Abstract
Polyadenosine RNA binding proteins (Pabs) play critical roles in regulating the polyadenylation, nuclear export, stability, and translation of cellular RNAs. Although most Pabs are ubiquitously expressed and are thought to play general roles in post-transcriptional regulation, mutations in genes encoding these factors have been linked to tissue-specific diseases including muscular dystrophy and now intellectual disability (ID). Our recent work defined this connection to ID, as we showed that mutations in the gene encoding the ubiquitously expressed Cys3His tandem zinc-finger (ZnF) Pab, ZC3H14 (Zinc finger protein, CCCH-type, number 14) are associated with non-syndromic autosomal recessive intellectual disability (NS-ARID). This study provided a first link between defects in Pab function and a brain disorder, suggesting that ZC3H14 plays a required role in regulating RNAs in nervous system cells. Here we highlight key questions raised by our study of ZC3H14 and its ortholog in the fruit fly Drosophila melanogaster, dNab2, and comment on future approaches that could provide insights into the cellular and molecular roles of this class of zinc finger-containing Pabs. We propose a summary model depicting how ZC3H14-type Pabs might play particularly important roles in neuronal RNA metabolism.
Keywords: intellectual disability, zinc finger polyadenosine RNA binding protein, Drosophila melanogaster, polyadenylation, post-transcriptional RNA processing
Introduction
Poly(A) RNA binding proteins (Pabs) play key roles in the post-transcriptional regulation of gene expression by virtue of their ability to bind to the polyadenosine tails of RNAs. We recently identified mutations in a zinc finger (ZnF)-containing Pab encoded by the ZC3H14 gene that are linked to non-syndromic autosomal recessive intellectual disability (NS-ARID) in humans.1 Moreover, the Drosophila melanogaster ortholog of ZC3H14, dNab2, is required in neurons for normal behavior.1 These two pieces of data provide strong evidence that ZC3H14/dNab2 is critically required for regulating gene expression within neurons in the central nervous system (CNS). In this point-of-view we provide a summary of work that is beginning to define this family of proteins, and also consider questions that have been raised by initial observations indicating that this class of proteins have critical functions in neurons.
Members of a family of ZnF-containing Pabs perform conserved essential functions
The ZC3H14 gene encodes an evolutionarily conserved Cys3His (CCCH) tandem ZnF polyadenosine RNA binding protein2 (Fig. 1). Previous functional studies of this class of Pabs focused on the budding yeast Saccharomyces cerevisiae Nuclear poly(A)-Binding protein 2 (Nab2).3-6 Nab2 is essential in budding yeast,3 and Nab2 mutant yeast cells display extended poly(A) tails and nuclear accumulation of poly(A) RNA.4,6 The Nab2 protein contains an N-terminal Proline-Tryptophan-Isoleucine (PWI)-like domain which is required for proper poly(A) RNA export from the nucleus,6,7 a nuclear targeting arginine-glycine rich (RGG) domain (denoted as NLS in Fig. 1A), and a C-terminal tandem CCCH ZnF domain which mediates binding to polyadenosine RNA3,6,8,9 (Fig. 1A). As shown in Figure 1A, key functional domains are conserved in Nab2 orthologs in multicellular eukaryotes, including D. melanogaster, mice, and humans.
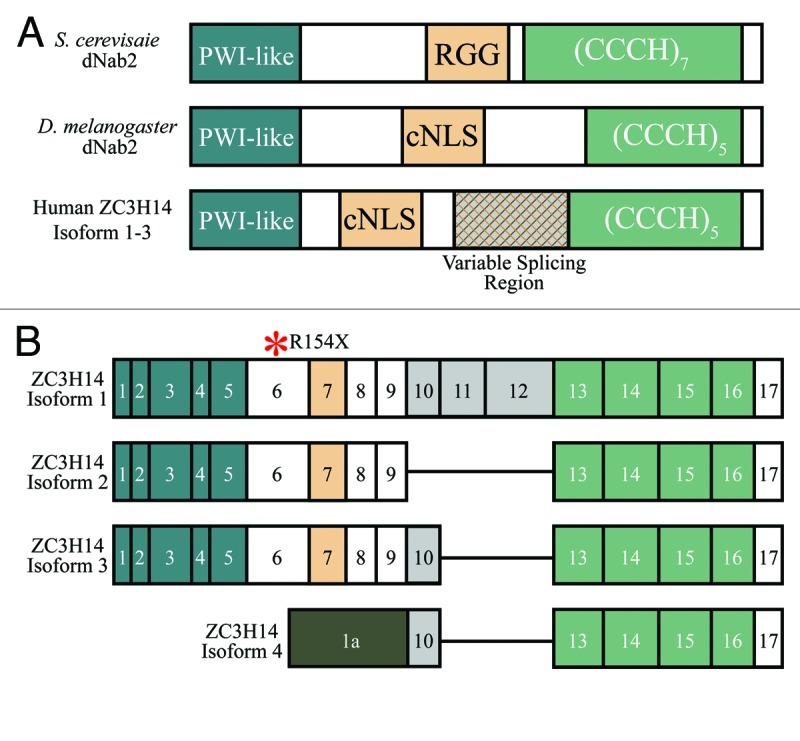
Figure 1. The Nab2/dNab2/ZC3H14 family of zinc finger polyadenosine RNA binding proteins (A) The Nab2/dNab2/ZC3H14 proteins share conserved domain architecture. The N-terminal domain of Saccharomyces cerevisiae Nab2 folds into a five α-helix bundle very similar to the three-dimensional structure of the Proline-Tryptophan-Isoleucine (PWI) domain (dark teal) found in other RNA binding proteins7. Key residues in this domain are conserved in the Drosophila melanogaster dNab2 and human ZC3H14 proteins. We term this region a “PWI-like” domain, however, because even though these proteins fold into a structure similar to that of a typical PWI domain, they lack the diagnostic Pro(P)-Trp(W)-Iso(I) residues for which this domain is named. Nab2/dNab2/ZC3H14 also contains a nuclear targeting signal in the central region of the protein (tan fill). While this signal in both dNab2 and ZC3H14 appears to be a classical bipartite NLS (cNLS)65, the nuclear targeting domain of yeast Nab2 is an arginine/glycine (RGG)-rich sequence bound by the importin β family import receptor, Kap10449. The C-terminus of each of these proteins contains a series of conserved tandem Cys-Cys-Cys-His (CCCH) zinc fingers (light green) required for polyadenosine RNA recognition8. Nab2 contains seven tandem CCCH zinc fingers while both dNab2 and ZC3H14 contain five. NAB2 and dNab2 encode a single protein isoform; however, alternative splicing of the human ZC3H14 gene results in transcripts that encode several protein isoforms. The variation among these isoforms arises from a central alternatively spliced region (exons 10–12) we have labeled the “variable splicing region” (hatched fill). (B) Human ZC3H14 encodes at least four splice variants. ZC3H14 isoforms 1–3 each encode a PWI-like domain (exons 1–5), a central cNLS (exon 6), and five tandem CCCH zinc fingers (exons 13–16). Isoforms 2 and 3 differ from isoform 1 in the exclusion of exons 10–12 or 11–12, respectively. Isoform 4 has an alternative transcriptional start site and first exon (Exon 1a). As isoform 4 lacks exon 7, it does not contain the predicted cNLS. A red asterisk above the isoform alignment denotes the recently identified human nonsense mutation in exon 6, R154X1.
