Abstract
With the advent of next generation sequencing methods and progress in transcriptome analysis, it became obvious that the human genome contains much more than just protein-coding genes. In fact, up to 70% of our genome is transcribed into RNA that does not serve as templates for proteins. In this review, we focus on the emerging roles of these long non-coding RNAs (lncRNAs) in the field of tumor biology. Long ncRNAs were found to be deregulated in several human cancers and show tissue-specific expression. Functional studies revealed a broad spectrum of mechanisms applied by lncRNAs such as HOTAIR, MALAT1, ANRIL or lincRNA-p21 to fulfill their functions. Here, we link the cellular processes influenced by long ncRNAs to the hallmarks of cancer and therefore provide an ncRNA point-of-view on tumor biology. This should stimulate new research directions and therapeutic options considering long ncRNAs as novel prognostic markers and therapeutic targets.
Keywords: non-coding RNA, ncRNA, lincRNA, gene expression, carcinogenesis, tumor biology, cancer therapy, MALAT1, HOTAIR, XIST, T-UCR
Introduction
In 2001, the human genome sequencing consortium released its final draft of the human genome.1 Based on this work, the complete human transcriptome could be identified and characterized in recent years mainly due to the development of two new techniques: deep sequencing and DNA tiling arrays. These methods revolutionized our view of genome organization and content as they revealed an unexpected finding: a much larger part of the human genome is pervasively transcribed into RNA than previously assumed. It is estimated that up to 70% of the genome is transcribed but only up to 2% of the human genome serve as blueprints for proteins.2-6 RNA molecules that lack protein-coding potential are collectively referred to as non-coding RNAs (ncRNAs) and well-known ncRNAs are classical “housekeeping” RNAs, such as tRNAs (tRNAs), rRNAs (rRNAs), small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs), which are constitutively expressed and play critical roles in protein biosynthesis.
Long ncRNAs: What, where and why?
According to their size, ncRNAs are subdivided into two groups: small ncRNAs (< 200 nt) and long ncRNAs. In recent years, small ncRNAs like microRNAs (miRNAs), small interfering RNAs (siRNAs) or PIWI-interacting RNAs (piRNAs) received most attention and especially miRNAs were shown to play many important roles in cancer.7-9 However, it has become increasingly clear that mammalian genomes encode also numerous long ncRNAs, defined as endogenous cellular RNAs of more than 200 nucleotides in length that lack an open reading frame of significant length (less than 100 amino acids).10-13 Therefore, long ncRNAs (lncRNAs) constitute a very heterogeneous group of RNA molecules that allows them to cover a broad spectrum of molecular and cellular functions by implementing different modes of action.
Originally, lncRNAs were discovered via large-scale sequencing of full-length cDNA libraries in the mouse.14 Other names such as large RNA, macroRNA and long intergenic ncRNA (lincRNA) are also used to refer to these. LncRNAs often overlap with or are interspersed between coding and non-coding transcripts.4,15,16 From a genetic point of view long, ncRNAs fall into one or more of five broad categories: (1) sense, or (2) antisense, when overlapping one or more exons of another transcript on the same or opposite strand, respectively; (3) bidirectional, when the expression of it and a neighboring coding transcript on the opposite strand is initiated in close genomic proximity, (4) intronic, when derived from an intron of a second transcript; or (5) intergenic, when it lies as an independent unit within the genomic interval between two genes.12
Several studies were conducted to identify lncRNAs in the human genome.17-24 A recent study identified 5446 lncRNA genes in the human genome and combined them with long ncRNAs from four published sources to derive 6736 long ncRNA genes.25 This study also investigated the protein-coding capacity of known genes overlapping with lncRNAs and revealed that 62% of known genes with “hypothetical protein” names lacked protein-coding capacity and this might even further enlarge the catalog of human lncRNAs.
