Abstract
Since its discovery in 1963, poly(ADP-ribose) (pADPr) has been shown to play important functions in the nucleus of multicellular eukaryotes. Each of these functions centers upon DNA metabolism, including DNA-damage repair, chromatin remodeling, transcription and telomere functions. We recently described two novel functions for pADPr in the cytoplasm, both of which involve RNA metabolism – 1) the assembly of cytoplasmic stress granules, cellular macrostructures that aggregate translationally stalled mRNA/protein complexes, and 2) modulation of microRNA activities. Multiple stress granule-localized, post-transcriptional gene regulators, including microRNA-binding argonaute family members, are substrates for pADPr modification and are increasingly modified by pADPr upon stress. Interestingly, the cytoplasmic RNA regulatory functions for PARPs are likely mediated through activities of catalytically inactive PARP-13/ARTD13/ZC3HAV1/ZAP and mono/poly(ADP-ribose)-synthesizing enzymes, including PARP-5a/ARTD5/TNKS1, PARP-12/ARTD12/ZC3HDC1 and PARP-15/ARTD7/BAL3. These data are consistent with other recent work, which suggests that mono(ADP-ribosyl)ated residues can be poly(ADP-ribosyl)ated by different enzymes.
Keywords: microRNA, PARG, PARP, PARP-13, stress, stress granule, ZAP
Introduction
Poly(ADP-ribose) (pADPr) is a macromolecular polymer consisting of 2–200 ADP-ribose subunits organized in linear or branched chains.1,2 It is a post-translational modification whose function is best understood in the context of the nucleus, where its synthesis is highly upregulated during DNA damage.3 The polymer acts as a scaffold to recruit repair factors to the sites of damage and helps relax chromatin structure to facilitate DNA repair.4 It was later recognized that pADPr has additional nuclear functions during non-stress conditions where it is involved in transcriptional regulation, chromatin remodeling and telomere functions.3
The poly(ADP-ribose) polymerase (PARP) family of proteins uses NAD+ as substrate to synthesize ADPr modifications onto acceptor proteins.1,2,5,6 PARP-1, which functions during DNA damage, is the founding member of the family. Using the PARP-1 catalytic domain, bioinformatics analyses identified 16 additional PARPs in the human genome, many with unknown function.5-7 A new nomenclature for this PARP family was thus recently proposed (Table 1),5 but their conventional names are used here for the sake of familiarity. A detailed examination of their conserved catalytic residues revealed that 11 out of 17 PARPs lack one ore more of three critical residues—histidine, tyrosine and glutamate (or HYE triad)—required for pADPr polymerization activities. Two PARPs, PARP-9 and -13, lack the histidine and glutamate, which are important for binding NAD+ and pADPr elongation, respectively, and were thus predicted to be catalytically inactive.8 On the other hand, the remaining 9 PARPs are predicted to transfer single ADP-ribose units, i.e., mono(ADP-ribose) or mADPr, onto acceptor proteins.8 Such mono(ADP-ribosyl)ating activities were confirmed recently for PARP-10, PARP-12, PARP-14 and PARP-15.8,9
Table 1. PARP family. ‘+’ denotes stress granule localization determined by antibody staining against endogenous proteins and GFP-tagging. The antibody staining of PARP-14 has yet to be verified by GFP-tagging.
| Alternative Names | Stress Granule Localization | |
|---|---|---|
|
PARP-1 |
ARTD1 |
|
|
PARP-2 |
ARTD2 |
|
|
PARP-3 |
ARTD3 |
|
|
PARP-4 |
ARTD4, vPARP |
|
|
PARP-5a |
ARTD5, TNKS1 |
+ |
|
PARP-5b |
ARTD6,TNKS2 |
|
|
PARP-6 |
ARTD17 |
|
|
PARP-7 |
ARTD14, tiPARP |
|
|
PARP-8 |
ARTD16 |
|
|
PARP-9 |
ARTD9, BAL1 |
|
|
PARP-10 |
ARTD10 |
|
|
PARP-11 |
ARTD11 |
|
|
PARP-12 |
ARTD12, ZC3HDC1 |
+ |
|
PARP-13.1 |
ARTD13, ZC3HAV1 |
+ |
|
PARP-13.2 |
ARTD13, ZC3HAV1, ZAP |
+ |
|
PARP-14 |
ARTD8, BAL2 |
(+) |
|
PARP-15 |
ARTD7, BAL3 |
+ |
| PARP-16 | ARTD15 |
The presence of PARPs with distinct catalytic activities—synthesis of mADPr or pADPr—suggests new possibilities. First, the effect of mADPr modification of an acceptor is likely quite different from addition of a long pADPr chain. Second, PARPs of distinct activities could potentially cooperate in order to modify a common acceptor protein.8,9 The physical association of mADPr- and pADPr-synthesizing enzymes was demonstrated by nuclear-localized PARP-1 and PARP-3,10 and, from our recent work, PARP-5a and PARP-12 in the cytoplasm.9 These observations suggest a novel mechanism of pADPr synthesis in which the initiation and elongation steps can be catalyzed by different PARPs.