Both the budding yeast and D. melanogaster genomes encode a single Nab2/dNab2 isoform. However, in mammals the ZC3H14 locus produces multiple transcripts that are alternatively spliced to encode distinct protein isoforms (Fig. 1B).2 ZC3H14 isoforms 1–3, which are ubiquitously expressed, have a very similar domain structure to Nab2/dNab2 (Fig. 1A). In ZC3H14 isoform 4 mRNA, an alternate 5′ exon (denoted as exon 1a in Fig. 1B) replaces the sequences encoding the PWI and NLS domains of isoforms 1–3, resulting in a shorter version of the ZC3H14 protein. Importantly, all four forms of the ZC3H14 protein share the same array of consecutive ZnFs in the C-terminal RNA binding domain, suggesting that they all bind polyadenosine RNA. At steady-state, ZC3H14 isoforms 1–3 localize primarily to the nucleus and are enriched in SC35-positive nuclear speckles,2 which are sites of RNA metabolism.10 In mouse brain sections, ZC3H14 isoforms 1–3 colocalize with bulk poly(A) RNA in the cell bodies of hippocampal neurons.1 In contrast, ZC3H14 isoform 4 protein localizes to the cytoplasm at steady-state and shows tissue-specific expression with enrichment in the brain and testes.2 Given the differential localization of these isoforms and the variability in tissue expression of isoform 4, these nuclear and cytoplasmic isoforms of the vertebrate ZC3H14 protein may play tissue-specific roles in modulating gene expression.
Given our recent finding that mutation of ZC3H14 is associated with intellectual disability, we assessed ZC3H14 expression patterns in the human brain using the Allen Brain Atlas.11 This resource indicates that ZC3H14 is ubiquitously expressed throughout the brain, but shows higher expression in regions such as the dentate gyrus, amygdala, cerebellum, and choroid plexus.11 Our published study also confirmed that ZC3H14 is expressed in the hippocampus, including the dentate gyrus.1 The high level of ZC3H14 expression in the dentate gyrus is intriguing given that this region of the hippocampus is directly implicated in learning and memory (for a review of dentate gyrus function, see ref. 12 and references therein). Interestingly, ZC3H14 and another nuclear Pab, PABPN1, have similar patterns of expression in the human brain cortex, while the cytoplasmic Pab, PABPC1, is more highly expressed in other areas of the brain, such as the striatum. The Fragile-X Syndrome gene FMR1, which also encodes an RNA binding protein implicated in intellectual disability,13-15 is expressed at lower levels in the hippocampus (and specifically the dentate gyrus), perhaps suggesting that disruption of ZC3H14 and FMR1 may impact different cell types in hippocampal circuitry.
The D. melanogaster ortholog of ZC3H14, dNab2, is expressed ubiquitously and resembles vertebrate isoforms 1–3 in domain structure (Fig. 1A).1 Like yeast Nab2 and ZC3H14, dNab2 binds polyadenosine RNA in vitro. As in S. cerevisiae, complete loss of dNab2 is incompatible with viability; however, flies with a maternal contribution of dNab2 can develop to adulthood but show a variety of organismal and molecular defects. Consistent with the extended poly(A) tails observed in yeast nab2 mutants, loss of D. melanogaster dNab2 causes extended poly(A) tails detected in bulk RNA from CNS-enriched samples of cellular RNAs, suggesting that dNab2 participates in poly(A) tail-length control of neuronal RNAs. These dNab2 mutant flies also show impaired flight and negative geotactic locomotor behavior.1 Significantly, RNAi-mediated depletion of dNab2 only in neurons recapitulates the locomotor phenotypes of dNab2 zygotic null animals and, reciprocally, re-expression of wild type dNab2 only in neurons rescues both viability and locomotor defects in animals otherwise completely lacking dNab2 (ref. 1). While we cannot currently rule out roles for dNab2 in other cell types such as glia, these data do suggest that dNab2 plays an important role in controlling neuronal RNAs that are required for neurodevelopment and/or function.
Mutations in the conserved ZnF Pab, ZC3H14, are linked to NS-ARID
One of the key pieces of evidence that ZC3H14 isoforms are likely to have non-redundant roles in the brain is the location of a mutation within ZC3H14 that has been linked to NS-ARID. This nonsense mutation replaces the sequence encoding arginine 154 with a premature termination codon (R154Stop). This termination codon occurs in ZC3H14 exon 6 and completely removes the nuclear isoforms 1–3, which are analogous to the essential protein in yeast and flies, while leaving the cytoplasmic isoform 4 intact1 (Fig. 1B). The fact that ID patients containing this mutation, and therefore lacking ZC3H14 isoforms 1–3, are still alive and only have cognitive defects argues that isoform 4 may be able to compensate for the absence of isoforms 1–3 in most human tissues but perhaps not in cells of the brain. In combination with the dNab2 data from flies, these observations indicate that a ubiquitously expressed Pab can nonetheless play critical tissue-specific roles in both flies and humans and raise several interrelated questions:
(1) Is ZC3H14 an essential gene in mammals?
