75mmol, 135mg) according to the general procedure described above (PyBop: 0.6mmol, 312mg; DIEA: 2mmol, 0.35ml) to give 7c as 2 diastereomers with an approximate ratio of 3: 2 as determined by 31P-NMR (138mg, 71.6%). 1H-NMR (CDCl3) δ 0.93 (complex, 6H) 1.44–1.64 (m, 2H) 1.65–1.80 (m, 1H) 2.75 & 2.77 (2 × d, J= 5Hz, 3H) 3.40–3.95 (complex, 7H) 5.10 (complex, 2H) 5.96 (br, 1H) 7.087& 7.17 (2 × t, J= 5Hz, 1H) 7.35 (m, 5H); 31P-NMR (CDCl3) δ 27.89 & 28.21 (approx. 3: 2). m/z (LCMS, ESI): found 408.1 [M + Na]+, C17H28N3O5P requires 385.2
N-(N-(benzyloxycarbonyl)-(R/S)-aminomethylphosphonyl)-N′-ethyl-L-leucinamide methyl ester (7d)
Compound 4 (0.5mmol, 130mg) was reacted with 6d (0.75mmol, 146mg) according to the general procedure described above (PyBop: 0.6mmol, 312mg; DIEA: 2mmol, 0.35ml) to give 7d as 2 diastereomers with an approximate ratio of 3: 2 as determined by 31P-NMR (93mg, 46.7%). 1H-NMR (CDCl3) δ 0.97 (complex, 6H) 1.13 & 1.16 (2 × d, J= 6.5Hz, 3H) 1.46–1.66 (m, 2H) 1.67–1.82 (m, 1H) 3.25 (complex, 2H) 3.40–4.1 (complex, 7H) 5.16 (complex, 2H) 6.05 (br, 1H) 7.14& 7.22 (2 × t, J= 5Hz, 1H) 7.38 (m, 5H); 31P-NMR (CDCl3) δ 27.80 & 28.20 (approx. 3: 2). m/z (LCMS, ESI): found 422.1 [M + Na]+, C18H30N3O5P requires 399.2
General procedure for the synthesis of compounds 8a-d
0.2mmol of the phosphonamidate ester (7a-d) was vigorously shaken at r.t. with 1–2ml 0.4M LiOH aqueous solution until all the solid dissolves (acetonitrile was used as a co-solvent whenever needed). The solution was stirred for 2–24hr and concentrated under reduced pressure. The final compound was then separated as a pure mono-, or di-lithium salt using semi-preparative HPLC.
N-(N-(N-(benzyloxycarbonyl)-aminomethylphosphonyl)-L-leucinyl)-glycinate di-lithium (8a)
Compound 7a (91mg, 0.2mmol) was reacted with 4eq LiOH (2ml of 0.4M aqueous solution) for 6hr to give 8a as di-lithium salt (72mg, 81.8%).1H-NMR (D2O) δ 0.73 (d, J= 6.5Hz, 3H) 0.74 (d, J= 6.5Hz, 3H) 1.33 (m, 2H) 1.55 (m, 1H) 3.13 (complex, 2H) 3.50–3.65 (complex, 3H) 4.97 (complex, 2H) 7.26 (m, 5H) 13C-NMR (D2O) δ 21.08, 22.42, 23.99 (3C, CH(CH3)2) 38.90& 40.71 (d, JC-P ~ 543Hz, 1C, CH2P) 43.15, 43.29 (2C, CH2CH(CH3)2 & CH2COO) 54.16 (1C, CHCO) 67.16 (1C, PhCH2O-) 127.82, 128.42, 128.83& 136.43 (6C, Ph) 158.31 (1C, Cbz C=O); 176.39 & 177.93 (2C, 2C=O); 31P-NMR (D2O) δ 17.56; m/z (LCMS, ESI): found 434.1[M + Li]+& 466.1 [M − Li + 2Na]+, C17H24N3O7PLi2 requires 427.2
N-(N-(N-(benzyloxycarbonyl)-aminomethylphosphonyl)-L-leucinyl)-alaninate di-Lithium (8b)
Compound 7b (91mg, 0.2mmol) was reacted with 4eq LiOH (2ml of 0.4M aqueous solution) for 5hr to give 8b as di-lithium salt (72mg, 81.8%).1H-NMR (D2O) δ 0.73 (complex, 6H) 1.17 (d, J= 7.5Hz, 3H) 1.26–1.36 (m, 2H) 1.53 (m, 1H) 3.13 (complex, 2H) 3.50 (q, J= 5Hz, 1H) 3.97 (q, J= 7Hz, 1H) 4.97 (complex, 2H) 7.28 (m, 5H) 31P-NMR (D2O) δ 17.57; m/z (LCMS, ESI): found 436.1[M−Li + 2H]+& 442.1 [M+ H]+, C18H26N3O7PLi2 requires 441.2