For the vast majority of these recently discovered lncRNAs, the cellular function needs to be elucidated. For each individual molecule, it needs to be established whether it executes important functions or whether it just represents “transcriptional noise” or background transcription. Very convincing arguments that support the idea of functional relevance are conservation and regulation. In fact, some lncRNAs show clear evolutionary conservation or strict regulation, implying that they are of functional importance.26-29 In addition, some transcripts are derived from ultra-conserved genomic regions (UCR) and these T-UCRs can be altered in human cancer.30,31
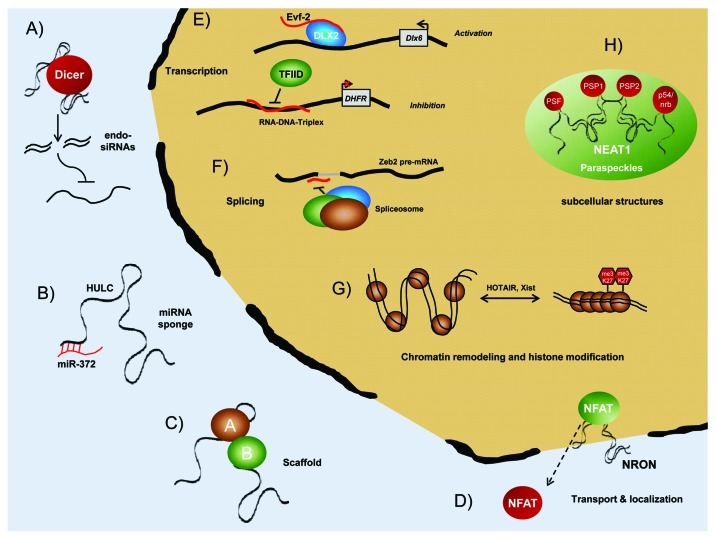
Also, long ncRNAs are often expressed in a disease-, tissue- or developmental stage-specific manner making these molecules attractive therapeutic targets and pointing toward specific functions for lncRNAs in development and diseases.32-34 Nevertheless, our knowledge of how lncRNAs can act in the cell and which roles they might play in diseases, e.g., cancer is still very limited. However, recently it became obvious that lncRNAs play an important role in regulating gene expression at various levels, including chromatin modification, transcription and post-transcriptional processing.16,35 For example, the lncRNAs Xist (X inactive-specific transcript) or HOTAIR (HOX Antisense Intergenic RNA) interact with chromatin remodeling complexes to induce heterochromatin formation in specific genomic loci leading to reduced target gene expression.23,36-38 Long ncRNAs can also function by regulating transcription through a variety of mechanisms that include interaction with RNA binding proteins, acting as a co-activator of transcription factors, or repressing a major promoter of their target gene.39-41 In addition to chromatin modification and transcriptional regulation, long ncRNAs can modulate gene expression at the post-transcriptional level or splicing level.42-44 In Figure 1 we provide an overview about lncRNA functions. However, this is only a snapshot of our current knowledge and more functional insights might be obtained in the future. Consequently, if we want to fully understand the complex mechanisms underlying malignant processes, e.g., carcinogenesis, metastasis and drug resistance, we need to consider this family of regulatory transcripts that add a new layer of complexity to biology.