Given that Nab2 and dNab2 are essential in budding yeast and Drosophila1,3 respectively, one might have predicted that the homologous ZC3H14 isoforms would be required for viability in mammals. However, patients with the allele of ZC3H14 that encodes the premature stop-codon lack the nuclear ZC3H14 isoforms analogous to the essential Nab2/dNab2 protein in yeast and flies. Surprisingly, these individuals show severe neuronal deficits, but no additional symptoms have been reported.1
(2) Why are neurons so sensitive to loss of ZC3H14 iso1–3/dNab2?
Neurons rely heavily on post-transcriptional RNA regulation mechanisms to compartmentalize gene expression in time and space.16 Perhaps this reliance makes these specialized cells more susceptible than other cells to loss of ZC3H14/dNab2.
(3) What are the molecular roles of the dNab2/ZC3H14 proteins?
Do all of the ZC3H14 isoforms have the same function? Could the requirement for a given isoform vary between cell types? If so, then the tissue-specific effects of ZC3H14 mutations could reflect cell type-specific roles of different ZC3H14 isoforms.
(4) What RNAs are affected by loss of ZC3H14/dNab2?
While Pab proteins are typically associated with regulation of mRNAs, there is growing evidence that other types of RNAs also require Pabs for proper regulation.17,18 The identity of the RNAs or classes of RNAs impacted by ZC3H14 could reveal much about the biological consequences of ZC3H14 mutations in humans.
(5) Are hyperadenylated RNAs causative of neuronal defects?
The primary molecular defect detected thus far in yeast, fly and human cells lacking Nab2/dNab2/ZC3H14 is extension of bulk poly(A) RNA tails. Remarkably, the functional consequences of the aberrantly long mRNA poly(A) tails are not well understood in any cell type, including neurons.
(6) How frequent are ZC3H14 alleles in the human population and could they be responsible for additional cases of NS-ARID or other forms of heritable ID?
In the following sections, we address each of these questions with particular reference to our published findings and the present state of published knowledge.
(1) Is ZC3H14 an essential gene in mammals?
As documented in our recent study, a nonsense mutation (R154Stop) in a 5′ exon leads to selective loss of ZC3H14 nuclear isoforms but leaves the cytoplasmic isoform intact (Fig. 1B). Patients lacking protein isoforms 1–3 display intellectual disability but no other symptoms, which contrasts with the essential requirement for the orthologs of these proteins in budding yeast and flies.1,3 There are several possibilities that could explain this surprising observation.
First, ZC3H14 may simply not be essential in humans except for proper nervous system function. Second, an unidentified functionally redundant protein could substitute for ZC3H14 function in non-neuronal tissues. Third, as the cytoplasmic isoform 4 is apparently unaffected in patients homozygous for the R154Stop allele, this form of ZC3H14 might be sufficient in every tissue except the brain. Initial assessment of the relative functional properties of ZC3H14 isoforms could be accomplished by nervous system-specific expression of human ZC3H14 in flies lacking dNab2. This human:fly trans-species neuronal rescue would logically be followed by genetic ablation of murine ZC3H14, ideally in a tissue-specific conditional approach that targets specific isoforms. Alternatively, a targeted knock-in of the R154Stop mutation could be generated to determine whether R154Stop homozygous mice exhibit brain-specific phenotypes similar to human patients.
A key advantage of the fly and mouse models is their ability to address the question of whether dNab2/ZC3H14 plays a developmental role in the nervous system, or is mainly required to support the physiologic function of neurons in the fully formed mature brain. A simple approach to this question would be to examine the brains of dNab2/ZC3H14 mutant flies and mice for evidence of developmental defects, which if present, would provide some justification to examine human patients for similar defects. However, dNab2/ZC3H14 may have important roles in brain development and physiology which each contribute to endpoint cognitive defects. Parsing the contribution of each of these to CNS phenotypes defects will require temporally controlled transgenic systems such as those readily available in the fly.19
(2) Why are neurons so sensitive to loss of ZC3H14 Iso1–3/dNab2?
As in the case of ZC3H14-associated NS-ARID, other diseases caused by mutations in ubiquitously expressed RNA processing factors can manifest with tissue- and cell type-specific defects.20 For example, the expansion of a polyalanine tract within the N-terminal end of the nuclear Pab, PABPN1, causes an autosomal dominant disease termed Oculopharyngeal Muscular Dystrophy (OPMD) which selectively impacts the muscles of the eyelids and pharynx.21 Similarly, mutations in the general splicing factor SMN1 affect only lower motor neurons,22 while the main physiologic features of defects in expression or function of Fragile-X Mental Retardation Protein (FMRP; also an RNA binding protein) are impaired higher-cognitive function and autism spectrum disorders.13,15,23 In each of these cases, all cells in the organism contain the disease-causing expansion, mutation, or chromosomal abnormality, but only certain cell types and tissues are sensitive to the resulting functional impairment of post-transcriptional RNA regulatory pathways. Thus a key unresolved issue in ZC3H14-mediated NS-ARID is why defects in the ubiquitously expressed RNA binding protein, ZC3H14, cause a neurological disorder.
