Abstract
Ripening-associated pectin disassembly in melon is characterized by a decrease in molecular mass and an increase in the solubilization of polyuronide, modifications that in other fruit have been attributed to the activity of polygalacturonase (PG). Although it has been reported that PG activity is absent during melon fruit ripening, a mechanism for PG-independent pectin disassembly has not been positively identified. Here we provide evidence that pectin disassembly in melon (Cucumis melo) may be PG mediated. Three melon cDNA clones with significant homology to other cloned PGs were isolated from the rapidly ripening cultivar Charentais (C. melo cv Reticulatus F1 Alpha) and were expressed at high levels during fruit ripening. The expression pattern correlated temporally with an increase in pectin-degrading activity and a decrease in the molecular mass of cell wall pectins, suggesting that these genes encode functional PGs. MPG1 and MPG2 were closely related to peach fruit and tomato abscission zone PGs, and MPG3 was closely related to tomato fruit PG. MPG1, the most abundant melon PG mRNA, was expressed in Aspergillus oryzae. The culture filtrate exponentially decreased the viscosity of a pectin solution and catalyzed the linear release of reducing groups, suggesting that MPG1 encodes an endo-PG with the potential to depolymerize melon fruit cell wall pectin. Because MPG1 belongs to a group of PGs divergent from the well-characterized tomato fruit PG, this supports the involvement of a second class of PGs in fruit ripening-associated pectin disassembly.
Fruit ripening is a genetically programmed event that is characterized by a number of biochemical and physiological processes that alter fruit color, flavor, aroma, and texture (Brady, 1987). Extensive cell wall modifications occur during ripening and are thought to underlie processes such as fruit softening, tissue deterioration, and pathogen susceptibility. These modifications are regulated at least in part by the expression of genes that encode cell wall-modifying enzymes (Fischer and Bennett, 1991). Pectins are a major class of cell wall polysaccharides that are degraded during ripening, undergoing both solubilization and depolymerization. In tomato the majority of ripening-associated pectin degradation is attributable to the cell wall hydrolase PG. Transgenic tomato plants with altered PG gene expression indicated that PG-dependent pectin degradation is neither required nor sufficient for tomato fruit softening to occur (Sheehy et al., 1988; Smith et al., 1988; Giovannoni et al., 1989). However, data from experiments using fruit of the same transgenic lines strongly suggested that PG-mediated pectin degradation is important in the later, deteriorative stages of ripening and in pathogen susceptibility of tomato fruit (Schuch et al., 1991; Kramer et al., 1992).
In melon (Cucumis melo) substantial amounts of pectin depolymerization and solubilization take place during ripening (McCollum et al., 1989; Ranwala et al., 1992; Rose et al., 1998), implicating a role for PG in ripening-associated cell wall disassembly in melons. However, melons have been reported to lack PG enzyme activity (Hobson, 1962; Lester and Dunlap, 1985; McCollum et al., 1989; Ranwala et al., 1992). The possibility exists that PG is present in melon but that it does not conform to the expected enzymic properties in terms of abundance and/or lability, a point illustrated by recent reports in apple and strawberry, which were previously reported to lack PG activity but that do in fact accumulate low amounts of protein and/or measurable activity (Nogata et al., 1993; Wu et al., 1993). In light of the unexplained discrepancy between ripening-associated pectin depolymerization and undetectable PG activity in melons, we have undertaken a study to reexamine the status of PG in melon using the rapidly ripening cv Charentais (C. melo cv Reticulatus F1 Alpha).
As reported for other cultivars, Charentais melons exhibit substantial solubilization and a downshift in the molecular-mass profile of water-soluble pectins, but this is associated with the later stages of ripening, after softening is initiated (Rose et al., 1998). By utilizing a molecular approach to analyze PG in melon, we have attempted to overcome some of the potential limitations of biochemical methods, such as low abundance of protein, reliance on other cell wall components, and unknown cofactors for activity and/or lability during extraction. In doing so, we have identified and characterized a multigene family encoding putative PGs from Charentais melon, including three PG homologs that are expressed abundantly during fruit ripening. The pattern of PG gene expression correlates temporally with the depolymerization of water-soluble pectins and an increase in pectin-degrading enzyme activity. Three additional PG homologs were also identified and shown to be expressed in mature anthers and fruit-abscission zones, tissues that, similar to ripening fruit, are undergoing cell separation. The most abundant ripening-associated putative PG mRNA, MPG1, was expressed in the filamentous fungus Aspergillus oryzae. The culture filtrate from the transformed A. oryzae strain XMPG1 exhibited endo-PG activity, further supporting a role for endo-PG in ripening-associated pectin disassembly in Charentais melon fruit.
MATERIALS AND METHODS
Plant Material
The Charentais melon (Cucumis melo cv Reticulatus F1 Alpha) fruit used in this study were harvested at six distinct developmental stages that included IG, MG, and R1 to R4. These stages are described in detail in Rose et al. (1998). Fruit-abscission zones were collected from the peduncle of field-grown R3 fruit. In melon the abscission zone is located immediately adjacent to the fruit and is characterized by the area that “slips” during the ripening of most cultivars. Anthers were collected from field-grown male flower explants on the day of flower opening, after they had begun to shed pollen. Pistils (consisting of ovary, style, and stigma) were collected from female flowers attached to the plant on the day of flower opening. Roots were collected from 9- and 10-d-old seedlings grown in vermiculite in a growth chamber at 25°C with a 16-h day/8-h night cycle. Stem and young leaf tissues were collected from 32-d-old greenhouse-grown plants. All tissues were frozen in liquid N2 immediately following harvest and stored at −80°C until use.
Protein Extraction and Enzyme Assay
Frozen mesocarp tissue (10 g) from fruit at each stage of development was homogenized in a mortar in 1 volume of low-salt extraction buffer (10 mm NaC2H3O2, pH 4.5, 5 mm β-mercaptoethanol, 0.5% [w/v] PVPP, 2 mm PMSF, 40 μm leupeptin, and 20 μg/mL chymostatin). The homogenates were centrifuged at 30,000g for 15 min, after which time the supernatants were filtered through two layers of Miracloth (Calbiochem) and designated the low-salt soluble fraction. The pellets were resuspended in 2 mL of high-salt extraction buffer (50 mm NaC2H3O2, pH 4.5, 1.5 m NaCl, 15 mm EDTA, 5 mm β-mercaptoethanol, 2 mm PMSF, 40 μm leupeptin, and 20 μg/mL chymostatin) and shaken at moderate speed for 2 h at 4°C. The homogenate was centrifuged at 30,000g for 15 min, after which the supernatant was filtered through two layers of Miracloth and designated the high-salt soluble fraction.
Pectin-degrading activity of melon protein extracts and culture filtrates from Aspergillus oryzae strains XMPG1 and BANe3 (see below) were assayed viscometrically in semi-micro viscometers (Canon-Manning, State College, PA). The reactions were initiated by adding 200 μL of melon protein extract representing 0.6 g fresh weight or 4.5 μg of culture-filtrate protein (brought to a final volume of 200 μL with 40 mm NaC2H3O2, pH 5.0) to 800 μL of 1.0% (w/v) pectin (Sigma, 10% esterified), 50 mm NaC2H3O2, pH 5.2, 100 mm EGTA, 150 mm NaCl, and 0.01% NaN3, and the reactions were incubated at 25°C for up to 6 h. One unit was defined as the amount of enzyme that reduced the viscosity by 1% per hour. The assays were conducted in triplicate. Protein extracts boiled for 30 min were also assayed in duplicate and did not cause a significant reduction in viscosity.