The possibility that poly(ADP-ribosyl)ation can be mediated by multiple enzymes is further supported by in vitro and “in-cell” experiments. For example, an existing single ADP-ribose could be poly(ADP-ribosyl)ated by incubating with NAD+ and bacterially expressed PARP-1 in vitro, where the initial and elongation steps were separated. In these cases, the initial ADP-ribose could be derived from the PARP-1 E988K mutant that can only auto-mono(ADP-ribosyl)-ate itself, or the ADP-ribose could be chemically synthesized and conjugated to peptides or agarose beads.10-12 Notably, mono(ADP-ribosyl)ated residues can be derived in cells from another family of NAD+-consuming enzymes known as sirtuins.13 This family is exemplified by the yeast member Sir2p, which catalyzes the removal of acetyl groups from histones and is involved in gene silencing, chromosomal stability and aging. However, quite a few members in yeast, Trypanosomes and mammals have also demonstrated mono(ADP-ribosyl)ating activities.14 For example, yeast Sir2p can catalyze the transfer of ADP-ribose to itself and histones. In mammals, two substrates were identified to date—SIRT4 ribosylates glutamate dehydrogenase to suppress insulin signaling in pancreatic β cells and, recently, SIRT6 mono(ADP-ribosyl)ates PARP-1, which then promotes PARP-1 automodification with further ADP-ribose units, upon DNA damage.15 This latter observation suggests that not only the two NAD+-dependent signaling pathways can crosstalk and cooperate, but also highlights the possibility that mono(ADP-ribosyl)ated residues can be poly(ADP-ribosyl)ated by different enzymes in cells. Similar sequential modification was also proposed for other macromolecular post-translational modifications, such as poly(ubiquitin)ation.16 Potentially, there are several advantages conferred by this type of conserved mechanism in post-translationally modifying substrates sequentially using different enzymes—(1) define specificity of substrates to poise for poly(ADP-ribosyl)-ation only at certain cellular conditions; (2) increase substrate diversity given a limited repertoire of enzymes; or (3) serve as a safeguard/crosstalk mechanism by adding an extra layer of regulation. So far, the functions of mono(ADP-ribosyl)-ation conferred by these subclasses of PARPs and SIRTs remain unclear. Given that there are ~1000 fold more amino acid residues that are mono(ADP-ribosyl)-ated than being poly(ADP-ribosyl)ated (as estimated in the rat liver),17 it is formally possible that these mono(ADP-ribosyl)-ated residues could serve as unique signals or, alternatively, as starting points for poly(ADP-ribosyl)ation.
