Abstract
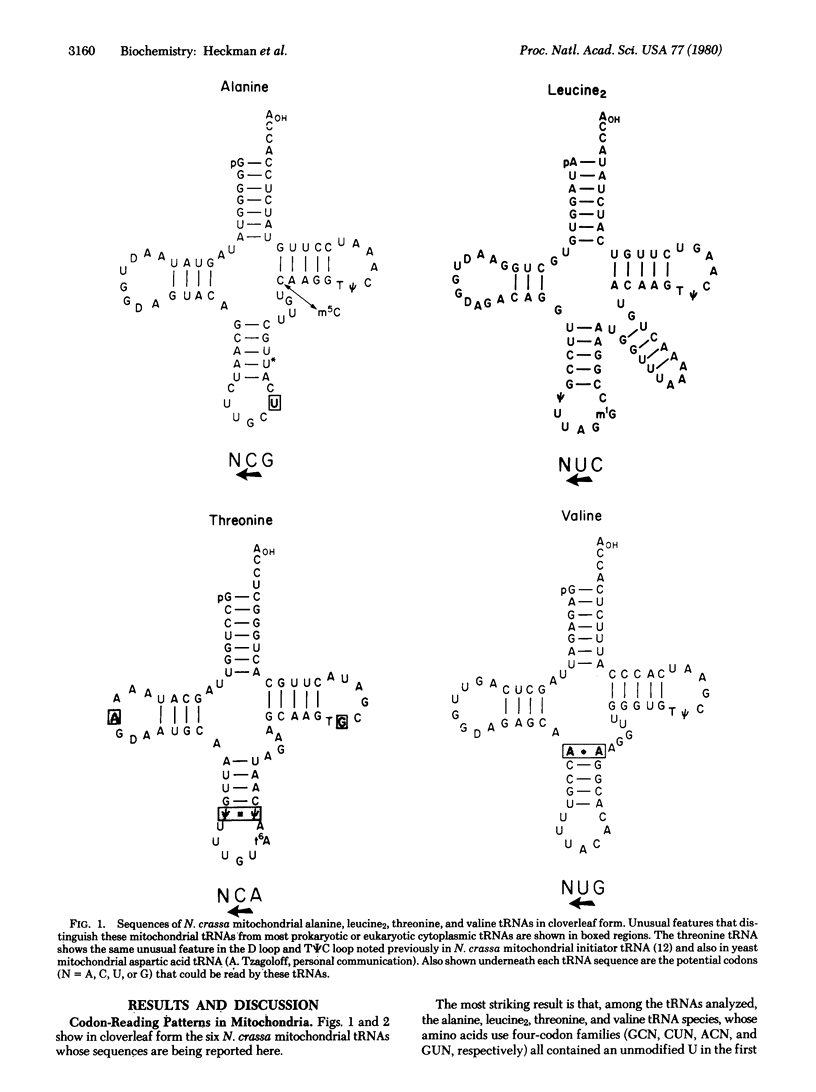
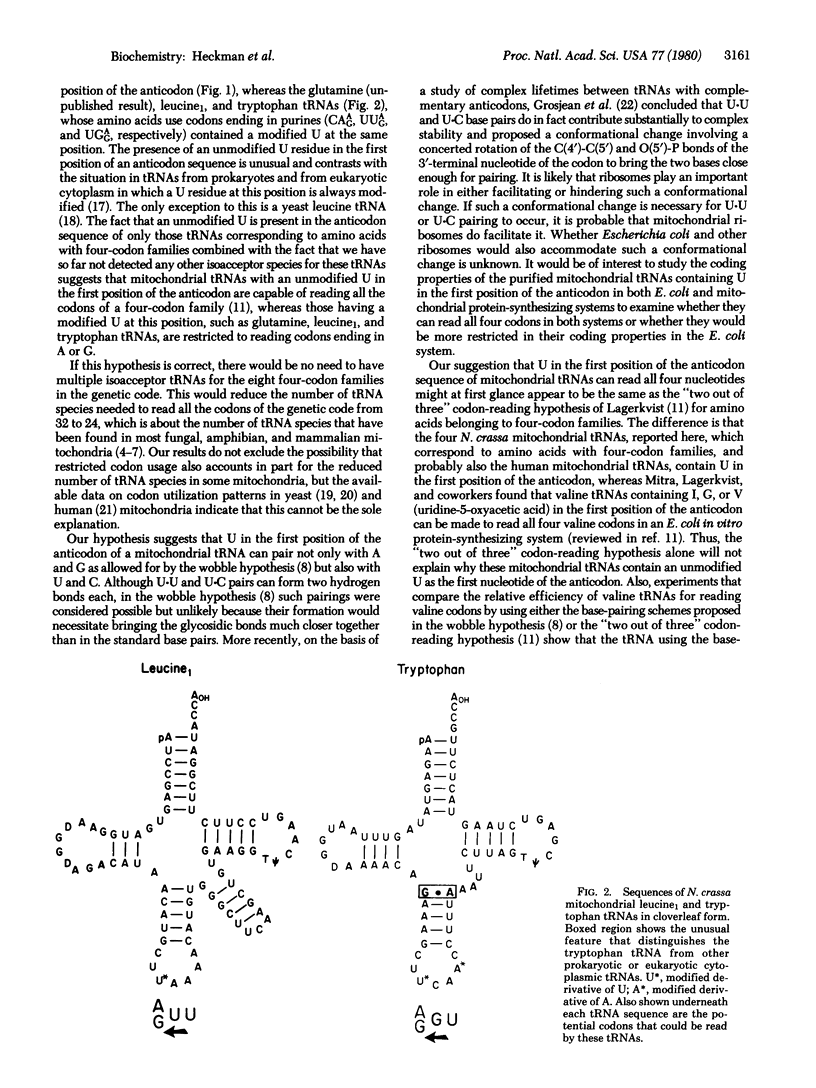
We report the sequences of Neurospora crassa mitochondrial alanine, leucine1, leucine2, threonine, tryptophan, and valine tRNAs. On the basis of the anticodon sequences of these tRNAs and of a glutamine tRNA, whose sequence analysis is nearly complete, we infer the following: (i) The N. crassa mitochondrial tRNA species for alanine, leucine2, threonine, and valine, amino acids that belong to four-codon families (GCN, CUN, ACN, and GUN, respectively; N = U, C, A, or G) all contain an unmodified U in the first position of the anticodon. In contrast, tRNA species for glutamine, leucine1, and tryptophan, amino acids that use codons ending in purines (CAGA, UUGA, and UGGA, respectively) contain a modified U derivative in the same position. These findings and the fact that we have not detected any other isoacceptor tRNAs for these amino acids suggest that N. crassa mitochondrial tRNAs containing U in the first position of the anticodon are capable of reading all four codons of a four-codon family whereas those containing a modified U are restricted to reading codons ending in A or G. Such an expanded codon-reading ability of certain mitochondrial tRNAs will explain how the mitochondrial protein-synthesizing system operates with a much lower number of tRNA species than do systems present in prokaryotes or in eukaryotic cytoplasm. (ii) The anticodon sequence of the N. crassa mitochondrial tryptophan tRNA is U*CA and not CCA or CmCA as is the case with tryptophan tRNAs from prokaryotes or from eukaryotic cytoplasm. Because a tRNA with U*CA in the anti-codon would be expected to read the codon UGA, as well as the normal tryptophan codon UGG, this suggests that in N. crassa mitochondria, as in yeast and in human mitochondria, UGA is a codon for tryptophan and not a signal for chain termination. (iii) The anticodon sequences of the two leucine tRNAs indicate that N. crassa mitochondria use both families of leucine codons (UUAG and CUN; N = U, C, A, or G) for leucine, in contrast to yeast mitochondria [Li, M. & Tzagoloff, A. (1979) Cell 18, 47-53] in which the CUA leucine codon and possibly the entire CUN family of leucine codons may be translated as threonine.
Keywords: UGA codon, mitochondrial evolution, codon-reading patterns
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angerer L., Davidson N., Murphy W., Lynch D., Attardi G. An electron microscope study of the relative positions of the 4S and ribosomal RNA genes in HeLa cells mitochondrial DNA. Cell. 1976 Sep;9(1):81–90. doi: 10.1016/0092-8674(76)90054-4. [DOI] [PubMed] [Google Scholar]