The pectin-degrading activity of culture filtrates from A. oryzae XMPG1 and BANe3 were determined over a time course of culture growth at 34°C in 100 mL of MY25 medium (Yaver et al., 1996) by gel diffusion. An aliquot of culture was removed at the time points shown in Figure 6B, filtered through two layers of Miracloth, and 280 ng of protein was assayed in a total volume of 20 μL in gel-diffusion plates containing 0.01% (w/v) polygalacturonic acid, 1 mm EDTA, 100 mm NaC2H3O2, pH 5.0, and 1% (w/v) agarose. The plates were incubated for 10 h at 34°C, stained with 0.05% (w/v) ruthenium red for 30 min, and destained with water. Culture filtrates from XMPG1 and BANe3 were also assayed for pectin-degrading activity by measuring the release of reducing sugars from pectin substrate. The composition of the reactions was 20 μg of ultrafiltered (YM10 membrane, Amicon, Beverly, MA) culture filtrate protein, 50 mm EGTA, 150 mm NaCl, 40 mm NaC2H3O2, pH 5.0, and 0.01% NaN3 in a total volume of 800 μL. The reactions were conducted in duplicate, and 200 μL was removed from each reaction at 0 min, 60 min, 3 h, and 6 h, and assayed for the presence of reducing groups using cyanoacetamide (Gross, 1982). The final values for activity of XMPG1 culture filtrate were calculated as the difference between the BANe3 and XMPG1 values.
Figure 6.
A, Gel-diffusion assay of culture filtrates from A. oryzae transformed with MPG1 (XMPG1) or untransformed. Aliquots were removed from cultures at 24, 34, 50, 58, and 72 h and filtered through two layers of Miracloth, and equal amounts of protein were assayed for PG activity. B, Viscometric and reducing sugar assays of XMPG1 and untransformed culture filtrates. Culture filtrates from one time point were incubated for up to 6.5 h and assayed at multiple time points for the ability to decrease the viscosity or release reducing groups of a pectin solution. •, Untransformed viscosity; ▪, XMPG1 viscosity; and ⋄, XMPG1 reducing sugar.
RNA Extraction
Polysomal RNA used for RT-PCR of fruit tissue was isolated as described by Larkins and Hurkman (1978), with some modifications: Frozen melon mesocarp tissues (200 g) were homogenized in an equal volume of ice-cold extraction buffer (0.2 m Tris, pH 8.5, 0.2 m Suc, 0.1 m KCl, 25 mm EGTA, 35 mm MgCl2, 1 mm DTT, 1 mm spermidine-HCl, 0.5% [w/v] deoxycholate, and 1% [v/v] Triton X-100) and filtered through three layers of Miracloth, and the cellular debris were pelleted by centrifugation at 12,000g for 10 min. Additional Triton X-100, 0.01% (v/v), was added to the supernatant, stirred at 4°C for 15 min, and the mixture centrifuged at 12,000g for 10 min. The polysomes were pelleted from the supernatant through a 5-mL layer of 1.8 m Suc (in 0.2 m Tris, pH 8.5, 0.1 m KCl, 25 mm EGTA, 35 mm MgCl2, and 1 mm DTT) at 257,000g for 3 h. The polysome pellet was resuspended in 1 mL of 40 mm Tris, pH 8.5, 20 mm KCl, and 10 mm MgCl2, and extracted twice with phenol equilibrated with NaC2H3O2, pH 4.0 and twice with chloroform:isoamyl alcohol (24:1, v/v). The RNA in the aqueous phase was precipitated by the addition of 0.5 volume of 7.5 m NH4-acetate and 2 volumes of 100% EtOH. The RNA was collected by centrifugation at 12,000g, washed with 70% EtOH, air-dried, and resuspended in RNase-free water.
RNA used for cDNA library construction was isolated from frozen melon tissue based on the protocol of Wadsworth et al. (1988) with all volumes scaled up to accommodate 30 g of tissue. Total RNA used for northern-blot analyses of all tissues and RT-PCR of anthers and abscission zones was isolated as described by Wan and Wilkins (1994). Poly(A+) RNA was selected using latex beads supplied in the Oligotex kit (Qiagen, Chatsworth, CA) and following the manufacturer's instructions (with the exception that the annealing time was increased to 45 min).
Oligonucleotide Design and RT-PCR
Degenerate oligonucleotides were designed based on regions of high homology between aligned PG-deduced amino acid sequences from tomato (Greirson et al., 1986), peach (Lee et al., 1990) and O. organensis (Brown and Crouch, 1990), and were synthesized by the Protein Structure Laboratory at the University of California (Davis). The sequence of the upstream primer, PG1.2, was 5′-ACI GGI GA(T/C) GA(TC) TG(TC) ATI UC 3′, and the sequence of the downstream primer, PG2.2, was 5′-CCA IGT (C/T)TT (A/G/T)AT IC(G/T) IAC ICC (A/G)TT-3′ (where I is inosine). The two oligonucleotides correspond to the amino acid sequences TGGDDCIS (PG1.2) and NGVRIKTW (PG2.2) at positions 267 to 274 and 323 to 330 of tomato fruit PG, respectively. These two regions flank an additional conserved region at amino acid position 286 to 297 of the tomato fruit protein that has the sequence CGPGHGISIGSL. The third sequence was used diagnostically to identify cDNAs that encode putative PGs due to its high level of conservation in all plant and microbial PGs cloned to date.
First-strand cDNAs were synthesized from 2 μg of total polysomal RNA from stage R3 Charentais fruit in three separate experiments. The first experiment used RNA that had not been treated with DNase. The other two experiments used RNA that had been DNase treated (fast-protein liquid chromatography-pure, Pharmacia) according to the manufacturer's instructions. First-strand cDNAs were also synthesized from 2 μg of total RNA treated with RNase-free DNase, from fruit-abscission zones, or from 40 ng of poly(A+) RNA from anthers. RNAs were incubated in 20 μL of 1× first-strand buffer (50 mm Tris-HCl, pH 8.3, 75 mm KCl, and 3 mm MgCl2; GIBCO-BRL), 0.5 mm each dATP, dCTP, dGTP, and deoxyribothymine triphosphate (Pharmacia), 100 ng of oligo dT12–18 (Pharmacia), 10 mm DTT (GIBCO-BRL), and 20 units of RNasin (Promega) at 65°C for 10 min and then placed on ice. Two microliters of Moloney murine leukemia virus RT (200 units/μL, GIBCO-BRL) was added and the reaction was incubated at 37°C for 1 h. Reactions were then heated to 95°C for 5 min, and then placed on ice or stored at −20°C until further use. For each RNA sample, a control reaction with 2 μL of 1× first-strand buffer added in place of 2 μL of RT was included.