PARP functions beyond the Nucleus
As more was discovered about the biology of these recently identified PARPs, it became clear that pADPr does have cellular functions beyond the nucleus. Most PARPs exhibit cytoplasmic localizations,18-22 yet relatively little is known about the function of pADPr in the cytoplasm. Early works by the founders of the field identified PARP activities in cytoplasmic, post-mitochondrial fractions.23 Such activities accounted for ~25% of total PARP activities in the cell. These cytoplasmic PARP activities were identified in multiple mammalian cell types (HeLa, macrophages, erythroblasts, plasma cells and neurons) and tissues (brain and liver extracts) suggesting that the modification is ubiquitous.24,25 Further functions for cytoplasmic pADPr were suggested by the activities of its degradative enzyme, pADPr glycohydrolase (PARG). More than 50% of total PARG activities are concentrated in the cytosol,23 suggesting that pADPr synthesis and function in the cytoplasm are tightly regulated. Hints regarding the functions of cytoplasmic pADPr came from an early observation of its activities enriched in free mRNP fractions in sucrose gradients. Such fractions were enriched in factors that regulate translation potential and decay of mRNAs.24,26-28 Such correlation led to the hypothesis that pADPr may be involved in post-transcriptional mRNA regulation. This hypothesis was further supported by the recent discovery of two PARPs, PARP-12 and -13, that localize to the cytoplasm and contain CCCH type zinc finger RNA-binding domains.9,29
While the biological function of PARP-12 remains poorly understood, PARP-13 is known for its roles in binding and regulating viral RNA transcripts.29-31 It was initially identified as Zinc finger Anti-viral Protein (ZAP) in a screen for host factors that confer resistance to the retrovirus moloney murine leukemia virus infection,29 and its anti-viral activities were later confirmed in other retroviruses such as HIV-1, or other RNA virus families, including Alphaviruses and Filoviruses.30-32 PARP-13 binding to viral RNAs correlates with their subsequent degradation and the inability of the virus to replicate efficiently.29 PARP-13 co-immunoprecipitates with two RNA helicases (p72 and DHX30) and the exosome complex, thought to be responsible for viral RNA unwinding and degradation, respectively.33-35 Knockdown of specific exosome components decreased the anti-viral activities of PARP-13, suggesting that PARP-13 mediates viral RNA degradation via an exosome dependent pathway.33-35
Moreover, PARP-13 has two isoforms; the shorter PARP-13.2 isoform has recently been shown to directly bind to RIG-I, a key factor of innate immune response, after viral infection.36 RIG-I itself is an RNA helicase and associates with viral RNAs. The PARP-13.2 binding to RIG-1 activates the type-I interferon signaling. As both RIG-1 and PARP-13.2 bind viral RNAs, such PARP-13.2-to-RIG-1 association stabilizes their binding to viral RNAs,36 which might further strengthen the anti-viral signaling pathway.
However, these anti-viral properties are not due to the ADP-ribosylating activities of PARP-13 because PARP-13 proteins are catalytically inactive, either lacking a PARP domain (PARP-13.2 isoform), or the critical HYE triad residues (PARP-13.1 contains YYV),5,8 where their lack of activities were confirmed by in vitro ADP-ribosylating assays.8,9 Yet the longer isoform, PARP-13.1, which contains the “catalytically inactive” PARP-like domain, confers stronger anti-viral activities.37 Despite the lack of catalytic activities, PARP-13 function may require ADPr modification by other PARPs instead, and we found that the modifications of these two isoforms change upon stress differently – pADPr modification of PARP-13.2 increases whereas PARP-13.1 remains unchanged.9 Such trans-modification is common among PARPs, including modification between PARP-1 and PARP-255 and between PARP-5a and PARP-5b.38 Thus, it is possible that trans-modification of PARP-13 by other cytoplasmic PARPs is important for mediating two novel roles of pADPr in the cytoplasm (Fig. 1): assembly of stress granules and regulating microRNA activities.9
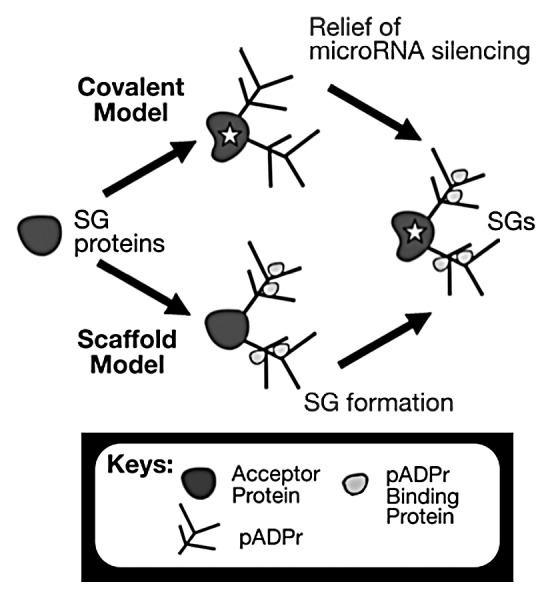
Figure 1. Two novel functions for pADPr in the cytoplasm. The covalent model indicates the alteration of protein properties inherently (highlighted with a star in the figure) whereas the caffold model indicates that the change of protein function relies on new non-covalent protein associations through pADPr.