Although dNab2/ZC3H14 could have as yet undefined roles in non-neuronal nervous system cells such as glia, the effects of neuron-specific expression and depletion of D. melanogaster dNab2 argues for a key requirement in this cell type.1 One explanation for this link may be that because neurons make extensive use of post-transcriptional RNA regulatory mechanisms to compartmentalize gene expression in time and space, they are more reliant than other cells on ZC3H14/dNab2 (ref. 16). A second explanation stems from the observation that RNAs harvested from the heads of dNab2 null flies show an extension in bulk poly(A) RNA tail length that occurs independent of an effect on poly(A) RNA nuclear export.1 As predicted for a protein that limits poly(A) tail length, overexpression of dNab2 shortens poly(A) tails among neuronal cells in the eye, and this effect can be reversed by overexpressing components of the nuclear polyadenylation machinery such as fly Pabp2 (ref. 1), arguing that dNab2/ZC3H14 proteins exert their effects on neural function at least in part by modulating polyadenylation of RNA transcripts. These data raise the possibility that neurons are reliant on specific dNab2/ZC3H14 RNA targets whose expression is more sensitive to changes in poly(A) tail length than the bulk of cellular RNAs. The ability of the cytoplasmic polyadenylation factor CPEB (see ref. 24 for review) to regulate the polyadenylation and translation of neuronal mRNAs at synapses provides precedent for such a hypothesis.
(3) What are the molecular roles of Nab2/dNab2/ZC3H14 proteins?
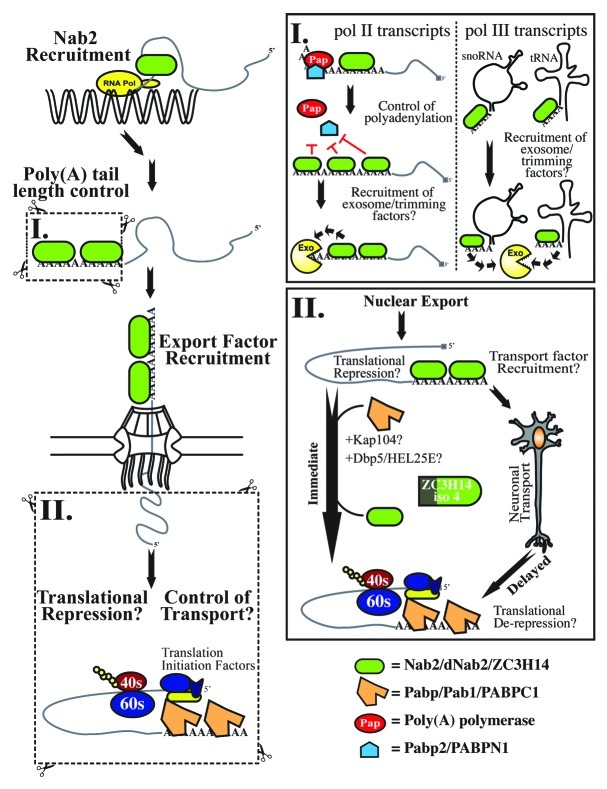
Our current understanding of the potential nuclear and cytoplasmic roles of Nab2/dNab2/ZC3H14 is based on studies performed in S. cerevisiae and D. melanogaster as well as in cultured mammalian cells. The available data support a theoretical consolidated model (Fig. 2) in which dNab2/Nab2/ZC3H14 initially controls the length of poly(A) tails on RNA transcripts in the nucleus and may subsequently recruit RNA export factors (such as the heterodimer Mex67:Mtr2 in yeast25) to facilitate the export of RNA transcripts from the nucleus. Following export from the nucleus, dNab2/ZC3H14 family members may also play a conserved role in translational regulatory mechanisms or RNA trafficking in the cytoplasm. Although dNab2 and ZC3H14 isoforms 1–3 proteins are predominantly nuclear1,2,9 at steady-state, we can also detect pools of these proteins in fly and vertebrate neurites (our unpublished data), suggesting that these proteins could have distinct molecular functions in the nucleus and the cytoplasm.
Figure 2. A consolidated model for Nab2/dNab2/ZC3H14 function. Nab2/dNab2/ZC3H14 (green oval) is initially recruited to the nascent RNA transcript via interactions with RNA itself or through an intermediate RNA binding protein. Following the completion of transcription, Nab2 family members help to dictate the final length of the poly(A) tail. For mRNAs (inset I, left), this mechanism could involve inhibition of poly(A) polymerase (red circle - PAP) or, in the case of fly dNab2 or human ZC3H14, competition with the poly(A) binding protein, Pabp2/PABPN1 (blue hexagon). Following termination of polyadenylation, Nab2/dNab2/ZC3H14 could recruit a ribonuclease such as the nuclear exosome (yellow ‘pac-man’ symbol) to modulate poly(A) tail length. In the case of non-coding RNAs (inset I, right), Nab2/dNab2/ZC3H14 might bind the short poly(A) tails to recruit the trimming and/or degradation activities of the nuclear exosome. Evidence from budding yeast demonstrates that Nab2 facilitates recruitment of mRNA export factors such as Mex67 that then mediate export of RNA from the nucleus to the cytoplasm. Once in the cytoplasm, numerous roles for Nab2/dNab2/ZC3H14 in translational control or RNA transport can be postulated (inset II). Following export from the nucleus, Nab2/dNab2/ZC3H14 could be removed immediately or this removal could be delayed until the transcript is transported to a sub-cellular location. The Nab2/dNab2/ZC3H14 protein could influence the fate of the RNA in the cytoplasm in a number of possible ways including modulating stability, impacting transport, or regulating translation. Regardless of the function, removal of Nab2/dNab2/ZC3H14 could involve a remodeling enzyme such as the DEAD-box helicase, Dbp5, or other proteins including transport receptors as demonstrated in yeast for the Kap104 import receptor49,66. Following removal of Nab2/dNab2/ZC3H14, interactions between the conventional cytoplasmic poly(A) binding protein (Pab1/Pabp/PABPC1) bound to the 3′-end poly(A) tail and mRNA translation initiation factors (blue and yellow symbols near the 5′–end of the transcript) can then stimulate translation by the ribosome.