Four microliters of each first-strand reaction or 1 ng of pBS1.9 (tomato fruit full-length PG cDNA; DellaPenna and Bennett, 1988) was used as a template in PCR. The reaction mixtures were composed of 10 mm Tris-HCl, pH 8.3, 50 mm KCl, 1 mm MgCl2, 0.01% (w/v) gelatin, 0.2 mm each dATP, dCTP, dGTP, and deoxyribothymine triphosphate (Pharmacia), 0.8 μm each PG1.2 and PG2.2 oligonucleotides, and 0.2 μL of Taq polymerase (Perkin-Elmer). The conditions for amplification were 25 or 40 cycles of 94°C for 1 min, 40°C for 1 min, and then 72°C for 2 min. The pBS1.9 amplification product served as a reference for gel purification of melon PCR products. Products were gel purified using Sephaglas (Pharmacia) and the products were cloned into pCRII using a cloning kit (Invitrogen, San Diego, CA), both according to the manufacturer's instructions. Cloned PCR products were sequenced by the dideoxynucleotide method (Sanger et al., 1977) using [35S]dATP and modified T7 DNA polymerase (Sequenase, United States Biochemical), according to the manufacturer's instructions. Sequence analysis was carried out using the MacDNASIS Pro 3.5 software package (Hitachi, San Bruno, CA).
RNA Gel-Blot Hybridization
Two micrograms of poly(A+) RNA from fruit tissues or 1 μg of poly(A+) RNA from root, stem, leaf, pistil, anther, and fruit-abscission zones was separated by 1.1% (w/v) formaldehyde/1.2% (w/v) agarose gel electrophoresis and transferred to nylon membranes (Hybond-N, Amersham), according to the manufacturer's instructions. Membranes were probed with gel-purified, [α-32P]dATP-labeled insert DNA from pMPG1, pMPG2, and pMPG3 (full-length clones, see below) and pPG1 to pPG14 (partial-length PCR clones). Probes were labeled by random-hexamer priming using Klenow DNA polymerase (United States Biochemical). The hybridizations were carried out for 16 h at 65°C in 2% (w/v) SDS, 1 m NaCl, 10% (w/v) dextran sulfate, and 100 μg mL−1 denatured salmon-sperm DNA, with approximately 50 ng of labeled probe. The blots were washed twice in 1× SSC (0.15 m NaCl and 15 mm sodium citrate) and 0.1% (w/v) SDS at 65°C and twice in 0.2× SSC and 0.1% SDS at 65°C. Blots were exposed to film with one intensifying screen (Reflection, DuPont) at −80°C. To estimate the relative abundance of mRNA encoded by MPG1, MPG2, and MPG3 during fruit ripening, the corresponding blots were probed with inserts that were labeled to approximately the same specific activity and exposed to film for 4 h. All other blots were exposed to film for 2 to 4 d, with the exception of the nonfruit blot probed with labeled insert from pPG4, which was exposed to film for 4 h.
cDNA Library Construction and Screening
A cDNA library was constructed using 3.3 μg of poly(A+) RNA prepared from stage R3 Charentais melon fruit using a λ-ZAP-cDNA synthesis kit (Stratagene). cDNAs were cloned into the Uni-XAP-XR λ-phage vector (Stratagene) and packaged in Gigapack Gold (Stratagene), and the primary library was amplified according to the manufacturer's protocols.
Duplicate plaque lifts of 1 × 106 (PG1) or 4 × 105 (PG2 and PG3) amplified recombinants were hybridized with gel-purified, radiolabeled inserts from pPG1, pPG2, or pPG3 using protocols described by Stratagene. Hybridized filters were washed twice in 1× SSC and 0.1% SDS at 65°C and twice in 0.2× SSC and 0.1% SDS at 65°C, and exposed to film with one intensifying screen (DuPont) at −80°C. Positive plaques were carried through two additional rounds of screening for purification and then in vivo excised to release the phagemid DNA. Both strands of positive cDNA clones corresponding to MPG1, MPG2, and MPG3 were sequenced in the Plant Genetics Facility at the University of California (Davis) using an automated DNA sequencer (model ABI 377, Perkin Elmer/Applied Biosystems), and gene-specific oligonucleotides synthesized by Genset (La Jolla, CA) or vector sequence oligonucleotides for sequencing the ends of the cDNAs. Sequence analyses were carried out using the MacDNASIS Pro 3.5 software package and Sequencher 3.0 (Genecode, Madison, WI). The cleavage site of the signal sequences were predicted using the rules of von Heijne (1983). All three of the cDNAs contained complete open reading frames. The deduced amino acid sequence alignments were generated using the Clustal V multiple-alignment software package (Higgins et al., 1992).
DNA Gel-Blot Hybridization
Total genomic DNA was isolated as previously described by Murray and Thompson (1980) as modified by Bernatzky and Tanksley (1986). Then, 5 μg was digested with the restriction enzymes EcoRI, BamHI, and HindIII (New England Biolabs), separated on 0.8% agarose gels, and transferred to Hybond nylon membranes according to the manufacturer's (Amersham) instructions. Membranes were probed with gel-purified, [α-32P]dATP-labeled insert DNA from pMPG1, pMPG2, and pMPG3 (full-length clones), and pPG4, pPG5, and pPG6 (partial-length RT-PCR clones) under the same conditions described above for RNA-blot hybridizations, washed in 5× SSC and 0.1% SDS at 65°C (Tm −32°C), and exposed to film with one intensifying screen (DuPont) at −80°C for 4 h. The blots were then washed twice in 0.2× SSC and 0.1% SDS at 65°C (Tm −8°C) and exposed to film as described above.
Phylogenetic Analysis
The deduced amino acid sequences of MPG1, MPG2, and MPG3 were aligned to 17 full-length deduced amino acid sequences of PG homologs using Clustal V multiple-sequence alignment software (Higgins et al., 1992). The sequences were: tomato fruit (pTOM6; Grierson et al., 1986) and abscission zone (TAPG1, TAPG2, and TAPG4; Kalaitzis et al., 1995, 1997), peach fruit (PRF5; Lester et al., 1994) and genomic clone (Lee et al., 1990), apple (Malus domestica) fruit (pGDP-1; Atkinson, 1994), kiwifruit (Actinidia deliciosa) genomic clone (Atkinson and Gardner, 1993), avocado (Persea americana) fruit (pAVOpg; Kutsunai et al., 1993), oilseed rape (Brassica napus) pod-dehiscence zone (SAC66; Jenkins et al., 1996) and pollen (Sta 44–4; Robert et al., 1993), maize (Zea mays) pollen (3C12; Rogers et al., 1991), Arabidopsis thaliana pollen (GenBank accession no. x73222), tobacco (Nicotiana tabacum) pollen (NPG1; Tebbutt et al., 1994), alfalfa (Medicago sativa) pollen (P73; Qiu and Erickson, 1996), cotton (Gossypium hirsutum) pollen (G9; John and Petersen, 1994), and the fungus Aspergillus flavus (Whitehead et al., 1995). Phylogenetic trees were inferred from the aligned sequences using the maximum parsimony algorithm of the PAUP software package, version 3.1 (Swofford, 1990). The aligned sequences were analyzed by a heuristic search with 100 replicates under the random stepwise addition option and global (tree bisection and reconnection) branch swapping, with the A. flavus sequence defined as the outgroup.