- Barrell B. G., Anderson S., Bankier A. T., de Bruijn M. H., Chen E., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A. Different pattern of codon recognition by mammalian mitochondrial tRNAs. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3164–3166. doi: 10.1073/pnas.77.6.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell B. G., Bankier A. T., Drouin J. A different genetic code in human mitochondria. Nature. 1979 Nov 8;282(5735):189–194. doi: 10.1038/282189a0. [DOI] [PubMed] [Google Scholar]
- Bonitz S. G., Berlani R., Coruzzi G., Li M., Macino G., Nobrega F. G., Nobrega M. P., Thalenfeld B. E., Tzagoloff A. Codon recognition rules in yeast mitochondria. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3167–3170. doi: 10.1073/pnas.77.6.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst P., Grivell L. A. The mitochondrial genome of yeast. Cell. 1978 Nov;15(3):705–723. doi: 10.1016/0092-8674(78)90257-x. [DOI] [PubMed] [Google Scholar]
- Chiu N., Chiu A., Suyama Y. Native and imported transfer RNA in mitochondria. J Mol Biol. 1975 Nov 25;99(1):37–50. doi: 10.1016/s0022-2836(75)80157-4. [DOI] [PubMed] [Google Scholar]
- Coruzzi G., Tzagoloff A. Assembly of the mitochondrial membrane system. DNA sequence of subunit 2 of yeast cytochrome oxidase. J Biol Chem. 1979 Sep 25;254(18):9324–9330. [PubMed] [Google Scholar]
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Fox T. D. Five TGA "stop" codons occur within the translated sequence of the yeast mitochondrial gene for cytochrome c oxidase subunit II. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6534–6538. doi: 10.1073/pnas.76.12.6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H. J., de Henau S., Crothers D. M. On the physical basis for ambiguity in genetic coding interactions. Proc Natl Acad Sci U S A. 1978 Feb;75(2):610–614. doi: 10.1073/pnas.75.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman J. E., Alzner-Deweerd B., RajBhandary U. L. Interesting and unusual features in the sequence of Neurospora crassa mitochondrial tyrosine transfer RNA. Proc Natl Acad Sci U S A. 1979 Feb;76(2):717–721. doi: 10.1073/pnas.76.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman J. E., Hecker L. I., Schwartzbach S. D., Barnett W. E., Baumstark B., RajBhandary U. L. Structure and function of initiator methionine tRNA from the mitochondria of Neurospora crassa. Cell. 1978 Jan;13(1):83–95. doi: 10.1016/0092-8674(78)90140-x. [DOI] [PubMed] [Google Scholar]
- Heckman J. E., RajBhandary U. L. Organization of tRNA and rRNA genes in N. crassa mitochondria: intervening sequence in the large rRNA gene and strand distribution of the RNA genes. Cell. 1979 Jul;17(3):583–595. doi: 10.1016/0092-8674(79)90266-6. [DOI] [PubMed] [Google Scholar]
- Heckman J. E., Yin S., Alzner-DeWeerd B., RajBhandary U. L. Mapping and cloning of Neurospora crassa mitochondrial transfer RNA genes. J Biol Chem. 1979 Dec 25;254(24):12694–12700. [PubMed] [Google Scholar]