Nuclear Roles
In one mechanistic model (Fig. 2, inset I), dNab2/ZC3H14 binds to poly(A) tails of RNAs in the nucleus and functionally interacts with other poly(A)-binding proteins, such as Pabp2/PABPN1 to properly dictate the length of the mature poly(A) tail on these RNA transcripts. Based on our genetic data in the fly eye1 dNab2/ZC3H14 function is likely to be antagonistic to that of the nuclear RRM-containing PABPN1/Pabp2 poly(A) RNA binding protein.26 Poly(A) tail length experiments suggest that while PABPN1/Pabp2 facilitates poly(A) tail length extension,26,27 dNab2/Nab2/ZC3H14 may help to limit the length of poly(A) tails in vivo.1 Given that PABPN1/Pabp2 helps to “tether” the poly(A) polymerase to nascent mRNA transcripts to increase processivity,26,28-30 and that dNab2 acts as a genetic antagonist of Pabp2 (ref. 1), one possibility is that Nab2 family members might bind elongating poly(A) tails, compete with Pabp2 for binding to polyadenosine RNA, and by displacing Pabp2 from polyadenosine RNA decrease poly(A) polymerase processivity (Fig. 2, inset I). However, the mechanistic details of this inhibition are not yet clear. Although the binding affinity of dNab2/ZC3H14 for polyadenosine RNA has not yet been determined, the affinity of yeast Nab2 for polyadenosine RNA in vitro is between 10.5 – 30 nM.4,9 Interestingly, the affinity of dPabp2/PABPN1 is similar, approximately 10 nM.29-31 Therefore, if the affinity of dNab2/ZC3H14 for polyadenosine RNA is similar to that of its yeast ortholog (and therefore similar to dPabp2/PABPN1 as well), this would imply that dNab2/ZC3H14 could not antagonize poly(A) tail length by merely outcompeting dPabp2/PABPN1 for binding to polyadenosine RNA and therefore decreasing the processivity of the poly(A) polymerase. One viable alternative to this “direct-competition” model is one in which addition of a certain length of adenosines to the extending poly(A) tail allows for multiple dNab2/ZC3H14 proteins to bind and induce a conformational change in the RNA that might then displace Pabp2/PABPN1 and inhibit the processivity of the poly(A) polymerase.32,33
Alternatively, Nab2/dNab2 could also recruit enzymes that actively trim the poly(A) tail. Yeast Nab2 and fly dNab2 both interact genetically with components of the nuclear exosome (our unpublished results and see ref. 18), which is a multi-protein complex required for the proper trimming and quality control of several classes of RNAs transcribed by RNA polymerase III, including rRNAs, tRNAs, and small nucleolar RNAs (snoRNAs).34-36 Recent data also suggest that the nuclear exosome may function in the quality control of mRNAs as well.37-41 Hence, one other hypothesis is that following addition of an extended poly(A) tail, Nab2/dNab2/ZC3H14 recruits a ribonuclease such as the nuclear exosome to trim poly(A) tails back to an appropriate length..
This “exosome recruitment” hypothesis would be particularly interesting since the short non-mRNAs mentioned above (rRNAs, tRNAs, and snoRNAs) can also receive short poly(A) tails when processed incorrectly. These short poly(A) tails are thought to target aberrant transcripts for degradation via the nuclear exosome42-45 and also promote normal non-coding transcript processing.17,46 The length of these poly(A) tails varies, but can be between 10 and 40 adenosines, depending on the transcript.47,48 Given that yeast Nab2 has a minimal binding footprint of roughly 10–15 adenosines (our unpublished data and see ref. 33), and antibodies targeting yeast Nab2 co-immunoprecipitate chromatin-containing genes transcribed by RNA polymerase II or RNA polymerase III,18 dNab2/ZC3H14 may be able to recognize these short poly(A) tails and trigger exosome recruitment and subsequent RNA turnover (Fig. 2, inset I). Experiments that probe potential links between dNab2/ZC3H14, the exosome, and turnover of various classes of RNAs could provide critical insight into the nuclear function of dNab2/ZC3H14 in RNA metabolism.
Cytoplasmic Roles
Several lines of evidence from experiments in yeast, flies, and vertebrate cells suggest that dNab2/ZC3H14 may be involved in modulating mRNA translation and/or RNA trafficking in the cytoplasm (Fig. 2, inset II). First, the yeast Nab2 nuclear import receptor, Kap104, which re-imports Nab2 into the nucleus during nucleocytoplasmic shuttling,49 has recently been implicated in the de-repression of translation.50,51 Since Kap104 may aid in dissociation of Nab2 from cytoplasmic RNAs,49 this reported role in translational activation could stem from removing Nab2 from cytoplasmic mRNPs.51 The same study also demonstrated that a small percentage of the DEAD-box containing RNA helicase Dbp5 is found with Kap104 at sites of translation at the yeast daughter cell bud site.50 Interestingly, Dbp5 can remove yeast Nab2 from RNA52 and is associated with polyribosomes,53 suggesting that perhaps Dbp5 may actively remove Nab2 from yeast transcripts that are transported to the daughter cell bud site. Following this removal, Nab2 could then be immediately bound by Kap104 and re-imported into the nucleus.51
Second, given that we detect a small pool of Nab2/dNab2/ZC3H14 localized to the cytoplasm and neurites of cultured mammalian and D. melanogaster neurons in vitro (our unpublished data), we additionally propose that members of this family of proteins may be associated with ribonucleoprotein (RNP) complexes responsible for the directional trafficking of RNAs to sites of active translation (Fig. 2, inset II). As ZC3H14-associated NS-ARID patients do not show motor defects consistent with loss of peripheral motor neurons, dNab2/ZC3H14 may not be absolutely required for RNP trafficking but rather may increase the efficiency or specificity of the process. Future studies that test this hypothesis could provide key insight into ZC3H14/dNab2 function in neurons.
(4) What RNAs are affected by loss of ZC3H14/dNab2?
Control of polyadenylation has classically been considered as a key step in mRNA transcript metabolism. However, recent studies demonstrate that many types of RNA transcripts are polyadenylated under certain circumstances and that poly(A) binding proteins can regulate the fate of non-mRNAs such as snoRNAs.17,46 Thus, a major question is whether dNab2 family members regulate particular RNAs or even classes of RNAs. The identity of the RNAs or classes of RNAs that are impacted by ZC3H14/dNab2 loss could reveal much about the biological roles of these proteins in the CNS.