Heterologous Expression of MPG1
Gene-specific oligonucleotides designed to introduce a SwaI restriction site at the 5′ end and a PacI restriction site at the 3′end of the MPG1-coding region were used to amplify this region from pMPG1. The reaction mixtures were composed of 10 mm Tris-HCl, pH 8.85, 25 mm KCl, 5 mm (NH4)2SO4, 2 mm MgSO4, 0.5 mm each dATP, dCTP, dGTP, and deoxyribothymine triphosphate (Pharmacia), 0.4 μm each oligonucleotide, and 2.5 units of Pwo DNA polymerase (Boehringer Mannheim). The conditions for amplification were one cycle of 94°C for 3 min; 25 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1.5 min; and one cycle of 72°C for 5 min. The product was gel purified using the Qiaquick gel-extraction kit (Qiagen) and cloned into pCR-Blunt using a PCR-cloning kit (Zero Blunt, Invitrogen).
Both strands of the cloned PCR product in the resulting plasmid, pP1, were sequenced using Taq polymerase cycle-sequencing and an automatic DNA sequencer (model 373A, version 1.2.0, Applied Biosystems). The SwaI/PacI insert of pP1 was ligated to SwaI/PacI digested pBANe15 to obtain the A. oryzae expression vector pXMPG1. The pBANe15 vector includes the promoter sequence from the A. oryzae α-amylase gene, the termination sequence from the Aspergillus niger glucoamylase gene and the Aspergillus nidulans acetamidase (amdS) gene used as a selectable marker (Christensen et al., 1988; Nelson et al., 1997). Protoplasts from A. oryzae strain BANe3 (A1560 [Christensen et al., 1988] and ΔamdS, ΔamyA, ΔamyB, and ΔamyC [Nelson et al., 1997]) were prepared and transformed with 10 μg of pXMPG1 DNA as previously described (Christensen et al., 1988), and transformants were selected on minimal-medium plates with acetamide as the sole N source (Cove, 1966). Culture filtrates of the primary transformants were screened for PG activity by gel-diffusion assays (described above), and transformants expressing high levels of activity were spore purified and rescreened. One strain expressing a high level of activity, XMPG1, was selected for further analysis.
RESULTS
Ethylene Production, Textural Changes, and PG Activity during Melon Fruit Ripening
Charentais melon fruit undergo a ripening-associated decrease in fruit firmness, which is accompanied by a dramatic increase in internal ethylene concentration (Rose et al., 1998). In the ripening fruit used in the present study, softening occurred rapidly between stages R1 and R2 and continued through R4. Softening beyond stage R3 represented overripe deterioration of the fruit. Protein was extracted from IG, MG, and R1 to R4 fruit and assayed for pectin-degrading activity viscometrically using commercially available citrus pectin. When high-salt extracts were assayed, a reduction in viscosity was detected at all time points but was highest at stage R3, after softening was initiated and during the later stages of ripening (Fig. 1). Low-salt extracts did not cause a significant reduction in viscosity (data not shown). The highest level of activity was correlated with the period of depolymerization of water-soluble pectins (Rose et al., 1998).
Figure 1.
Pectin-degrading activity in high-salt protein extracts from developing melon fruit. Extracts from fruit at six developmental stages of ripening (IG, MG, and R1–R4) were assayed viscometrically using 10% esterified citrus pectin. One unit was defined as the amount of enzyme that reduced the viscosity by 1% per hour. Each value is an average ± sd of three independent measurements. Low-salt and boiled (30 min) high-salt protein extracts did not cause a significant reduction in viscosity (data not shown). gfw, Grams fresh weight.
Cloning of a Gene Family Encoding Putative PGs
Degenerate oligonucleotides were designed based on regions of homology between aligned PG deduced amino acid sequences from tomato, peach, and O. organensis and were used to amplify partial-length melon cDNAs from reverse-transcribed RNA of ripe fruit mesocarp, ripe fruit-abscission zones, and mature anthers. The amplified product was 190 bp, as predicted from the sequences of known PGs.
A total of 27 individual RT-PCR clones were sequenced from ripe fruit, resulting in the identification of 12 unique sequences, designated pPG1 to pPG3, pPG5 to pPG6, and pPG8 to pPG14. All of the sequences were represented only once except pPG1 (seven times), pPG5 (five times), pPG8 (two times), pPG9 (three times), and pPG10 (three times).
Ten individual PG cDNA clones were sequenced from anthers and found to be identical. This sequence was unique compared with the melon fruit clones and was designated pPG4. Seven individual RT-PCR clones from fruit-abscission zones were sequenced, five of which were identical to pPG5 and two of which defined a second gene, designated pPG7. In total, 14 distinct, partial-length cDNAs encoding putative PGs were identified in melon.
Genome Structure of PGs
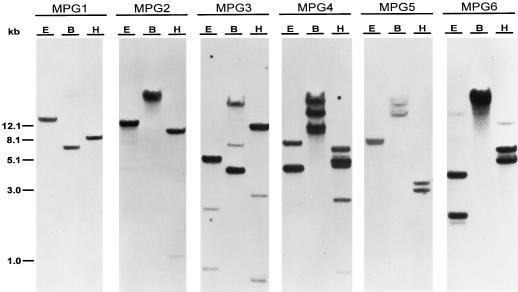
The full-length cDNAs MPG1, MPG2, and MPG3 and the partial length cDNAs pPG4, pPG5, and pPG6 were used to probe genomic DNA blots to determine the organization of these genes in the melon genome. MPG1, MPG2, MPG3, and pPG5 all hybridized to a small number of distinct genomic fragments at both low stringency (Tm −32°C; data not shown) and high stringency (Tm −8°C; Fig. 2), indicating that they are transcribed from divergent genes of low copy number. pPG4 and pPG6, however, hybridized to a common set of genomic fragments at low stringency (Tm −33°C; data not shown) but each to a distinct subset at high stringency (Tm −8°C; Fig. 2), indicating that they belong to the same genomic subfamily but that they are transcribed from distinct genes.
Figure 2.
Melon genomic DNA gel-blot analysis of PG. Genomic DNA (5 μg/lane) was digested with EcoRI (E), BamHI (B), or HindIII (H). The blots were probed with the MPG1, MPG2, or MPG3 full-length cDNA or the PG4, PG5, or PG6 PCR fragments, and washed at a stringency of Tm −8°C.
Tissue-Specific Expression of PG Genes in Melon
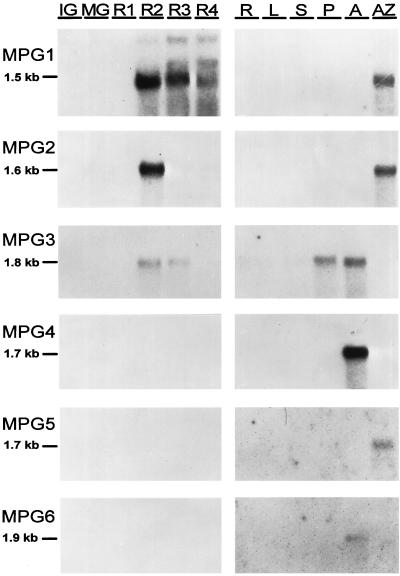
Gel blots of RNA from preripe and ripening fruit and other nonfruit tissues were probed with the 14 partial length cDNAs described above to determine their patterns of expression. The expression of six of the corresponding PG genes was detected in a variety of tissues; three of the PG genes, MPG1, MPG2, and MPG3, were expressed in ripening fruit. The full-length clones corresponding to these genes were isolated from a ripe-fruit cDNA library (see below). Figure 3 shows the results of hybridization with labeled inserts from the full-length clones MPG1, MPG2, and MPG3 and labeled inserts from the partial length clones pPG4, pPG5, and pPG6. pPG4 hybridized strongly to a 1.7-kb mRNA present in anthers, and was not detected in any other tissue examined. pPG6 also hybridized to a mRNA from anthers but was larger, 1.9 kb, and much less abundant compared with pPG4. The sequences of these two cDNAs were also very similar to each other compared with other melon PGs over the same region. pPG5 hybridized to a 1.7-kb mRNA in fruit-abscission zones, but did not hybridize to RNA from any other tissue.