Multiple cytoplasmic PARPs regulate SG assembly
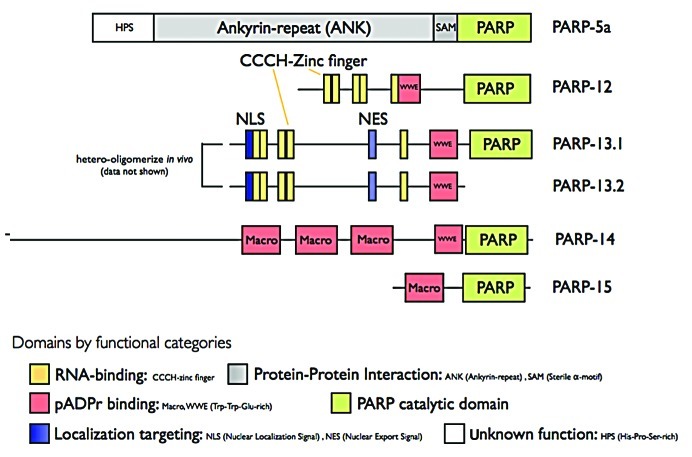
Cells respond to physiological stresses, such as heat shock, oxidative stress, ischemia and viral infection by assembling large multi-protein complexes in the cytoplasm called stress granules (SGs).39 SGs are found in many pathological conditions, including hypoxic tumor cores40 and neurons in animal models of ischemia stroke41 and can be induced by anti-cancer agents, such as arsenite42 and pateamine A.43 SGs contain poly(A)+ mRNAs, stalled translation initiation complexes and multiple RNA-binding proteins, and are thought to regulate the stability and translation potential of mRNAs. We have recently shown that SGs also comprise pADPr, 2 PARG isoforms (PARG99 and PARG102) and 5 PARPs (PARP-5a, -12, -13.1, -13.2, and -15; localization of PARP-14 was also identified by antibody staining but yet to be confirmed by GFP tagging; Table 1 and Fig. 2).9 Overexpression of these PARPs resulted in the de novo nucleation of SGs, identifying them as core SG components. In contrast, overexpression of the PARG isoforms result in inhibition of SG assembly and knockdown of PARG delays disassembly of SG.9 Together, these data suggest that pADPr concentration in the cytoplasm is locally regulated for the assembly and maintenance of SG structure.
Figure 2. Domain structure of PARPs localized in SGs.
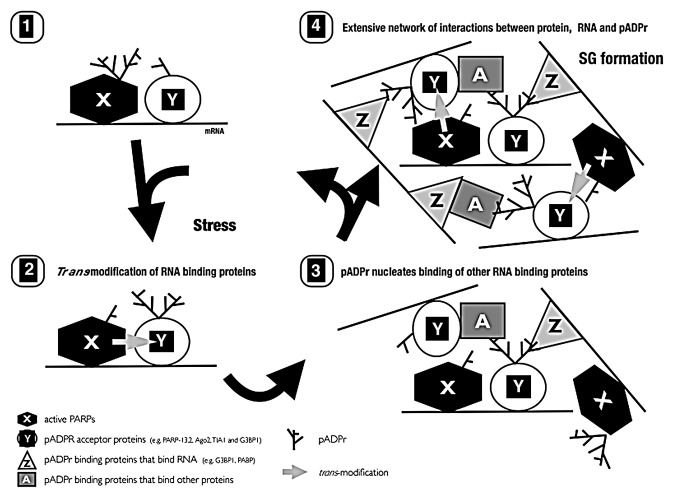
How does pADPr mediate the assembly and disassembly of the mRNP-enriched SG? We propose that pADPr functions as a scaffold to bridge diverse mRNA/protein complexes together. Such scaffolding properties of pADPr have also been observed in DNA repair complexes,4 the mitotic spindle44,45 and another RNA organelle Cajal bodies.46 To scaffold, two classes of proteins appear to be required: proteins that are covalently modified by pADPr and proteins that bind to pADPr non-covalently. During stress, select sets of RNA-binding SG components, AGO1–4, TIA-1, G3BP1 and PARP-13.1/2 complex are increasingly modified by pADPr.9 On the other hand, poly(A)-binding protein PABP and heterogeneous ribonucleoprotein hnRNP A1 both bind to but are not modified by pADPr.47 In addition, a subset of proteins such as the endoribonuclease G3BP1, both bind to and are modified by pADPr.9,47 In assembling SGs, proteins that are modified by pADPr become cross-linked to pADPr binding proteins via protein-pADPr binding interactions. The increasing chains of pADPr emanating from modified RNA-binding proteins then recruit additional RNA binding proteins via non-covalent pADPr-protein interactions. Thus pADPr could function as a focal nucleator of SGs through multiple cycles of modification, recruitment and cross-linking (Fig. 3). Consistent with this hypothesis, previously reported dominant negative mutants of G3BP148 and TIA-149 that disrupt SG assembly are not modified by pADPr in stressed cells.9
Figure 3. Proposed model of SG assembly.