- Hensgens L. A., Grivell L. A., Borst P., Bos J. L. Nucleotide sequence of the mitochondrial structural gene for subunit 9 of yeast ATPase complex. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1663–1667. doi: 10.1073/pnas.76.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes T. H. Possibilities for the evolution of the genetic code from a preceding form. Nature. 1973 Nov 2;246(5427):22–26. doi: 10.1038/246022a0. [DOI] [PubMed] [Google Scholar]
- Lagerkvist U. "Two out of three": an alternative method for codon reading. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1759–1762. doi: 10.1073/pnas.75.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Tzagoloff A. Assembly of the mitochondrial membrane system: sequences of yeast mitochondrial valine and an unusual threonine tRNA gene. Cell. 1979 Sep;18(1):47–53. doi: 10.1016/0092-8674(79)90352-0. [DOI] [PubMed] [Google Scholar]
- Macino G., Coruzzi G., Nobrega F. G., Li M., Tzagoloff A. Use of the UGA terminator as a tryptophan codon in yeast mitochondria. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3784–3785. doi: 10.1073/pnas.76.8.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macino G., Tzagoloff A. Assembly of the mitochondrial membrane system: partial sequence of a mitochondrial ATPase gene in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1979 Jan;76(1):131–135. doi: 10.1073/pnas.76.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N. C., rabinowitz M. Mitochondrial transfer RNAs in yeast: identification of isoaccepting transfer RNAs. Biochemistry. 1978 May 2;17(9):1628–1634. doi: 10.1021/bi00602a008. [DOI] [PubMed] [Google Scholar]
- Randerath K., Chia L. S., Gupta R. C., Randerath E. Structural analysis of nonradioactive RNA by postlabeling: the primary structure of baker's yeast tRNA Leu/CUA. Biochem Biophys Res Commun. 1975 Mar 3;63(1):157–163. doi: 10.1016/s0006-291x(75)80024-6. [DOI] [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. Use of in vitro 32P labeling in the sequence analysis of nonradioactive tRNAs. Methods Enzymol. 1979;59:58–109. doi: 10.1016/0076-6879(79)59072-7. [DOI] [PubMed] [Google Scholar]
- Simsek M., Ziegenmeyer J., Heckman J., Rajbhandary U. L. Absence of the sequence G-T-psi-C-G(A)- in several eukaryotic cytoplasmic initiator transfer RNAs. Proc Natl Acad Sci U S A. 1973 Apr;70(4):1041–1045. doi: 10.1073/pnas.70.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Grueter F., Spelzhaus A., Gauss D. H. Compilation of tRNA sequences. Nucleic Acids Res. 1980 Jan 11;8(1):r1–r22. [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Macino G., Sebald W. Mitochondrial genes and translation products. Annu Rev Biochem. 1979;48:419–441. doi: 10.1146/annurev.bi.48.070179.002223. [DOI] [PubMed] [Google Scholar]