Figure 3.
RNA-blot analysis of PG RNA in developing melon fruit and nonfruit tissues. Each lane was loaded with 2 μg of poly(A+) RNA isolated from fruit tissues at six stages of development (IG, MG, and R1–R4) or 1 μg of poly(A+) RNA from roots (R), young leaves (L), stems (S), pistils (P), anthers (A), and fruit-abscission zones (AZ). The blots were probed with the MPG1, MPG2, or MPG3 full-length cDNA or the PG4, PG5, or PG6 PCR fragments, and washed at a stringency of Tm −8°C.
To estimate the relative abundance of MPG1, MPG2, and MPG3 mRNA during fruit ripening, the full-length clones were labeled to the same specific activity and the fruit RNA blots were exposed to film for an equal length of time. The genes corresponding to MPG1, MPG2, and MPG3 were all expressed during fruit ripening but differed in the relative abundance of mRNA accumulation, the temporal pattern of expression, and in the size of mRNAs they encode. MPG1 hybridized predominantly to a 1.5-kb mRNA but also detected a 1.9- and a 2.6-kb mRNA. The abundance of the predominant 1.5-kb mRNA was highest at stage R2 and then decreased slightly at stages R3 and R4. The largest transcript was least abundant at stage R2, increased at stage R3, and remained at the same level at stage R4. The intermediate-sized transcript appeared at stage R3 and then increased. The relative abundance of the two larger transcripts hybridizing to MPG1 were approximately the same at stage R4, each about one-half as abundant as the 1.5-kb transcript. MPG2 hybridized to a 1.6-kb transcript and was expressed very abundantly at stage R2 and at much reduced levels at stages R3 and R4. When the MPG2 blot was exposed to film for a longer time (data not shown), a 3.0-kb transcript became evident at stage R2. MPG3 hybridized to a 1.8-kb transcript and was highest at stages R2 and R3, but the overall abundance of this transcript was much lower in fruit compared with MPG1 and MPG2.
After extended exposure times, hybridization of MPG1, MPG2, and MPG3 to mRNA from nonfruit tissues was also detected at moderate levels. MPG1 hybridized to a 1.5-kb mRNA from fruit-abscission zones, but was not detected in any other tissue examined. MPG2 hybridized moderately to a 1.6-kb transcript in fruit-abscission zones, and MPG3 hybridized moderately to 1.8-kb transcripts in pistil and anther RNA and very weakly to transcripts in all of the other tissues examined.
Isolation and Characterization of cDNAs Encoding MPG1, MPG2, and MPG3
A cDNA library was constructed from ripe melon fruit RNA and screened with inserts from pPG1, pPG2, and pPG3. The resulting cDNA clones MPG1, MPG2, and MPG3 were 1521, 1641, and 1767 bp in length, respectively, and each contained a complete open reading frame (Fig. 4). The length of the cDNA clones corresponded to the size of the most abundant corresponding mRNA, and it is assumed that they represent full-length mRNAs.
Figure 4.
Sequence analysis of the MPG1, MPG2, and MPG3 cDNAs and alignment of their deduced amino acid sequences. Asterisks and dots indicate identical and conserved amino acid residues, respectively, between the MPG1, MPG2, and MPG3 sequences. Arrows indicate predicted signal sequence cleavage sites. Potential Asn glycosylation sites (N-X-S/T) in each sequence are underlined.
Analysis of the deduced amino acid sequences revealed several structural features of interest (Fig. 4). MPG1, MPG2, and MPG3 encoded predicted mature proteins of 40, 43, and 47 kD, respectively, with basic pIs of 7.9, 8.5, and 7.5, respectively. All three contained an N-terminal hydrophobic signal sequence characteristic of proteins that are translocated into the lumen of the ER, the point of entry into the secretory system for proteins targeted to various cellular compartments, including the cell wall. Additionally, the three mature proteins contained potential N-glycosylation sites (N-X-S/T); one, located at amino acid position 259 to 261 in MPG1, is conserved between MPG1, MPG2, and many other plant PGs. There was a high level of conservation of Cys residues and short domains between the melon sequences and other plant PGs, suggesting that these regions may be critical to activity. A Gly-rich region in the carboxyl-half of the sequences (position 237–249 in MPG1) is highly conserved, and a His residue (position 241 in MPG1) found in this region is present in all PGs sequenced to date and has been ascribed a catalytic function (Scott-Craig et al., 1990; Caprari et al., 1996). In addition, a Tyr residue at amino acid position 310 in MPG1 is strictly conserved and has been shown to be essential for the activity of PG from Aspergillus spp. (Stratilova et al., 1996). The three deduced sequences differed in their N termini of the predicted mature (after cleavage of the signal peptide) protein. MPG3 encoded for an additional 47 amino acids and MPG2 for an additional 16 compared with MPG1. This region of MPG3 is reminiscent of the 47-amino acid prosequence that is present in tomato fruit PG and cleaved during transport through the secretory system (DellaPenna and Bennett, 1988). MPG1, MPG2, and MPG3 show differences at the level of amino acid identity as well. MPG3 was less than 20% identical to MPG1 and MPG2, but was 40% identical to tomato fruit PG. MPG1 and MPG2 shared 40% identity with each other, and were also similar to peach fruit and tomato leaf-abscission zone (TAPG1) PGs, sharing 60 and 48% identity, respectively.
A phylogram was generated using an alignment of the deduced amino acid sequences of MPG1, MPG2, and MPG3 and of 17 PG homologs from other plant and fungal species, described in Methods (Fig. 5). The phylogram groups the PGs into three major clades. PG gene family members from a single species segregate into different clades, suggesting that the structural divergence of plant PGs occurred prior to the divergence of the major angiosperm families. Clade A includes PGs that are expressed in fruit and/or abscission zones and includes the peach fruit-specific PG, tomato abscission zone-specific PGs, and MPG1 and MPG2. Clade B includes PGs that are expressed in fruit or dehiscence zones and includes the tomato fruit-specific and ripening-regulated PG and MPG3. Clade C is comprised primarily of PGs that are expressed in pollen or anthers, tissues in which exo-PG enzyme activity is prevalent, suggesting that the clade C PGs are likely to encode exo-PGs. When a phylogram was generated from an alignment of the region encoded by the partial-length melon PGs, pPG4 and pPG6 were placed in clade C and pPG5 was placed in clade A (data not shown), as expected based on their pattern of expression.
Figure 5.
Phylogram for 19 plant and 1 fungal PG cDNA and genomic clones. The phylogram was generated from the alignment of the full-length deduced amino acid sequences described in Methods and was created using the PAUP software package (Swofford et al., 1990). The PG sequences segregate into three major clades that we have designated A, B, and C.