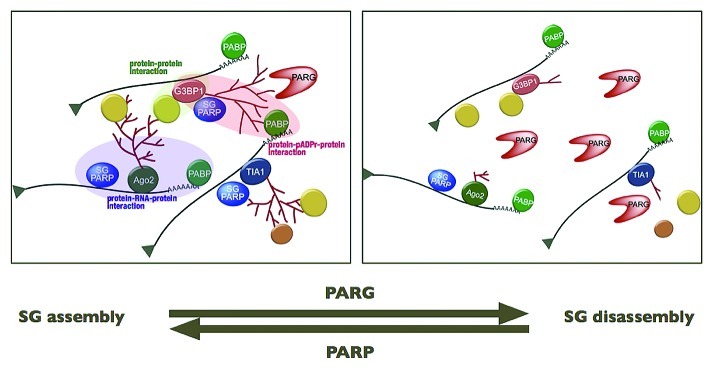
In our working model, SGs are a physical manifestation of the extensive networks formed by interactions between pADPr modification on RNA-binding proteins, proteins that bind pADPr and proteins that bind to RNA. In fact, those PARPs localized in SGs, including the CCCH zinc finger PARPs, could initiate part of this interaction network through their various binding domains for protein, RNA and pADPr (Fig. 2). The pADPr–SG scaffold appears to be highly dynamic and, depending on cell stress state, disassembled or assembled by regulating PARP/PARG activities. These opposing enzymatic activities could thus provide a rapid, regulatory mechanism for SG assembly or disassembly, while keeping individual RNA/protein complexes and their functions intact (Fig. 4). The preservation of RNA/protein complex function could be critical to a rapid cellular recovery from stress. Furthermore, one notable characteristic of pADPr, distinctive from other macromolecular post-translational modifications, such as ubiquitin and SUMO, is its non-proteinaceous nature, which theoretically renders the pADPr scaffold resistant to the activities of proteases, such as caspases, during stress.
Figure 4. Proposed model of SG disassembly.
PARP-13 together with other PARPs regulate microRNA silencing
In addition to a function in the assembly of macrostructures, pADPr also plays important roles in regulating mRNA/protein interactions at the molecular scale. To better understand this role, we focused on examining pADPr modification of AGO2 and its effect on microRNA silencing.9 This was due to its pervasive roles in regulating > 60% of mammalian mRNAs.50 AGO2 modification is mediated by multiple PARPs that bind to AGO2, each of which exhibits distinct catalytic activities: PARP-13 is catalytically inactive, PARP-5a has demonstrated poly(ADP-ribosyl)ating activities and PARP-12 has mono(ADP-ribosyl)ating activities. We also observed that other family members of argonaute (AGO1–4) are modified by pADPr. Not surprisingly, AGO2 immunoprecipitates exhibited ADP-ribosylating activities upon NAD+ addition. Addition of the general PARP inhibitor 3-aminobenzamide, which can potentially bind to PARP domains of all catalytically active PARPs, completely abrogated ADPr synthesis in the AGO2 immunoprecipitates. The pattern of AGO2 modification and the concentration-dependent incorporation of NAD+ confirmed that the AGO2 precipitates contain pADPr synthesizing activities and that it is directly modified by mADPr and/or pADPr. However, it remains to be investigated whether the poly(ADP-ribosyl)ation is mediated in a sequential manner – as in the case of SIRT-6 mediated poly(ADP-ribosyl)-ation of PARP-115 – that is, first by the mono(ADP-ribosyl)ating PARP-12, followed by poly(ADP-ribosyl)ating PARP-5a, or directly through PARP-5a alone.
What is the effect of ADPr modification on argonaute activities? The amount of pADPr modification of argonaute was inversely correlated with microRNA silencing. Upon stress, argonaute modification by pADPr increased, and microRNA silencing was relieved. Notably, under the same conditions, the global translation rate was reduced; however the expression of microRNA targets decreased to a lesser extent than other mRNAs, consistent with selective regulation of argonaute activities. Similarly, we observed an increase in pADPr modification of argonaute and a decrease in microRNA silencing when PARG was knocked down, consistent with a function for pADPr modification of argonaute in regulating microRNA activities.
How is argonaute modification by pADPr regulated? We are just beginning to study this aspect of microRNA/ADPr function, however critical data has emerged. Poly(ADP-ribosyl)ation of AGO2 is dependent on its ability to bind RNA. AGO2 mutants lacking the mRNA binding PIWI domain are unable to be poly(ADP-ribosyl)ated. Interestingly, a similar phenomenon was observed for other post-transcriptional regulators TIA-1 and G3BP1, where their respective mutants lacking RNA-binding motifs were also not poly(ADP-ribosyl)-ated. These data suggest that either RNA-binding is required for modification, or that the RNA-binding domain is the site of pADPr modification.