N-(N-(benzyloxycarbonyl)-aminomethylphosphonyl)-N′-methyl-L-leucinamide lithium (8c)
Compound 7c (77mg, 0.2mmol) was reacted with 2eq LiOH (1ml of 0.4M aqueous solution) for 2hr to give 8c as mono-lithium salt (68.5mg, 90.7%). 1H-NMR (D2O) δ 0.72 (complex, 6H) 1.2–1.4 (m, 2H) 1.49 (m, 1H) 2.55 (s, 3H) 3.12 (complex, 2H) 3.47 (m, 1H) 4.98 (complex, 2H) 7.29 (m, 5H) 13C-NMR (D2O) δ 22.599, 23.78 & 25.52 (3C, CH(CH3)2) 27.21 (1C, -NHCH3) 40.29 & 42.09 (2 × d, JC-P ~ 540Hz, 1C, CH2P) 44.63 & 44.69 (1C, CH2CH(CH3)2) 55.65 (1C: CHCONH-) 68.59 (1C, PhCH2O-) 129.28, 129.85, 130.23 & 137.87 (6C, Ph) 159.72 (1C, Cbz C=O); 180.11 (1C, C=O); 31P-NMR (D2O) δ 17.40; m/z (LCMS, ESI): found 384.2 [M + Li]+& 761.3 [2M + Li]+, C16H25N3O5PLi requires 377.2
N-(N-(benzyloxycarbonyl)-aminomethylphosphonyl)-N′-ethyl-L-leucinamide lithium (8d)
Compound 7d (80mg, 0.2mmol) was reacted with 2eq LiOH (1ml of 0.4M aqueous solution) for 24hr to give 8d as mono-lithium salt (47mg, 60%). 1H-NMR (D2O) δ 0.73 (complex, 6H) 0.93 (t, J= 7Hz, 3H) 1.2–1.4 (m, 2H) 1.48 (m, 1H) 3.00 (complex, 2H) 3.11 (complex, 2H) 3.46 (m, 1H) 4.97 (complex, 2H) 7.28 (m, 5H) 31P-NMR (D2O) δ 17.29; m/z (LCMS, ESI): found 408.1[M−Li+ Na+ H]+, C17H27N3O5PLi requires 391.2
Biological assay
The inhibition constants of the thermolysin phosphonamidate inhibitors 8a-d were determined photometrically at 345nm using 2-furanacryloyl-Gly-Leu-NH2 as a substrate24. The assay was carried out on a Cary 100/300 UV/VIS spectrophotometer at 25.0± 0.2 °C. A 0.05M Tris buffer containing 0.02M CaCl2, 2.5M NaBr16 and 1.25% DMF, was used in all measurements. Buffer pH was adjusted to 7.3±0.5 at room temperature prior to use. The concentration of the enzyme stock solution was determined by UV absorbance at 280nm (ε1% =17.65 cm−1)59. The concentrations of the stock solutions of the substrate and the inhibitors were determined from accurately weighed samples. The enzyme concentration in all the final assay solutions was approximately 8nM, the substrate concentration in the final assay solutions was 0.8M, and the inhibitors’ concentrations were in the range of 0.5Ki–10Ki for each inhibitor. The inhibition constant (Ki) for the inhibitor was taken to be the average of at least three Ki determinations, each of which was calculated from the experimentally determined IC50 using Cheng-Prusoff equation60 (Km= 3.9± 0.6 mM). The IC50 values were determined from υ0/υi vs. [I] plots16, 61 for inhibitors 8a, 8c and 8d (υ0/υi = [I]/IC50 + 1), or Henderson plots62 for inhibitor 8b (At least six different inhibitor concentrations [I] were used to construct each plot).