The recruitment of dNab2/ZC3H14 to RNAs is likely to be an important step in determining target specificity. As a polyadenosine RNA binding protein, the null hypothesis is that dNab2 and ZC3H14 bind the poly(A) tails of all RNAs in cells; however, this hypothesis has yet to be properly tested in multicellular organisms and presents interesting challenges to explanations of the neuronal-specific defects associated with loss of these proteins. The reciprocal hypothesis that dNab2/ZC3H14 only binds a subset of RNAs also remains untested. At present, very little is known about how members of the dNab2/ZC3H14 family of proteins are recruited to nascent transcripts. As mentioned above, a recent study found that yeast Nab2 associates with RNA polymerase II genes in a transcription-dependent manner,18 and studies in yeast also suggest that Nab2 can interact with the vast majority of poly(A) RNAs.54 Although most mRNA transcripts should be polyadenylated and therefore theoretically bound by dNab2/ZC3H14, other proteins bound to sequences within the 3′UTR of specific transcripts could also preferentially recruit dNab2/ZC3H14 to specific transcripts. In the future, it will be important to revisit the question of dNab2/ZC3H14 target specificity in fly/vertebrate neurons using RNA-immunoprecipitation/RNA-sequencing (RNA-Seq) to identify transcripts that physically associate with dNab2/ZC3H14, in combination with biochemical experiments55 to identify RNAs whose poly(A) tails are most affected by loss of these proteins. Overlap between these data sets would presumably be enriched for RNAs directly regulated by dNab2/ZC3H14. These kinds of target-specificity experiments could provide important clues to the physiologic defect(s) in ZC3H14 mutant neurons.
(5) Are hyperadenylated RNAs causative of neuronal defects?
Traditionally, deadenylation and the shortening of poly(A) tails are thought to be the first step in transcript degradation by the quality control machinery.56 However, the fate of hyperadenylated transcripts is unclear. These hyperadenylated transcripts may simply be more abundant57 and therefore translated more often, leading to elevated levels of their encoded proteins. Alternatively, hyperadenylated transcripts could be removed from cells via the RNA degradation machinery. However, experimental evidence suggests that the answer to this question may not be straightforward. Mutations in several genes in addition to NAB2/dNab2 lead to an increase in bulk poly(A) tail length, including for example, poly(A) RNA export mutants in budding yeast.38,58 In Drosophila, mutants in the deadenylation machinery lead to hyperadenylated transcripts,59 as does overexpression of poly(A) polymerase (PAP).60 However, in the case of PAP overexpression, the resultant effect on translation depends on the developmental context.60 Hyperadenylation could also impair cytoplasmic polyadenylation-mediated translational regulation (e.g., via CBEP) of mRNA transcripts critical for normal brain function, or it could titrate other poly(A) RNA-binding proteins. Beyond the few reports cited here, the functional consequence of hyperadenylated transcripts has not been studied and is thus an area that invites future analysis.
6) How frequent are ZC3H14 alleles in the human population, and could they be responsible for additional cases of NS-ARID or other forms of heritable ID?
As the ZC3H14 mutations we discovered1 were found in a geographically restricted location, an additional key question is whether ID-causing mutations in ZC3H14 are population-specific or are also present in patients with similar ID features from other populations (e.g., in the Western world). One approach to answering this question would be to sequence the ZC3H14 gene in patient collectives from different human populations around the world. In view of the genetic heterogeneity of non-syndromic ID, this kind of candidate-based investigation of NS-ID genes has until recently been time consuming, costly, and inefficient. However, given the advent of new sequencing technologies, performing mutation screening in complete exomes of large cohorts can be accomplished with increasing ease at rapidly decreasing costs,61 and an expeditious answer to this question seems to be within reach.
Another factor to consider in these human sequencing studies is the spectrum of clinical features caused by ZC3H14 mutations, as abnormalities in addition to ID might emerge as additional patients are identified. A prominent example of this type of shifting diagnostic criteria is Fragile-X syndrome, which was originally described as a non-syndromic form of ID.62,63 Furthermore, some mutations can give rise to both non-syndromic and syndromic ID, depending on their nature or location within the gene (for examples see e.g., Ropers and Hamel64). One line of investigation could examine correlations between different mutations in ZC3H14 and the severity of ID or specific accompanying clinical features. This analysis would give further valuable clues as to the specific aspects of ZC3H14 function, aid functional investigations in animal or cellular models, and place ZC3H14 within a larger molecular network in neurons that underlies cognition.
A final perspective
In summary, data from yeast, flies, and mammalian cells are beginning to provide a glimpse into the sort of RNA regulatory roles dNab2/ZC3H14 may play in cells generally, and in neurons specifically. RNA targets of these proteins in the nuclear and cytoplasmic compartments are currently unknown, but they presumably include transcripts that are required for neurodevelopment and/or neuronal physiology. Identifying these RNAs, and characterizing their dependence on nuclear and cytoplasmic pools of dNab2/ZC3H14 will not only shed light on cellular defects in ZC3H14-associated NS-ARID patients, but will also expand our current understanding of the central roles that Nab/dNab2/ZC3H14 proteins play in poly(A) tail length control and RNA metabolism.
Like the majority of the genes that have been implicated in the etiology of ID in humans, ZC3H14 is evolutionarily conserved from man to single-celled eukaryotes. As cognition represents a comparatively recently evolved feature of the metazoan nervous system, ZC3H14 mutations are thus likely to impair either a newly acquired molecular function of the Nab2/dNab2/ZC3H14 protein, or to perturb a molecular network or pathway that has recently incorporated it. The lack of functional redundancy in evolutionarily young systems often means that loss of individual components elicits strong effects in the organism, which in the case of ZC3H14 reveals potentially novel aspects of RNA regulation in the nervous system and opens up interesting new avenues of research into the central nervous system in health and disease.