Figure 1. Cellular functions of long ncRNAs. LncRNAs can act in diverse ways in the cell. In general, they can regulate gene expression, influence protein localization (D) and are important for the formation of cellular substructures or protein complexes, where they fulfill scaffolding functions (C; H).199 Regulating gene expression is one of the best studied functions of lncRNA and multiple mechanisms are applied by lncRNAs. (A) LncRNAs coul be processed into small, single- or double-stranded RNAs that could act as endo-siRNAs targeting other RNAs, which subsequently leads to target degradation. (B) LncRNAs can act as “miRNA sponge” and sequester miRNAs to inactivate these small regulatory RNAs. This influences the expression of miRNA target genes.200 (D) The interaction of lncRNAs with proteins they can modulate protein activity and localization. For example, the lncRNA NRON (non-coding repressor of NFAT) binds to the cellular transcription factor NFAT (nuclear factor of activated T cells). This regulates nuclear-cytoplasmic trafficking of NFAT and finally leading to an repression of NFAT target gene expression.201 (E) Furthermore, lncRNA regulate gene transcription via recruiting transcription factors to their target gene promoters, therefore activating gene expression.39 However, they can also block binding of general transcription factors, potentially via formation of RNA-DNA-Triplexes.40 (F) LncRNAs contribute to transcriptome complexity, as they can regulate alternative splicing of pre-mRNAs.42 (G) The balance between transcriptional active euchromatin and silent heterochromatin is controlled by lncRNAs. They can interact with chromatin remodeling complexes and induce local or global changes in chromatin packaging.23,202
Hallmarks of cancer: The basics
Cancer is one of the leading causes of death worldwide and accounted for 7.6 million (13% of all deaths) in 2008.45 In the US, for example, lifetime probability of developing cancer is ~44% for men or ~38% for women, respectively.46 Determining the causes of cancer is a complex issue, but well-known risk factors are alcohol and tobacco abuse, infections, radiation, obesity and a lack of physical activity.
Although “cancer” comprises a heterogeneous group of diseases, one characteristic and unifying feature is the creation of abnormal cells that grow beyond their natural boundaries. In 2000, Hanahan and Weinberg proposed six hallmarks of cancer that all together form the fundamental principle of this malignant transformation.47 Since tumor formation is a multistep process, normal cells evolve progressively to the neoplastic stage and along their way they acquire particular capacities that enable them to become tumorigenic. These basic hallmark capabilities, distinct and supplementary, are: (1) sustaining proliferative signaling; (2) evading growth suppressors; (3) enabling replicative immortality; (4) activating invasion and metastasis; (5) inducing angiogenesis and (6) resisting cell death. Over the last decade, remarkable progress was made in the field of cancer research which led to a better understanding of these hallmark capabilities, but also led to modifications and, ultimately, expansions of the original concept.48
In this review, we would like to further expand our thinking of the causes and consequences of cancer by introducing long ncRNAs into cancer biology. Thus, we will first summarize the conceptual basis of each hallmark and, second, highlight latest findings that connect lncRNAs with these fundamental capacities. Moreover, we will point out new areas of cancer research by including lncRNAs into basic scientific questions. Finally, we will introduce new therapeutic concepts that focus on lncRNAs as treatment targets.
Hallmarks of cancer: Adding long ncRNAs
Sustaining proliferative signaling
One of the most prominent characteristics of a cancer cell is its ability to proliferate constantly and in the absence of external stimuli. Normal cells carefully manage the production of growth promoting or inhibiting factors to ensure a tight control of cell number, tissue architecture and function. In contrast, tumor cells show deregulated signaling cascades that enable them to be more or less independent of proliferation signals, which results in unlimited growth. To achieve this independency, tumor cells acquired the capability to sustain proliferative signaling in multiple ways: (1) They produce their own growth factors and the corresponding receptor molecules themselves resulting in autocrine stimulation. (2) They might also produce paracrine signals to stimulate normal, tumor-associated cells (tumor stroma) which in turn produce various growth factors to support the cancer cells. (3) Moreover, growth factor receptor levels could be elevated or the receptor signaling cascade could be altered, making cancer cells hyperresponsive. (4) Last but not least, cancer cells could become completely independent from exogenous growth factors, because of constitutive activation of downstream signaling pathways or the disruption of negative-feedback mechanisms.