Heterologous Expression of MPG1
The most abundant putative PG mRNA in ripening melon fruit was MPG1, whereas mRNA corresponding to the apparent homolog of the tomato fruit-ripening PG, MPG3, was expressed at much lower levels. To test whether MPG1 encoded a functional PG enzyme with the potential to catalyze pectin disassembly in ripening melon fruit, the cDNA was expressed in A. oryzae and the resulting enzyme was tested for PG activity (Fig. 6). Several strains of A. oryzae were tested for production of PG activity, and one strain (XMPG1) was selected for further characterization. The XMPG1 strain was shown to have the MPG1 cDNA integrated into the A. oryzae genome and was shown to accumulate full-length MPG1 transcript. Samples of the culture filtrate from XMPG1 or an untransformed control were collected over a 72-h culture period and assayed for PG activity using a gel-diffusion assay to determine the optimal growth period for heterologous protein production and to test for the presence of endogenous PGs expressed in A. oryzae.
There was no detectable PG activity in culture filtrates of the untransformed strain of A. oryzae under the growth conditions used, whereas strain XMPG1 produced high levels of PG activity that were detectable after 34 h of growth and accumulated to high levels throughout the culture period (Fig. 6A). To determine whether MPG1 encoded an endo- or exo-acting PG, the XMPG1 culture filtrate was further characterized for pectinase activity by both viscometric and reducing sugar assays (Fig. 6B). Compared with the untransformed control, the PG present in the culture filtrate of XMPG1 exponentially reduced the viscosity of a pectin solution but resulted in the linear production of reducing groups (Fig. 6B), indicating that MPG1 encodes an endo-PG with the potential to depolymerize pectin substrates in muro.
DISCUSSION
Charentais melon is a typical climacteric fruit, exhibiting an increase in respiration and autocatalytic ethylene production that accompanies ripening (Hadfield et al., 1995). During ripening there is a shift to a lower-molecular-mass distribution of hemicellulose polymers and a substantial solubilization and depolymerization of pectins, particularly the water-soluble pectins (Rose et al., 1998). Previous attempts to detect PG activity in ripening melon have been unsuccessful, and it has been suggested that ripening-associated pectin degradation is PG independent (Hobson, 1962; Lester and Dunlap, 1985; McCollum et al., 1989; Ranwala et al., 1992) and that pectin depolymerization may result primarily from the activity of β-galactosidase in melon (Ranwala et al., 1992). We present data here that support a role for PG in pectin depolymerization in melon.
Pectin-degrading enzyme activity was measured viscometrically in tissue extracts from all stages of fruit maturity. The highest level of activity was at R3 and correlates with data from cell wall analysis showing the greatest reduction in the molecular size of water-soluble pectins occurring at the same developmental time (Rose et al., 1998). The activity observed in the earlier time points was largely unexpected based on the cell wall analysis, but suggests that other constraints on PG activity are operative in vivo. In tomato there is ample evidence that PG-dependent pectin degradation may be limited by factors such as Ca2+ or substrate accessibility (Brady et al., 1987). Alternatively, the pectin-degrading activity observed in preripe fruit may have resulted from glycosidase activity. It has been previously reported that partially purified extracts of β-galactosidase from melon are able to cause a decrease in the molecular mass of pectins, although this decrease does not closely resemble that occurring endogenously during ripening (Ranwala et al., 1992).
Using sensitive molecular techniques we have identified three genes that are expressed abundantly during melon fruit ripening and that encode putative PGs. The similarity of the deduced amino acid sequences to other plant PGs and the temporal correlation between the appearance of their mRNAs, in vitro pectin-degrading activity, and the decrease in molecular mass of cell wall pectins suggests that the proteins encoded by these genes are PGs. Expression of the most abundant fruit-ripening PG, MPG1, in the heterologous host A. oryzae and analysis of culture filtrate from strain XMPG1 showed that MPG1 encodes an endo-PG and has the potential to depolymerize pectin in muro. The overlapping expression of MPG1, MPG2, and MPG3 suggests a potential cooperative role of these enzymes in degrading pectin, perhaps reflecting their mode of action as endo- or exo-PGs. A similar case exists in peach, in which three separate PG activities are present, two of which are exo- and one of which is endo-acting (Lester et al., 1994).
The patterns of hybridization shown on genomic Southern analysis indicate that the melon PG gene family is composed of members that are highly divergent. This has been shown in other species such as tomato, in which the fruit-ripening PG is encoded by a single gene that does not hybridize to any other genomic fragment, even at low stringency (DellaPenna et al., 1986). Similarly, in melon, MPG1 and MPG2 each hybridized strongly to a single genomic fragment, and MPG3 to a small number of genomic fragments at low and high stringency, indicating that they are encoded by single genes that are divergent from each other. (The presence of multiple fragments that hybridize to MPG3 at high stringency can be partially accounted for by restriction sites known to exist in the cDNA sequence.)
pPG5 hybridizes to a unique set of fragments at low stringency and to a subset of these fragments at high stringency, indicating that pPG5 belongs to a small subset of closely related PGs that does not include the fruit-ripening PGs. A similar result was reported for a PG gene expressed in tomato-abscission zones, which hybridizes to a set of genomic fragments distinct from that of the fruit-ripening PG (Kalaitzis et al., 1995). pPG4 and pPG6 hybridize to a common set of genomic restriction fragments at low stringency but to distinct subsets at high stringency, indicating that they are members of yet another subfamily of PG genes, this one possibly encoding PGs that are all expressed in pollen. It has been shown in maize that pollen PG is encoded by a small family of closely related genes that differ in nucleotide sequence by only 1% (Niogret et al., 1991).
Evolutionary relationships were inferred from a phylogenetic tree generated from an alignment of the deduced amino acid sequences of MPG1, MPG2, and MPG3, with one fungal homolog and 16 plant PG homologs. The PGs group into three major families, which we designated clades A, B, and C. The PGs in clade C are probably related in terms of function and tissue specificity because they are all found in pollen or anthers and are presumably exo-PGs. Exo-PGs do not, however, appear to group with clade C exclusively. A gene expressed during tomato seed germination that encodes a putative exo-PG is a member of clade B (B. Downie, Y. Sitrit, K.A. Hadfield, A.B. Bennett, and K.J. Bradford, personal communication).
The two most abundant ripening-induced PG genes in melon, MPG1 and MPG2, are members of clade A, and the least abundant, MPG3, is a member of clade B. A distinction based on mode of action or tissue specificity between members of clade A and B is not evident, but one feature that distinguishes clade B from the other two clades is the presence of a predicted prosequence immediately following the N-terminal hydrophobic signal sequence. In tomato fruit PG, the propeptide is cleaved from the mature protein during transport through the secretory system en route to the cell wall (DellaPenna and Bennett, 1988). The purpose of the prosequence is not known, but it has been hypothesized that it functions to keep proteins inactive as they travel to their ultimate destination in the cell, or that it is involved in subcellular localization. The same three clades were formed when a phylogenetic tree was generated using the 190-bp conserved region defined by the RT-PCR primers, indicating that the presence of the prosequence is not the basis of the divergence of the members of clade B. The members of clades A and B appear to share a common ancestor and are more closely related to each other than to members of clade C. It remains to be seen whether detailed analyses of a variety of PGs will eventually reveal a biochemical and/or tissue-specific basis for their divergence into three major clades.