PARP-13 appears to be critical for microRNA silencing. Upon stress, modification of argonaute family members and PARP-13.1/2 complex by pADPr significantly increases. Overexpression of the catalytically inactive PARP-13.1 or PARP-13.2 relieves microRNA silencing. While the mechanism of PARP-13 function in AGO2 regulation is not currently clear, one possibility is that the poly(ADP-ribosyl)ation of argonaute is mediated by catalytically active PARPs (PARP-5a and PARP-12) via PARP-13.1/2. In fact, all of these PARPs bind to PARP-13 family members. This is reminiscent of the activation of tyrosine kinase pathway where erbB-3, which, though itself has no active kinase domain, can mediate signaling through heterodimerization with active EGF family kinases like Her2.51 Consistent with this, AGO2 associates with PARP-13 in an RNA-dependent manner. Perhaps, because of their ability to bind mRNA, the function of the inactive PARP-13 isoforms is to anchor the activities of the catalytically active PARPs to the mRNP complex. This might partially explain why the mRNA-binding domain of AGO2 is required for its poly(ADP-ribosyl)ation.
Based on these observations, we propose that the relief of microRNA silencing occurs due to a decrease in the accessibility of the argonaute/microRNA complex to target mRNAs. This decrease likely results from the increased pADPr modification of proteins within the complex, such as those microRNA-binding argonaute family members and PARP-13. Such modification could change the protein structure of target proteins resulting in altered accessibility to mRNA. Alternatively, considering that one ADP-ribose unit is ~0.5kDa in size (equivalent to ~5 amino acids) and has two negatively-charged phosphate groups, increased concentrations of pADPr near the RNA binding site in the argonaute/microRNA complex could disrupt electrostatic interactions or present steric hindrances between argonaute/microRNA and mRNA target. Finally, the pADPr might provide a scaffold for recruiting repressor(s) of microRNA activities to target mRNAs.
Conclusion
In summary, we propose that pADPr functions as a general mediator of stress and can specifically regulate post-transcriptional gene expression in the cytoplasm: stress granule assembly and microRNA activities. These results further extend its well-characterized DNA metabolism roles mediated by PARP-1 in the nucleus. Small molecule inhibitors targeted against PARP-1, which mediates DNA damage repair in the nucleus, are already in phase II/III human trials for cancer therapy.52 These inhibitors have also shown promise for treatment of inflammation, ischemia, and degenerative vascular diseases.53,54 Our results suggest that these therapeutic effects may be due to partial inhibition of PARPs in the cytoplasm. (Note added in proof) Wahlberg et al. (2012)56 recently profiled 185 existing PARP inhibitors and found that some existing PARP-1 inhibitors can also bind catalytic domains of other PARPs with comparable affinity. Therefore, designing specific inhibitors to PARPs may potentially result in highly effective treatments for these stress-related diseases.
Acknowledgments
P.C. is a Rita Allen Foundation Scholar and a Kimmel Foundation for Cancer Research Scholar. This work is supported by NIH Award R01-GM087465–01A2 to P.C and DOD Idea Award BC101881 to A.K.L.L. We thank Phillip A. Sharp for his insights on the project and comments on this manuscript.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/19899
References
- 1.Schreiber V, Dantzer F, Ame J-C, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–28. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 2.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–82. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 3.Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahel D, Horejsí Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–3. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35:208–19. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Amé J-C, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–93. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 7.Otto H, Reche PA, Bazan F, Dittmar K, Haag F, Koch-Nolte F. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs) BMC Genomics. 2005;6:139. doi: 10.1186/1471-2164-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, et al. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Leung AKL, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell. 2011;42:489–99. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loseva O, Jemth A-S, Bryant HE, Schüler H, Lehtiö L, Karlberg T, et al. PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J Biol Chem. 2010;285:8054–60. doi: 10.1074/jbc.M109.077834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moyle PM, Muir TW. Method for the synthesis of mono-ADP-ribose conjugated peptides. J Am Chem Soc. 2010;132:15878–80. doi: 10.1021/ja1064312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panzeter PL, Zweifel B, Althaus FR. Synthesis of poly(ADP-ribose)-agarose beads: an affinity resin for studying (ADP-ribose)n-protein interactions. Anal Biochem. 1992;207:157–62. doi: 10.1016/0003-2697(92)90517-B. [DOI] [PubMed] [Google Scholar]