Quantum Mechanical Calculations
Two model systems were built from the X-ray crystal structures of 8c-TLN and 8a-TLN complexes. Each of these model systems consisted of the terminal part of the ligand, the part of the protein that directly interacts with the chosen part of the ligand and a number of the crystallographic water molecules that solvate the part of the ligand-protein complex that was included in the model. Water molecules were chosen in such a way that important water polygonal and chain structures were kept intact. The two water molecules that are displaced by the Me group were included. Each model consisted of about 100 atoms in total. The positions of the hydrogen atoms were added manually and further optimized with DFT/B3LYP method47–49 with 3-21G basis. The positions of all the heavy atoms except the two water molecules replaced by methyl were constrained in the original positions of the crystal structure during the DFT optimization. After geometry optimizations, a duplicate pdb file was generated for each model, and the two water molecules displaced by the Me group were removed from that second file. The total energies of the four models and their Mulliken electronic charges were calculated with B3LYP functional with dispersion correction using exchange-dipole moment’ method63,64 and triple-zeta cc-pVTZ basis50. The total energies of the removed water molecules were similarly calculated. Values of the calculated total energies and interaction energies are provided in table 2 in the supporting information. Calculations were done using Q-Chem Program65.
ITC and x-ray crystallography data
Experimental details of ITC and x-ray crystallography will be published elsewhere by Biela, A. et al.25
Supplementary Material
Acknowledgments
Funding Sources: The computational part of the work was supported by National Institute of Health through a grant (R44GM084555).
We kindly acknowledge Adam Biela, Michael Betz, Andreas Heine, and Prof. Gerhard Klebe, the Department of Pharmaceutical Chemistry, Philipps University Marburg, Germany for making the ITC values and the crystal structures 3T8G, 3T74, 3T73 and 3T8F available to use prior to publication. This project was designed, and the hypothesis to be tested was put forth by N N and D H. The compounds were synthesized, and the kinetic assay was run by N N under the supervision of D H. These compounds were sent to Prof. G. Klebe’s lab for ITC and crystal structures. The complete set of data, including ITC and crystal structures, was initially and independently analyzed by N N and D H, and initial drafts of this manuscript, all of which proposed the involvement of water in the observed cooperativity, were shared with Prof. G. Klebe’s lab prior to their analysis of the data, and prior to the preparation of their manuscript. Although, Prof. Klebe’s lab was in agreement with our overall conclusions, they analyzed the data differently, and it was therefore mutually agreed that the two research groups should publish separately. Both groups provided each other with their submitted manuscripts in the spirit of a good collaboration. H T and J K contributed the computational data and their interpretation. The use of the computational data to support the conclusions of this paper was mutually done by all authors. The computational part of the work was supported by National Institute of Health through a grant (R44GM084555).
ABBREVIATIONS
- Boc
tert-Butyloxycarbonyl
- DCM
Dichloromethane
- DIEA
diisoproplyethylamine
- DMF
N,N-dimethylformamide
- EDCI
1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride
- HOBT
Hydroxybenzotriazole
- HCl
hydrochloric acid
- HPLC
high pressure liquid chromatography
- ITC
Isothermal Titration Calorimetry
- TLN
Thermolysin
- NMR
Nuclear Magnetic Resonance
- ZGPLG
N-carbobenzyloxy-Glyp-L-Leu-Gly
- ZGPLG
N-carbobenzyloxy-Glyp-L-Leu-L-Leu
Footnotes
Unlike disordered waters whose displacement may produce an enthalpically favorable thermodynamic signal if these waters experience stronger binding in the bulk solvent (binding of disordered waters in the active site could be weak, and their existence there could be entropically driven).
The water bridges counted are those having 4 or less water molecules bridging two ligand/protein polar atoms.
While this paper was in the peer-review stage, this reference appeared in ChemMedChem 2012; 7: 1423–1434
Author Contributions
N N and D H contributed the design of the project, the synthesis of the compounds, the kinetic assay and the interpretation of the experimental data. N N was the first to propose the dissection of the differential thermodynamic parameters through theoretical mutations. H T and J K contributed the computational data and their interpretation. The use of the computational data to support the conclusions of this paper was mutually done by all authors.