With this in mind, we can now ask the question: Which roles do long ncRNAs play here? A first answer to this arises from hormone signaling, especially sex steroid hormone signaling. Estrogen, progesterone and androgen are well-known hormones targeting female mammary glands, ovary and uterus or the male testis and prostate gland. They exert their functions in these tissues through the specific interaction with their intracellular receptors, the estrogen receptor (ER), the progesterone receptor (PR) and the androgen receptor (AR). This regulates target gene expression, as these receptors function as transcription factors and the abnormal expression or function of these receptors has been implicated in tumors of reproductive organs in both, males and females.49 As for many other transcription factors, the activity of these receptors is influenced by additional factors, so called coactivators or corepressors. While coactivators enhance the transcriptional activity, corepressors block their activity resulting in a complex interplay underlying coordinated gene expression. Changes in the expression level or pattern of these coactivators and corepressors can affect transcriptional activity of the steroid hormones and subsequently cause disorders of their target tissues.50-52
A unique coactivator for the steroid receptors PR, ER, GR (glucocorticoid receptor) and AR is the steroid receptor RNA activator (SRA). SRA acts as an ncRNA.53 It is part of an RNA-protein complex containing also SRC-1 and it fulfills its transactivation function through the AF-1 domain of the nuclear receptors. SRA expression can be detected in normal and malignant human mammary tissues. Interestingly, elevated levels of SRA are found in breast tumors and the increased SRA levels might contribute to the altered ER/PR action that occurs during breast tumorigenesis.54 However, recent progress in this field has revealed a more complex situation: the SRA1 gene might not only act as an ncRNA but also produces a protein that acts as a coactivator or corepressor, as well.55,56 Alternative splicing balances the ratio of non-coding and coding transcripts derived from the SRA1 gene.57 This balance of transcripts not only characterizes specific tumor phenotypes but might also be involved in breast tumorigenesis and tumor progression by regulating the expression of specific genes.58 This duality of RNA transcripts and the concept of coding and non-coding functions add another level of complexity and should be considered to gain deeper insights into complex regulatory circuits. In line with this concept, a recent report presented a novel, coding-independent function for the p53 mRNA.59 Usually, the E3 ubiquitin ligase Mdm2 is a negative regulator of p53 protein expression. However, Mdm2 bound to p53 mRNA shows a different activity: it promotes p53 expression following genotoxic stress. This is achieved because the p53 mRNA binding to Mdm2 controls Mdm2 SUMOylation and nuclear trafficking and the accumulation of Mdm2 in nucleoli. This plays an important role in p53’s capacity to respond to DNA damage.60,61 These two examples emphasize the importance of being open-minded and reflect RNA as a multi-functional molecule and not only an intermediate for protein synthesis.
In addition to SRA, there are several other long ncRNAs recently discovered which have a role in cell proliferation. In an exemplary study, Prensner et al.22 applied RNA-Seq technology and identified 121 differentially expressed long ncRNAs in prostate cancer whose expression patterns distinguished benign, localized and metastatic prostate cancer. Moreover, they characterized one long ncRNA, PCAT-1 (prostate cancer associated transcript 1), in more detail. PCAT-1 was highly upregulated in a subset of metastatic and high-grade localized prostate cancers. To further explore the functional role of this novel ncRNA, overexpression and knockdown experiments were performed, which resulted in a modest increase in cell proliferation in case of stable overexpression and consistently a reduced proliferation rate (25–50%) after siRNA-mediated depletion. Gene expression profiling after knockdown of PCAT-1 in LNCaP cells identified 255 genes upregulated and 115 genes downregulated by the loss of PCAT-1. Gene ontology analysis of the upregulated genes showed enrichment for gene sets associated with mitosis and cell cycle, whereas the downregulated genes had no significant associations. Taken together, these results suggest that PCAT-1 functions as transcriptional repressor for a subset of genes and thereby might contribute to prostate cancer progression.
A further example of an lncRNA impacting cell proliferation is introduced by a novel role for the well-known small nuclear RNA 7SK, also known as RN7SK. A key function of this ncRNAs is the regulation of transcription elongation via binding to the positive transcription elongation factor b (P-TEFb) which abolishes its positive effect on RNA Polymerase II transcription elongation.62,63 Now, HMGA1, a transcription factor and chromatin regulator, was identified as a novel 7SK interaction partner.64 HMGA1 (high mobility group AT-hook 1) itself shows high expression levels in both, embryonic and transformed neoplastic cells.65,66 In this recent study, 7SK RNA was shown to interact with HMGA1 and compete with its binding to DNA. This, in turn, has an impact on HMGA1 target gene expression affecting also growth-related genes. This again shows the diverse mechanistic functions of lncRNAs and underlines the need to develop new methods to identify and analyze these transcripts in more detail.