Acknowledgments
We wish to thank members of our laboratories for helpful discussion and advice. We are also very thankful to the ZC3H14-associated NS-ARID patients and their families for their contributions to this work. Financial support: NIH 5K12 GM000680–12 (SMK) and NIH GM058728–09A1S109 (AHC and KHM).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/20187
References
- 1.Pak C, Garshasbi M, Kahrizi K, Gross C, Apponi LH, Noto JJ, et al. Mutation of the conserved polyadenosine RNA binding protein, ZC3H14/dNab2, impairs neural function in Drosophila and humans. Proc Natl Acad Sci U S A. 2011;108:12390–5. doi: 10.1073/pnas.1107103108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leung SW, Apponi LH, Cornejo OE, Kitchen CM, Valentini SR, Pavlath GK, et al. Splice variants of the human ZC3H14 gene generate multiple isoforms of a zinc finger polyadenosine RNA binding protein. Gene. 2009;439:71–8. doi: 10.1016/j.gene.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson JT, Wilson SM, Datar KV, Swanson MS. NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol Cell Biol. 1993;13:2730–41. doi: 10.1128/mcb.13.5.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hector RE, Nykamp KR, Dheur S, Anderson JT, Non PJ, Urbinati CR, et al. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 2002;21:1800–10. doi: 10.1093/emboj/21.7.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green DM, Marfatia KA, Crafton EB, Zhang X, Cheng X, Corbett AH. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J Biol Chem. 2002;277:7752–60. doi: 10.1074/jbc.M110053200. [DOI] [PubMed] [Google Scholar]
- 6.Marfatia KA, Crafton EB, Green DM, Corbett AH. Domain analysis of the Saccharomyces cerevisiae heterogeneous nuclear ribonucleoprotein, Nab2p. Dissecting the requirements for Nab2p-facilitated poly(A) RNA export. J Biol Chem. 2003;278:6731–40. doi: 10.1074/jbc.M207571200. [DOI] [PubMed] [Google Scholar]
- 7.Grant RP, Marshall NJ, Yang JC, Fasken MB, Kelly SM, Harreman MT, et al. Structure of the N-terminal Mlp1-binding domain of the Saccharomyces cerevisiae mRNA-binding protein, Nab2. J Mol Biol. 2008;376:1048–59. doi: 10.1016/j.jmb.2007.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelly SM, Leung SW, Apponi LH, Bramley AM, Tran EJ, Chekanova JA, et al. Recognition of polyadenosine RNA by the zinc finger domain of nuclear poly(A) RNA-binding protein 2 (Nab2) is required for correct mRNA 3′-end formation. J Biol Chem. 2010;285:26022–32. doi: 10.1074/jbc.M110.141127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly SM, Pabit SA, Kitchen CM, Guo P, Marfatia KA, Murphy TJ, et al. Recognition of polyadenosine RNA by zinc finger proteins. Proc Natl Acad Sci U S A. 2007;104:12306–11. doi: 10.1073/pnas.0701244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–12. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 11.Allen Brain Atlas Resources [Internet]. Seattle (WA): Allen Institute for Brain Science. ©2009. Available from: http://www.brain-map.org
- 12.Treves A, Tashiro A, Witter ME, Moser EI. What is the mammalian dentate gyrus good for? Neuroscience. 2008;154:1155–72. doi: 10.1016/j.neuroscience.2008.04.073. [DOI] [PubMed] [Google Scholar]
- 13.Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–14. doi: 10.1016/0092-8674(91)90397-H. [DOI] [PubMed] [Google Scholar]
- 14.Oberle I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, et al. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 15.Barbe B, Franke P, Maier W, Leboyer M. Fragile X syndrome. I. An overview on its genetic mechanism. Eur Psychiatry. 1996;11:227–32. doi: 10.1016/0924-9338(96)82328-5. [DOI] [PubMed] [Google Scholar]
- 16.Goldie BJ, Cairns MJ. Post-transcriptional trafficking and regulation of neuronal gene expression. Mol Neurobiol. 2012;45:99–108. doi: 10.1007/s12035-011-8222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemay JF, D’Amours A, Lemieux C, Lackner DH, St-Sauveur VG, Bähler J, et al. The nuclear poly(A)-binding protein interacts with the exosome to promote synthesis of noncoding small nucleolar RNAs. Mol Cell. 2010;37:34–45. doi: 10.1016/j.molcel.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 18.González-Aguilera C, Tous C, Babiano R, de la Cruz J, Luna R, Aguilera A. Nab2 functions in the metabolism of RNA driven by polymerases II and III. Mol Biol Cell. 2011;22:2729–40. doi: 10.1091/mbc.E11-01-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–8. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 20.Lage K, Hansen NT, Karlberg EO, Eklund AC, Roque FS, Donahoe PK, et al. A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes. Proc Natl Acad Sci U S A. 2008;105:20870–5. doi: 10.1073/pnas.0810772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brais B. Oculopharyngeal muscular dystrophy: a late-onset polyalanine disease. Cytogenet Genome Res. 2003;100:252–60. doi: 10.1159/000072861. [DOI] [PubMed] [Google Scholar]
- 22.Kolb SJ, Kissel JT. Spinal muscular atrophy: a timely review. Arch Neurol. 2011;68:979–84. doi: 10.1001/archneurol.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pirozzi F, Tabolacci E, Neri G. The FRAXopathies: definition, overview, and update. Am J Med Genet A. 2011;155A:1803–16. doi: 10.1002/ajmg.a.34113. [DOI] [PubMed] [Google Scholar]
- 24.Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–85. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Iglesias N, Tutucci E, Gwizdek C, Vinciguerra P, Von Dach E, Corbett AH, et al. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 2010;24:1927–38. doi: 10.1101/gad.583310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kühn U, Wahle E. Structure and function of poly(A) binding proteins. Biochim Biophys Acta. 2004;1678:67–84. doi: 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Kerwitz Y, Kühn U, Lilie H, Knoth A, Scheuermann T, Friedrich H, et al. Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A) binding protein allosterically regulated by RNA. EMBO J. 2003;22:3705–14. doi: 10.1093/emboj/cdg347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wahle E. A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell. 1991;66:759–68. doi: 10.1016/0092-8674(91)90119-J. [DOI] [PubMed] [Google Scholar]