- 13.Finkel T, Deng C-X, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–91. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Girolamo M, Dani N, Stilla A, Corda D. Physiological relevance of the endogenous mono(ADP-ribosyl)ation of cellular proteins. FEBS J. 2005;272:4565–75. doi: 10.1111/j.1742-4658.2005.04876.x. [DOI] [PubMed] [Google Scholar]
- 15.Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–6. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen PL, Xu F, Xiao W. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 2008;18:162–73. doi: 10.1038/cr.2007.114. [DOI] [PubMed] [Google Scholar]
- 17.Bredehorst R, Wielckens K, Adamietz P, Steinhagen-Thiessen E, Hilz H. Mono(ADP-ribosyl)ation and poly(ADP-ribosyl)ation of proteins in developing liver and in hepatomas: relation of conjugate subfractions to metabolic competence and proliferation rates. Eur J Biochem. 1981;120:267–74. doi: 10.1111/j.1432-1033.1981.tb05699.x. [DOI] [PubMed] [Google Scholar]
- 18.Juszczynski P, Kutok JL, Li C, Mitra J, Aguiar RCT, Shipp MA. BAL1 and BBAP are regulated by a gamma interferon-responsive bidirectional promoter and are overexpressed in diffuse large B-cell lymphomas with a prominent inflammatory infiltrate. Mol Cell Biol. 2006;26:5348–59. doi: 10.1128/MCB.02351-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith S, de Lange T. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J Cell Sci. 1999;112:3649–56. doi: 10.1242/jcs.112.21.3649. [DOI] [PubMed] [Google Scholar]
- 20.Kickhoefer VA, Siva AC, Kedersha NL, Inman EM, Ruland C, Streuli M, et al. The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J Cell Biol. 1999;146:917–28. doi: 10.1083/jcb.146.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu M, Schreek S, Cerni C, Schamberger C, Lesniewicz K, Poreba E, et al. PARP-10, a novel Myc-interacting protein with poly(ADP-ribose) polymerase activity, inhibits transformation. Oncogene. 2005;24:1982–93. doi: 10.1038/sj.onc.1208410. [DOI] [PubMed] [Google Scholar]
- 22.Liu L, Chen G, Ji X, Gao G. ZAP is a CRM1-dependent nucleocytoplasmic shuttling protein. Biochem Biophys Res Commun. 2004;321:517–23. doi: 10.1016/j.bbrc.2004.06.174. [DOI] [PubMed] [Google Scholar]
- 23.Thomassin H, Martins de Sa C, Scherrer K, Maniez C, Mandel P. Cytoplasmic poly(ADP-ribose) polymerase and poly(ADP-ribose) glycohydrolase in AEV-transformed chicken erythroblasts. Mol Biol Rep. 1988;13:35–44. doi: 10.1007/BF00805637. [DOI] [PubMed] [Google Scholar]
- 24.Jesser M, Chypre C, Hog F, Mandel P. Cytoplasmic poly(ADP-ribose)polymerase from mouse plasmacytoma free messenger ribonucleoprotein particles: purification and characterization. Biochem Biophys Res Commun. 1993;195:558–64. doi: 10.1006/bbrc.1993.2082. [DOI] [PubMed] [Google Scholar]
- 25.Roberts JH, Stard P, Giri CP, Smulson M. Cytoplasmic poly(ADP-ribose) polymerase during the HeLa cell cycle. Arch Biochem Biophys. 1975;171:305–15. doi: 10.1016/0003-9861(75)90037-5. [DOI] [PubMed] [Google Scholar]
- 26.Thomassin H, Niedergang C, Mandel P. Characterization of the poly(ADP-ribose) polymerase associated with free cytoplasmic mRNA-protein particles. Biochem Biophys Res Commun. 1985;133:654–61. doi: 10.1016/0006-291X(85)90955-6. [DOI] [PubMed] [Google Scholar]
- 27.Elkaim R, Thomassin H, Niedergang C, Egly JM, Kempf J, Mandel P. Adenosine diphosphate ribosyltransferase and protein acceptors associated with cytoplasmic free messenger ribonucleoprotein particles. Biochimie. 1984;65:653–9. doi: 10.1016/S0300-9084(84)80029-2. [DOI] [PubMed] [Google Scholar]
- 28.Jesser M, Hog F, Chypre C, Leterrier JF, Mandel P. ADP-ribosylation of neurofilaments by a cytoplasmic ADP-ribose transferase associated with free mRNP. Biochem Biophys Res Commun. 1993;194:916–22. doi: 10.1006/bbrc.1993.1908. [DOI] [PubMed] [Google Scholar]