As more sensitive techniques are used to detect PGs in tissues undergoing pectin degradation, it is becoming evident that PG is consistently associated with this process, not only during fruit maturation, but in other tissues where cell separation is taking place, such as abscission and pod-dehiscence zones, germinating pollen, and growing pollen tubes (Brown and Crouch, 1990; Taylor et al., 1990; Bonghi et al., 1992; Allen and Lonsdale, 1993; Kalaitzis et al., 1995, 1997; Petersen et al., 1996). Because of its high abundance, tomato-fruit PG is the most widely studied in relation to pectin disassembly. However, experiments with transgenic plants indicate that suppression of PG gene expression by 80% has little effect on pectin disassembly (Smith et al., 1990), suggesting that the enzyme is present in at least 5-fold excess. Low PG levels, such as those found in apple and melon, may be more typical in fruit undergoing ripening-associated pectin disassembly. In addition, PGs divergent from tomato-fruit PG, such as MPG1, may also be important in the ripening process of other fruit. Suppression of MPG1 gene expression in transgenic melons will provide a critical assessment of this possibility.
ACKNOWLEDGMENTS
We would like to thank Dr. John Labavitch for his careful reading of this manuscript and Aubrey Jones for providing A. oryzae protoplasts.
Abbreviations:
- IG
immature-green ripening stage
- MG
mature-green ripening stage
- PG
polygalacturonase
- R1 to R4
ripening stages 1 to 4
- RT
reverse transcriptase
Footnotes
This research was supported in part by a grant from Zeneca Plant Science, Jealotts' Hill, UK.
LITERATURE CITED
- Allen RL, Lonsdale DM. Molecular characterization of one of the maize polygalacturonase gene family members which are expressed during late pollen development. Plant J. 1993;3:261–271. doi: 10.1111/j.1365-313x.1993.tb00177.x. [DOI] [PubMed] [Google Scholar]
- Atkinson RG. A cDNA clone for endopolygalacturonase from apple. Plant Physiol. 1994;105:1437–1438. doi: 10.1104/pp.105.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson RG, Gardner RC. A polygalacturonase gene from kiwifruit (Actinidia deliciosa) Plant Physiol. 1993;103:669–670. doi: 10.1104/pp.103.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatzky R, Tanksley SD. Genetics of actin-related sequences in tomato. Theor Appl Genet. 1986;72:314–321. doi: 10.1007/BF00288567. [DOI] [PubMed] [Google Scholar]
- Bonghi C, Rascio N, Ramina A, Casadoro G. Cellulase and polygalacturonase involvement in the abscission of leaf and fruit explants of peach. Plant Mol Biol. 1992;20:839–848. doi: 10.1007/BF00027155. [DOI] [PubMed] [Google Scholar]
- Brady CJ. Fruit ripening. Annu Rev Plant Physiol. 1987;38:155–179. [Google Scholar]
- Brady CJ, McGlasson WB, Speirs J (1987) The biochemistry of fruit ripening. In D Nevins, R Jones, eds, Tomato Biotechnology. Alan R. Liss, New York, pp 279–288
- Brown SM, Crouch ML. Characterization of a gene family abundantly expressed in Oenothera organensis pollen that shows sequence similarity to polygalacturonase. Plant Cell. 1990;2:263–274. doi: 10.1105/tpc.2.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprari C, Mattei B, Basile ML, Salvi G, Crescenzi V, De Lorenzo G, Cervone F. Mutagenesis of endopolygalacturonase from Fusariummoniliforme: histidine residue 234 is critical for enzymatic and macerating activities and not for binding to polygalacturonase-inhibiting protein (PGIP) Mol Plant-Microbe Interact. 1996;9:617–624. doi: 10.1094/mpmi-9-0617. [DOI] [PubMed] [Google Scholar]
- Christensen T, Woeldike H, Boel E, Mortensen SB, Hjortshoej K, Thim L, Hansen MT. High level of expression of recombinant genes in Aspergillus oryzae. Biotechnology. 1988;6:1419–1422. [Google Scholar]
- Cove DJ. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta. 1966;113:51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- DellaPenna D, Alexander DC, Bennett AB. Molecular cloning of tomato fruit polygalacturonase: analysis of polygalacturonase mRNA levels during ripening. Proc Natl Acad Sci USA. 1986;83:6420–6424. doi: 10.1073/pnas.83.17.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D, Bennett AB. In vitro synthesis and processing of tomato fruit polygalacturonase. Plant Physiol. 1988;86:1057–1063. doi: 10.1104/pp.86.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer RL, Bennett AB. Role of cell wall hydrolases in fruit ripening. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:675–703. [Google Scholar]
- Giovannoni JJ, DellaPenna D, Bennett AB, Fischer RL. Expression of a chimeric polygalacturonase gene in transgenic rin (ripening inhibitor) tomato fruit results in polyuronide degradation but not fruit softening. Plant Cell. 1989;1:53–63. doi: 10.1105/tpc.1.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson D, Tucker GA, Keen J, Ray J, Bird CR, Schuch W. Sequencing and identification of a cDNA clone for tomato polygalacturonase. Nucleic Acids Res. 1986;14:8595–8601. doi: 10.1093/nar/14.21.8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross KC. A rapid and sensitive spectrophotometric method for assaying polygalacturonase using 2-cyanoacetamide. Hortscience. 1982;17:933–944. [Google Scholar]
- Hadfield KA, Rose JKC, Bennett AB. The respiratory climacteric is present in Charentais (Cucumis melo cv. Reticulatus F1 Alpha) melons ripened on or off the plant. J Exp Bot. 1995;46:1923–1925. [Google Scholar]
- Higgins DG, Bleasby AJ, Fuchs R. Clustal V. Improved software for multiple sequence alignment. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Hobson GE. Determination of polygalacturonase in fruits. Nature. 1962;195:804–805. doi: 10.1038/195804a0. [DOI] [PubMed] [Google Scholar]
- Jenkins ES, Paul W, Coupe SA, Bell SJ, Davies EC, Roberts JA. Characterization of an mRNA encoding a polygalacturonase expressed during pod development in oilseed rape (Brassica napus L.) J Exp Bot. 1996;47:111–115. [Google Scholar]
- John ME, Petersen MW. Cotton (Gossypium hirsutum L.) pollen-specific polygalacturonase mRNA: tissue and temporal specificity of its promoter in transgenic tobacco. Plant Mol Biol. 1994;26:1989–1993. doi: 10.1007/BF00019509. [DOI] [PubMed] [Google Scholar]
- Kalaitzis P, Koehler SM, Tucker ML. Cloning of a tomato polygalacturonase expressed in abscission. Plant Mol Biol. 1995;28:647–656. doi: 10.1007/BF00021190. [DOI] [PubMed] [Google Scholar]
- Kalaitzis P, Solomos T, Tucker ML. Three different polygalacturonases are expressed in tomato leaf and flower abscission, each with a different temporal expression pattern. Plant Physiol. 1997;113:1303–1308. doi: 10.1104/pp.113.4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer M, Sanders R, Bolkan H, Waters C, Sheehy RE, Hiatt WR. Postharvest evaluation of transgenic tomatoes with reduced levels of polygalacturonase: processing, firmness and disease resistance. Post Biol Tech. 1992;1:241–255. [Google Scholar]