Supporting Information available: A table containing the computational data used to calculate the interaction energies of the S2′ water molecules in the complex before their displacement, another table containing the crystallographic data used to calculate the water gain/loss indexes of the mutations discussed in this paper, and a figure corresponding to Fig 6c and Fig 7c are included in the supporting information file. Mathematical derivations for equations “6” are also provided. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Brandt T, Holzmann N, Muley L, Khayat M, Wegscheid-Gerlach C, Baum B, Heine A, Hangauer D, Klebe G. Congeneric but still distinct: How closely related ligands exhibit different thermodynamic and structural properties. J Mol Biol. 2011;404:1170–1187. doi: 10.1016/j.jmb.2010.11.038. [DOI] [PubMed] [Google Scholar]
- 2.Ladbury JE, Klebe G, Freire E. Adding calorimetric data to decision making in lead discovery: a hot tip. Nature Rev Drug Discovery. 2010;9:23–27. doi: 10.1038/nrd3054. [DOI] [PubMed] [Google Scholar]
- 3.Whitesides GM, Krishnamurthy VM. Designing ligands to bind proteins. Q Rev Biophys. 2005;38:385–395. doi: 10.1017/S0033583506004240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernandez A, Rogale K, Scott R, Scheraga HA. Inhibitor design by wrapping packing defects in HIV-1 proteins. PNAS. 2004;101:11640–11645. doi: 10.1073/pnas.0404641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernández A, Sanguino A, Peng Z, Ozturk E, Chen J, Crespo A, Wulf S, Shavrin A, Qin C, Ma J, Trent J, Lin Y, Han H, Mangala LS, Bankson JA, Gelovani J, Samarel A, Bornmann W, Sood AK, Lopez-Berestein G. An anticancer C-Kit kinase inhibitor is reengineered to make it more active and less cardiotoxic. J Clin Invest. 2007;117:4044–4054. doi: 10.1172/JCI32373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young T, Abel R, Kim B, Berne BJ, Friesner RA. Motifs for molecular recognition exploiting hydrophobic enclosure in protein-ligand binding; PNAS. 2007;104:808–813. doi: 10.1073/pnas.0610202104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abel R, Young T, Farid R, Berne BJ, Friesner RA. Role of the active site solvent in the thermodynamics of Factor Xa ligand binding. J Am Chem Soc. 2008;130:2817–2831. doi: 10.1021/ja0771033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Berne BJ, Friesner RA. Ligand-binding to protein binding pockets with wet and dry regions. PNAS. 2011;108:1326–1330. doi: 10.1073/pnas.1016793108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snyder PW, Mecinovic J, Moustakas DT, Tomas SW, III, Harder M, Mack ET, Lockett MR, Heroux A, Sherman W, Whitesides GM. Mechanism of the hydrophobic effect in the biomolecular recognition of arylsulfonamides by carbonic anhydrase. PNAS. 2011;108:17889–17894. doi: 10.1073/pnas.1114107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharrow SD, Edmonds KA, Goodman MA, Novotny MV, Stone MJ. Thermodynamic consequences of disrupting a water-mediated hydrogen bond network in a protein: pheromone complex. Protein Science. 2005;14:249–256. doi: 10.1110/ps.04912605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muley L, Baum B, Smolinski M, Freindorf M, Heine A, Klebe G, Hangauer D. Enhancement of hydrophobic interactions and hydrogen bond strength by cooperativity: Synthesis, modeling and molecular dynamics simulations of a congeneric series of thrombin inhibitors. J Med Chem. 2010;53:2126–2135. doi: 10.1021/jm9016416. [DOI] [PubMed] [Google Scholar]