- 29.Bienroth S, Keller W, Wahle E. Assembly of a processive messenger RNA polyadenylation complex. EMBO J. 1993;12:585–94. doi: 10.1002/j.1460-2075.1993.tb05690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benoit B, Nemeth A, Aulner N, Kühn U, Simonelig M, Wahle E, et al. The Drosophila poly(A)-binding protein II is ubiquitous throughout Drosophila development and has the same function in mRNA polyadenylation as its bovine homolog in vitro. Nucleic Acids Res. 1999;27:3771–8. doi: 10.1093/nar/27.19.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wahle E, Rüegsegger U. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev. 1999;23:277–95. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 32.Wahle E. Poly(A) tail length control is caused by termination of processive synthesis. J Biol Chem. 1995;270:2800–8. doi: 10.1074/jbc.270.6.2800. [DOI] [PubMed] [Google Scholar]
- 33.Viphakone N, Voisinet-Hakil F, Minvielle-Sebastia L. Molecular dissection of mRNA poly(A) tail length control in yeast. Nucleic Acids Res. 2008;36:2418–33. doi: 10.1093/nar/gkn080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell. 1997;91:457–66. doi: 10.1016/S0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 35.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–39. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 36.Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci. 2008;33:501–10. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 37.Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–75. doi: 10.1016/S0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- 38.Hilleren P, Parker R. Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3′-end formation of nascent transcripts. RNA. 2001;7:753–64. doi: 10.1017/S1355838201010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roth KM, Wolf MK, Rossi M, Butler JS. The nuclear exosome contributes to autogenous control of NAB2 mRNA levels. Mol Cell Biol. 2005;25:1577–85. doi: 10.1128/MCB.25.5.1577-1585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milligan L, Torchet C, Allmang C, Shipman T, Tollervey D. A nuclear surveillance pathway for mRNAs with defective polyadenylation. Mol Cell Biol. 2005;25:9996–10004. doi: 10.1128/MCB.25.22.9996-10004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Almeida SF, García-Sacristán A, Custódio N, Carmo-Fonseca M. A link between nuclear RNA surveillance, the human exosome and RNA polymerase II transcriptional termination. Nucleic Acids Res. 2010;38:8015–26. doi: 10.1093/nar/gkq703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–24. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 43.Vanácová S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, et al. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–37. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 45.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–76. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 46.Grzechnik P, Kufel J. Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol Cell. 2008;32:247–58. doi: 10.1016/j.molcel.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuai L, Fang F, Butler JS, Sherman F. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2004;101:8581–6. doi: 10.1073/pnas.0402888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slomovic S, Laufer D, Geiger D, Schuster G. Polyadenylation of ribosomal RNA in human cells. Nucleic Acids Res. 2006;34:2966–75. doi: 10.1093/nar/gkl357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee DC, Aitchison JD. Kap104p-mediated nuclear import. Nuclear localization signals in mRNA-binding proteins and the role of Ran and Rna. J Biol Chem. 1999;274:29031–7. doi: 10.1074/jbc.274.41.29031. [DOI] [PubMed] [Google Scholar]
- 50.van den Bogaart G, Meinema AC, Krasnikov V, Veenhoff LM, Poolman B. Nuclear transport factor directs localization of protein synthesis during mitosis. Nat Cell Biol. 2009;11:350–6. doi: 10.1038/ncb1844. [DOI] [PubMed] [Google Scholar]
- 51.Goldfarb DS. How to grow a bud: an importin acts in asymmetric division. Nat Cell Biol. 2009;11:243–5. doi: 10.1038/ncb0309-243. [DOI] [PubMed] [Google Scholar]
- 52.Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell. 2007;28:850–9. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 53.Gross T, Siepmann A, Sturm D, Windgassen M, Scarcelli JJ, Seedorf M, et al. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007;315:646–9. doi: 10.1126/science.1134641. [DOI] [PubMed] [Google Scholar]
- 54.Batisse J, Batisse C, Budd A, Böttcher B, Hurt E. Purification of nuclear poly(A)-binding protein Nab2 reveals association with the yeast transcriptome and a messenger ribonucleoprotein core structure. J Biol Chem. 2009;284:34911–7. doi: 10.1074/jbc.M109.062034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meijer HA, Bushell M, Hill K, Gant TW, Willis AE, Jones P, et al. A novel method for poly(A) fractionation reveals a large population of mRNAs with a short poly(A) tail in mammalian cells. Nucleic Acids Res. 2007;35:e132. doi: 10.1093/nar/gkm830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–26. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 57.Beilharz TH, Preiss T. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA. 2007;13:982–97. doi: 10.1261/rna.569407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jensen TH, Patricio K, McCarthy T, Rosbash M. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol Cell. 2001;7:887–98. doi: 10.1016/S1097-2765(01)00232-5. [DOI] [PubMed] [Google Scholar]
- 59.Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit Rev Biochem Mol Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- 60.Juge F, Zaessinger S, Temme C, Wahle E, Simonelig M. Control of poly(A) polymerase level is essential to cytoplasmic polyadenylation and early development in Drosophila. EMBO J. 2002;21:6603–13. doi: 10.1093/emboj/cdf633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–55. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 62.Lubs HA. A marker X chromosome. Am J Hum Genet. 1969;21:231–44. [PMC free article] [PubMed] [Google Scholar]
- 63.Lubs HA, Watson M, Breg R, Lujan E. Restudy of the original marker X family. Am J Med Genet. 1984;17:133–44. doi: 10.1002/ajmg.1320170108. [DOI] [PubMed] [Google Scholar]
- 64.Ropers HH, Hamel BC. X-linked mental retardation. Nat Rev Genet. 2005;6:46–57. doi: 10.1038/nrg1501. [DOI] [PubMed] [Google Scholar]
- 65.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bolger TA, Folkmann AW, Tran EJ, Wente SR. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell. 2008;134:624–33. doi: 10.1016/j.cell.2008.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]