- Kutsunai SY, Lin A-C, Percival FW, Laties GG, Christoffersen RE. Ripening-related polygalacturonase cDNA from avocado. Plant Physiol. 1993;103:289–290. doi: 10.1104/pp.103.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkins BA, Hurkman WJ. Synthesis and deposition of zein in protein bodies of maize endosperm. Plant Physiol. 1978;62:256–263. doi: 10.1104/pp.62.2.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Speirs J, Gray J, Brady CJ. Homologies to the tomato endopolygalacturonase in the peach genome. Plant Cell Environ. 1990;13:513–521. [Google Scholar]
- Lester DR, Speirs J, Orr G, Brady CJ. Peach (Prunus persica) endopolygalacturonase cDNA isolation and mRNA analysis in melting and nonmelting peach cultivars. Plant Physiol. 1994;105:225–231. doi: 10.1104/pp.105.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester GE, Dunlap JR. Physiological changes during development and ripening of ‘Perlita’ muskmelon fruits. Sci Hortic. 1985;26:323–331. [Google Scholar]
- McCollum TG, Huber DJ, Cantliffe DJ. Modification of polyuronides and hemicelluloses during muskmelon fruit softening. Physiol Plant. 1989;76:303–308. [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BA, Shuster JR, Rey MW, Yaver DS (1997) Improved enzyme expression in Aspergillus oryzae due to vector construction and chromosomal amdS deletion (abstract no. 101). 19th Fungal Genetics Conference, Asilomar, Pacific Grove, CA, March 18–23, 1997. Fungal Stock Center, Kansas City, KS
- Niogret M-F, Dubald M, Mandaron P, Mache R. Characterization of pollen polygalacturonase encoded by several cDNA clones in maize. Plant Mol Biol. 1991;17:1155–1164. doi: 10.1007/BF00028732. [DOI] [PubMed] [Google Scholar]
- Nogata Y, Ohta H, Voragen AGJ. Polygalacturonase in strawberry fruit. Phytochemistry. 1993;34:617–620. [Google Scholar]
- Petersen M, Sander L, Child R, Onekelen HV, Ulvskov P, Borkhardt B. Isolation and characterisation of a pod dehiscence zone-specific polygalacturonase from Brassica napus. Plant Mol Biol. 1996;31:517–527. doi: 10.1007/BF00042225. [DOI] [PubMed] [Google Scholar]
- Qiu X, Erickson L. A pollen-specific polygalacturonase-like cDNA from alfalfa. Sex Plant Rep. 1996;9:123–124. [Google Scholar]
- Ranwala AP, Suematsu C, Masuda H. The role of β-galactosidases in the modification of cell wall components during muskmelon fruit ripening. Plant Physiol. 1992;100:1318–1325. doi: 10.1104/pp.100.3.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert LS, Allard S, Gerster JL, Cass L, Simmonds J. Isolation and characterization of a polygalacturonase gene highly expressed in Brassica napus pollen. Plant Mol Biol. 1993;23:1273–1278. doi: 10.1007/BF00042360. [DOI] [PubMed] [Google Scholar]
- Rogers HJ, Allen RL, Hamilton WDO, Lonsdale DM. Pollen-specific cDNA clones from Zea mays. Biochim Biophys Acta. 1991;1089:411–413. doi: 10.1016/0167-4781(91)90188-r. [DOI] [PubMed] [Google Scholar]
- Rose JKC, Hadfield KA, Bennett AB. Temporal sequence of cell wall disassembly in rapidly ripening melon fruit. Plant Physiol. 1998;117:345–361. doi: 10.1104/pp.117.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuch W, Kanczler J, Robertson D, Hobson G, Tucker G, Grierson D, Bright S, Bird C. Fruit quality characteristics of transgenic tomato fruit with altered polygalacturonase activity. Hortscience. 1991;26:1517–1520. [Google Scholar]
- Scott-Craig JS, Panaccione DG, Cervone F, Walton JD. Endopolygalacturonase is not required for pathogenicity of Cochliobolus carbonum on maize. Plant Cell. 1990;2:1191–1200. doi: 10.1105/tpc.2.12.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy RE, Kramer M, Hiatt WR. Reduction of polygalacturonase activity in tomato fruit by antisense RNA. Proc Natl Acad Sci USA. 1988;85:8805–8809. doi: 10.1073/pnas.85.23.8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJS, Watson CF, Morris PC, Bird CR, Seymour GB, Gray JE, Arnold C, Tucker GA, Schuch W, Harding S and others. Inheritance and effect on ripening of antisense polygalacturonase genes in transgenic tomatoes. Plant Mol Biol. 1990;14:369–379. doi: 10.1007/BF00028773. [DOI] [PubMed] [Google Scholar]
- Smith CJS, Watson CF, Ray J, Bird CR, Morris PC, Schuch W, Grierson D. Antisense RNA inhibition of polygalacturo--nase gene expression in transgenic tomatoes. Nature. 1988;334:724–726. [Google Scholar]
- Stratilova E, Dzurova M, Markovic O, Jornvall H. An essential tyrosine residue of Aspergillus polygalacturonase. FEBS Lett. 1996;382:164–166. doi: 10.1016/0014-5793(96)00146-9. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP: Phylogenetic Analysis Using Parsimony Version 3.0. Champaign, IL: Illinois Natural History Survey; 1990. [Google Scholar]
- Taylor JE, Tucker GA, Lasslett Y, Smith CJS, Arnold CM, Watson CF, Schuch W, Grierson D, Roberts JA. Polygalacturonase expression during leaf abscission of normal and transgenic plants. Planta. 1990;183:133–138. doi: 10.1007/BF00197577. [DOI] [PubMed] [Google Scholar]
- Tebbutt SJ, Rogers HJ, Lonsdale DM. Characterization of a tobacco gene encoding a pollen-specific polygalacturonase. Plant Mol Biol. 1994;25:283–297. doi: 10.1007/BF00023244. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983;133:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- Wadsworth GJ, Redinbaugh MG, Scandalios JG. A procedure for the small scale isolation of plant RNA suitable for RNA blot analysis. Anal Biochem. 1988;172:279–283. doi: 10.1016/0003-2697(88)90443-5. [DOI] [PubMed] [Google Scholar]
- Wan C, Wilkins TA. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.) Anal Biochem. 1994;223:7–12. doi: 10.1006/abio.1994.1538. [DOI] [PubMed] [Google Scholar]
- Whitehead MP, Shieh MT, Cleveland TE, Cary JW, Dean RA. Isolation and characterization of polygalacturonase genes (peca and pecb) from Aspergillus flavus. Appl Environ Microbiol. 1995;61:3316–3322. doi: 10.1128/aem.61.9.3316-3322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Szakacs-Dobozi M, Hemmat M, Hrazdina G. Endopolygalacturonase in apples (Malus domestica) and its expression during fruit ripening. Plant Physiol. 1993;102:219–225. doi: 10.1104/pp.102.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaver DS, Xu F, Golightly EJ, Brown KM, Brown SH, Rey MW, Schneider P, Haliker T, Mondorf K, Dalboge H. Purification, characterization, molecular cloning, and expression of two laccase genes from the white rot basidiomycete Trametes villosa. Appl Environ Microbiol. 1996;62:834–841. doi: 10.1128/aem.62.3.834-841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]