- 12.Kawai SH, Bailey MD, Halmos T, Forgione P, LaPlante SR, Llinas-Brunet M, Naud J, Goudreau N. The use of chemical double-mutant cycles in biomolecular recognition studies: Application to HCV NS3 Protease Inhibitors. ChemMedChem. 2008;3:1654–1657. doi: 10.1002/cmdc.200800214. [DOI] [PubMed] [Google Scholar]
- 13.Horovitz A, Fersht AR. Strategy for analyzing the co-operativity of intramolecular interactions in peptides and proteins. J Mol Biol. 1990;214:613–617. doi: 10.1016/0022-2836(90)90275-Q. [DOI] [PubMed] [Google Scholar]
- 14.Cera ED. Site-specific thermodynamics: understanding cooperativity in molecular recognition. Chem Rev. 1998;98:1563–1591. doi: 10.1021/cr960135g. [DOI] [PubMed] [Google Scholar]
- 15.Cockroft SL, Hunter CA. Chemical double-mutant cycles: dissecting non-covalent interactions. Chem Soc Rev. 2007;36:172–188. doi: 10.1039/b603842p. [DOI] [PubMed] [Google Scholar]
- 16.Morgan BP, Scholtz JM, Ballinger MD, Zipkin ID, Bartlett PA. Differential binding energy: A detailed evaluation of the influence of hydrogen-bonding and hydrophobic groups on the inhibition of Thermolysin by phosphorus-containing inhibitors. J Am Chem Soc. 1991;113:297–307. [Google Scholar]
- 17.Williams DH, Stephens E, O’Brien DP, Zhou M. Understanding noncovalent interactions: Ligand binding energy and catalytic efficiency from ligand-induced reductions in motion within receptors and enzymes. Angew Chem Int Ed. 2004;43:6596–6616. doi: 10.1002/anie.200300644. [DOI] [PubMed] [Google Scholar]
- 18.Latt SA, Holmquist B, Vallee BL. Thermolysin: a zinc metalloenzyme. Biochem Biophys Res Commun. 1969;37:333–339. doi: 10.1016/0006-291x(69)90739-6. [DOI] [PubMed] [Google Scholar]
- 19.Morihara K, Tsuzuki H. Thermolysin: kinetic study with oligopeptides. Eur J Biochem. 1970;15:374–380. doi: 10.1111/j.1432-1033.1970.tb01018.x. [DOI] [PubMed] [Google Scholar]
- 20.Hangauer DG, Monzingo AF, Matthews BW. An interactive computer graphics study of thermolysin-catalyzed peptide cleavage and inhibition by N-carboxymethyl dipeptides. Biochemistry. 1984;23:5730–5741. doi: 10.1021/bi00319a011. [DOI] [PubMed] [Google Scholar]
- 21.Holden HM, Tronrud DE, Monzingo AF, Weaver LH, Matthews BW. Slow-and fast-binding inhibitors of thermolysin display different modes of binding: crystallographic analysis of extended phosphonamidate transition-state analogues. Biochemistry. 1987;26:8542–8553. doi: 10.1021/bi00400a008. [DOI] [PubMed] [Google Scholar]
- 22.Englert L, Biela A, Zayed M, Heine A, Hangauer D, Klebe G. Displacement of disordered water molecules from hydrophobic pocket creates enthalpic signature: Binding of phosphonamidate to the S1′-pocket of thermolysin. Biochem et Biophys Acta. 2010;1800 (11):1192–1202. doi: 10.1016/j.bbagen.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Vo-Quang Y, Gravey AM, Simonneau R, Vo-Quang L, Lacoste AM, Le Goffic F. Towards new inhibitors of D-alanine: D-alanine ligase: the synthesis of 3-amino butenylphosphonic and aminophosphonamidic acids. Tetrahedron Letters. 1987;28 (49):6167–6170. [Google Scholar]
- 24.Feder J, Schuck JM. Studies on the Bacillus subtilis neutral-protease- and Bacillus thermoproteolyticus thermolysin-catalyzed hydrolysis of dipeptide substrates; Biochemistry. 1970;9:2784–2791. doi: 10.1021/bi00816a005. [DOI] [PubMed] [Google Scholar]
- 25.Details of the ITC and the crystallographic data will be published by Biela, A. et al elsewhere**.