- 29.Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science. 2002;297:1703–6. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 30.Bick MJ, Carroll J-WN, Gao G, Goff SP, Rice CM, MacDonald MR. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol. 2003;77:11555–62. doi: 10.1128/JVI.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller S, Möller P, Bick MJ, Wurr S, Becker S, Günther S, et al. Inhibition of filovirus replication by the zinc finger antiviral protein. J Virol. 2007;81:2391–400. doi: 10.1128/JVI.01601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Y, Chen G, Lv F, Wang X, Ji X, Xu Y, et al. Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc Natl Acad Sci U S A. 2011;108:15834–9. doi: 10.1073/pnas.1101676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo X, Ma J, Sun J, Gao G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci U S A. 2007;104:151–6. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen G, Guo X, Lv F, Xu Y, Gao G. p72 DEAD box RNA helicase is required for optimal function of the zinc-finger antiviral protein. Proc Natl Acad Sci U S A. 2008;105:4352–7. doi: 10.1073/pnas.0712276105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye P, Liu S, Zhu Y, Chen G, Gao G. DEXH-Box protein DHX30 is required for optimal function of the zinc-finger antiviral protein. Protein Cell. 2010;1:956–64. doi: 10.1007/s13238-010-0117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayakawa S, Shiratori S, Yamato H, Kameyama T, Kitatsuji C, Kashigi F, et al. ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat Immunol. 2011;12:37–44. doi: 10.1038/ni.1963. [DOI] [PubMed] [Google Scholar]
- 37.Kerns JA, Emerman M, Malik HS. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet. 2008;4:e21. doi: 10.1371/journal.pgen.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sbodio JI, Lodish HF, Chi N-W. Tankyrase-2 oligomerizes with tankyrase-1 and binds to both TRF1 (telomere-repeat-binding factor 1) and IRAP (insulin-responsive aminopeptidase) Biochem J. 2002;361:451–9. doi: 10.1042/0264-6021:3610451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–50. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 40.Moeller BJ, Cao Y, Li CY, Dewhirst MW. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer Cell. 2004;5:429–41. doi: 10.1016/S1535-6108(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 41.Kayali F, Montie HL, Rafols JA, DeGracia DJ. Prolonged translation arrest in reperfused hippocampal cornu Ammonis 1 is mediated by stress granules. Neuroscience. 2005;134:1223–45. doi: 10.1016/j.neuroscience.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 42.Ivanov VN, Zhou H, Hei TK. Sequential treatment by ionizing radiation and sodium arsenite dramatically accelerates TRAIL-mediated apoptosis of human melanoma cells. Cancer Res. 2007;67:5397–407. doi: 10.1158/0008-5472.CAN-07-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuznetsov G, Xu Q, Rudolph-Owen L, Tendyke K, Liu J, Towle M, et al. Potent in vitro and in vivo anticancer activities of des-methyl, des-amino pateamine A, a synthetic analogue of marine natural product pateamine A. Mol Cancer Ther. 2009;8:1250–60. doi: 10.1158/1535-7163.MCT-08-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–9. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- 45.Chang W, Dynek JN, Smith S. NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis. Biochem J. 2005;391:177–84. doi: 10.1042/BJ20050885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kotova E, Jarnik M, Tulin AV. Poly (ADP-ribose) polymerase 1 is required for protein localization to Cajal body. PLoS Genet. 2009;5:e1000387. doi: 10.1371/journal.pgen.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gagné J-P, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–76. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tourrière H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160:823–31. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–42. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2008;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holbro T, Beerli RR, Maurer F, Koziczak M, Barbas CF, 3rd, Hynes NE. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci U S A. 2003;100:8933–8. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rios J, Puhalla S. PARP inhibitors in breast cancer: BRCA and beyond. Oncology (Williston Park) 2011;25:1014–25. [PubMed] [Google Scholar]
- 53.Jagtap P, Szabó C. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Discov. 2005;4:421–40. doi: 10.1038/nrd1718. [DOI] [PubMed] [Google Scholar]
- 54.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schreiber V, Amé J-C, Dollé P, Schultz I, Rinaldi B, Fraulob V, et al. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem. 2002;277:23028–36. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 56.Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell AG, et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotechnol. 2012;30:283–8. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]