- 26.Beres L, Sturtevant JM. Calorimetric studies of the activation of Chymotrypsinogen A. Biochemistry. 1971;10:2120–2126. doi: 10.1021/bi00787a025. [DOI] [PubMed] [Google Scholar]
- 27.Fersht AR. Relationships between apparent binding energies measured in site-directed mutagenesis experiments and energetics of binding and catalysis; Biochemistry. 1988;27:1577–1580. doi: 10.1021/bi00405a027. [DOI] [PubMed] [Google Scholar]
- 28.Serrano L, Horovitz A, Avron B, Bycroft M, Fersht AR. Estimating the contribution of engineered surface electrostatic interactions to protein stability by using double-mutant cycle. Biochemistry. 1990;29:9343–9352. doi: 10.1021/bi00492a006. [DOI] [PubMed] [Google Scholar]
- 29.Carugoa O, Bordo D. How many water molecules can be detected by protein crystallography? Acta Cryst. 1999;D55:479–483. doi: 10.1107/s0907444998012086. [DOI] [PubMed] [Google Scholar]
- 30.Ernst JA, Clubb RT, Zhou H-X, Gronenborn AM, Clore GM. Demonstration of positionally disordered water within a protein hydrophobic cavity by NMR. Science. 1995;267:1813–1817. doi: 10.1126/science.7892604. [DOI] [PubMed] [Google Scholar]
- 31.Goldbeck RA, Pillsbury ML, Jensen RA, Mendoza JL, Nguyen RL, Olson JS, Soman J, Klinger DS, Esquerra RM. Optical detection of disordered water within a protein cavity. J Am Chem Soc. 2009;131:12265–12272. doi: 10.1021/ja903409j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denisov VP, Venu K, Peters J, Horlein HD, Halle B. Orientational disorder and entropy of water in protein cavites. J Phys Chem B. 1997;101:9380–9389. [Google Scholar]
- 33.Mecinovic J, Snyder PW, Mirica KA, Bai S, Mack ET, Kwant RL, Moustakas DT, Heroux A, Whitesides GM. Fluoroalkyl and alkyl chains have similar hydrophobicities in binding to the “hydrophobic wall” of carbonic anhydrase. J Am Chem Soc. 2011;133:14017–14026. doi: 10.1021/ja2045293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthews BW, Liu Lijun. A review about nothing: Are apolar cavities in proteins really empty? Protein Science. 2009;18:494–502. doi: 10.1002/pro.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wade RC, Mazor MH, McCammon JA, Quiocho FA. A molecular dynamics study of thermodynamic and structural aspects of the hydration of cavities in proteins; Biopolymers. 1991;31:919–931. doi: 10.1002/bip.360310802. [DOI] [PubMed] [Google Scholar]
- 36.Olano LR, Rick SW. Hydration free energies and entropies for water in protein interiors. J Am Chem Soc. 2004;126:7991–8000. doi: 10.1021/ja049701c. [DOI] [PubMed] [Google Scholar]
- 37.Lee J, Kim SH. Water polydons in high-resolution protein crystal structures; Protein Science. 2009;18:1370–1376. doi: 10.1002/pro.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilli G, Bellucci F, Ferretti V, Bertolasi V. Evidence for resonance-assisted Hydrogen bonding from crystal-structure correlations on the Enol Form of the β-diketone fragment. J Am Chem Soc. 1989;111:1023–1028. [Google Scholar]
- 39.Bertolasi V, Pretto L, Ferretti V, Gilli G, Gilli P. π Bond cooperativity and anticooperativity effects in resonance assisted hydrogen bonds (RAHBs) Acta Cryst. 2006;B62:850–863. doi: 10.1107/S0108768106020933. [DOI] [PubMed] [Google Scholar]
- 40.Sobczyk L, Grabowski SJ, Krygowski TM. Interrelation between H-bond and pi-electron delocalization. Chem Rev. 2005;105:3513–3560. doi: 10.1021/cr030083c. [DOI] [PubMed] [Google Scholar]
- 41.Π-bond cooperativity is the mutual reinforcement of the H-bonds formed by electronegative atoms which are linked by bonds that have π-electron character. One of these electronegative atoms should act as an H-bond donor and the other as an H-bond acceptor. If both atoms are H-bond donors or both are H-bond acceptors, the H-bonds formed by these atoms are weakened due to π-anticooperativity. Σ-bond cooperativity involves the mutual reinforcement of two H-bonds formed by a OH group which participates in one of these H-bonds as a donor and in the other as acceptor (σ-anticooperativity arises among H-bonds simultaneously accepted or simultaneously donated by this group). A H-bond formed by a OH group tends to polarize the OH σ-bond causing it to have greater tendency to form another H-bond of the opposite donor/acceptor type and lower tendency to form H-bonds of the same type.
- 42.Del Bene J, Pople JA. Theory of molecular interactions. I. Molecular orbital studies of water polymers using a minimal Slater-type basis. J Chem Phys. 1970;52:4858–4866. [Google Scholar]
- 43.Del Bene J, Pople JA. Theory of molecular interactions. III. A comparison of studies of H2O polymers using different molecular-orbital basis sets. J Chem Phys. 1973;58:3605–3608. [Google Scholar]
- 44.Jeffrey GA. Hydrogen bonds and molecular recognition. Food Chem. 1996;56:241–246. [Google Scholar]
- 45.Saenger W. Circular hydrogen bonds. Nature. 1979;279:343–344. [Google Scholar]
- 46.Saenger W, Lindner K. OH clusters with homodromic circular arrangement of hydrogen bonds. Angew Chem Int Ed. 1980;19:398–399. [Google Scholar]
- 47.Becke AD. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys Rev A. 1988;38:3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 48.Lee C, Weitao Y, Parr RG. Development of the Colle-Salvetti conelation energy formula into a functional of the electron density. Phys Rev B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 49.Becke AD. Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys. 1993;98:5648. [Google Scholar]
- 50.Dunning TH., Jr Gaussian Basis Sets for Use in Correlated Molecular Calculations. I. The Atoms Boron through Neon Hydrogen. J Chem Phys. 1989;90:1007–1023. [Google Scholar]
- 51.Tan H, Qu W, Chen G, Liu R. The role of charge transfer in the hydrogen bond cooperative effect of cis-N-methylformamide oligomers. J Phys Chem A. 2005;109:6303–6308. doi: 10.1021/jp051444q. [DOI] [PubMed] [Google Scholar]
- 52.Blake CCF, Pulford WCA, Artymiuk PJ, Huber R. X-ray studies of water in crystals of lysozyme. J Mol Biol. 1983;167:693–723. doi: 10.1016/s0022-2836(83)80105-3. [DOI] [PubMed] [Google Scholar]
- 53.Baker T, Dodson E, Dodson G, Hodgkin D, Hubbsrd R. The water structure in 2 Zn insulin crystals. In: Moras D, Drenth J, Strandberg B, Suck D, Wilson K, editors. Crystallography in molecular biology. Plenum Press; New York: pp. 179–192. [Google Scholar]
- 54.Nasief NN, et al. Unpublished data [Google Scholar]
- 55.Southall NT, Dill KA. The mechanism of hydrophobic solvation depends on solute radius. J Phys Chem B. 2000;104:1326–1331. [Google Scholar]
- 56.Graziano G. Hydration entropy of polar, nonpolar and charged species. Chem Phys Lett. 2009;479:56–59. [Google Scholar]
- 57.Graziano G. Scaled particle theory study of the length scale dependence of cavity thermodynamics in different liquids. J Phys Chem B. 2006;110:11421–11426. doi: 10.1021/jp0571269. [DOI] [PubMed] [Google Scholar]
- 58.Graziano G, Lee B. Entropy convergence in hydrophobic hydration: a scaled particle theory analysis. Biophys Chem. 2003;105:241–250. doi: 10.1016/s0301-4622(03)00073-5. [DOI] [PubMed] [Google Scholar]
- 59.Morgan G, Fruton JS. Kinetics of the action of thermolysin on peptide substrates. Biochemistry. 1978;17:3562–3568. doi: 10.1021/bi00610a022. [DOI] [PubMed] [Google Scholar]
- 60.Cheng Y, Prusoff WH. Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 61.Feder J, Brougham L, Wildi B. Inhibition of Thermolysin by dipeptides. Biochemistry. 1974;13:1186–1189. doi: 10.1021/bi00703a600. [DOI] [PubMed] [Google Scholar]
- 62.Henderson PJF. A linear equation that describes the steady-state kinetics of enzymes and subcellular particles interacting with tightly bound inhibitors. Biochem J. 1972;127:321–333. doi: 10.1042/bj1270321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johnson ER, Becke AD. A post-Hartree-Fock model of intermolecular interactions: Inclusion of higher-order corrections. J Chem Phys. 2006;124:174104. doi: 10.1063/1.2190220. [DOI] [PubMed] [Google Scholar]
- 64.Kong J, Gan Z, Proynov E, Freindorf M, Furlani TR. Efficient computation of the dispersion interaction with density functional theory. Phys Rev A. 2009;79:042510. [Google Scholar]
- 65.Shao Y, Fusti-Molnar L, et al. Advances in methods and algorithms in a modern quantum chemistry program package. Phys Chem Chem Phys. 2006;8:3172–3191. doi: 10.1039/b517914a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.