Finally, a recent study identified 216 putative long ncRNAs derived from promoter regions of cell cycle genes.67 Many of these transcripts showed periodic expression during the cell cycle and an altered expression in human cancers. Their expression is regulated by specific oncogenic stimuli, stem cell differentiation or DNA damage and future work will elucidate their molecular functions and their role in cancer cell proliferation.
Taken together, the newly discovered long ncRNAs are more and more recognized as active molecules instead of “transcriptional noise” and evidence is accumulating that some of them have critical roles in carcinogenesis by influencing tumor cell proliferation.
Evading growth suppressors
A highly complementary hallmark capability for sustaining proliferative signaling in cancer cells is the ability to evade growth suppression. Several tumor suppressive protein-coding genes that operate in diverse ways to inhibit cellular growth and proliferation had been discovered, e.g., TP53, PTEN or RB. Activation of these tumor suppressor genes depends on external or internal stimuli and can either lead to cell cycle arrest or might induce senescence and even apoptosis in cells. Therefore, tumor cells must find a way to prevent their activation or expression. One mechanism how cancer cells deal with this is the complete loss of the tumor suppressor gene or the accumulation of mutations that render this gene inactive. In fact, more than 50% of human tumors contain a mutation or deletion of the TP53 gene and people who inherited only one functional copy of the TP53 gene will most likely develop tumors in early adulthood, a disease known as Li-Fraumeni syndrome.68 Alternatively, tumor suppressor genes could also bind to other proteins expressed in tumor cells and therefore become inactivated or rapidly degraded. One such example is given by the human papilloma virus (HPV) oncogenes E6 and E7, which are preferentially expressed in human cervical cancers. They interact with TP53 and RB, which leads to their inactivation.69 Also, Mdm2 as mentioned earlier is the predominant E3 ubiquitin ligase of TP53 and therefore induces its degradation via the proteasome.70
In addition to these mechanisms, cancer cells have developed alternative ways to inhibit tumor suppressor functions with the help of long ncRNAs. For instance, five human ncRNA fragments interact with the tumor suppressor PSF.71 The PSF protein represses transcription of proto-oncogenes via binding to their regulatory regions. It contains a DNA-binding domain and two RNA-binding domains that bind VL30–1 RNA in mice, which leads to the release of PSF from a repressed proto-oncogene and activates transcription of the latter ones.72-74 However, VL30–1 RNA is not encoded in the human genome and Li et al. aimed at identifying human RNA counterparts that interact with PSF protein.71 In their screen, they found five RNA fragments associated with PSF and releasing it from the human proto-oncogene GAGE6 regulatory region resulting in an activation of GAGE6 expression. Overexpression of these RNA fragments in a human melanoma cell line led to an enhanced tumorigenic phenotype. In an additional experiment, the relative amounts of the PSF-binding RNAs were determined in two human fibroblast cell lines and nine human tumor cells. Each tumor cell line expressed higher levels of PSF-binding RNAs compared with normal human fibroblasts. The pattern of expression differed among the tumor lines, suggesting that different groups of PSF-binding RNAs contribute to the tumorigenicity of each tumor cell line. Interestingly, the amount of PSF mRNA was not changed in eight out of nine tumor cell lines compared with fibroblasts, suggesting that not a decrease of PSF protein contributes to the tumorigenicity of human tumor cells but rather the increased expression of its interacting RNAs. Future work should therefore focus on protein-RNA interactions and reveal their molecular consequences.
A completely different mode of action is executed by the long ncRNA ANRIL (antisense non-coding RNA in the INK4 locus) to block the activity of tumor suppressor genes.75 Instead of competing with DNA for binding to the repressor protein, ANRIL interacts with SUZ12 (suppressor of zeste 12 homolog), a subunit of the polycomb repression complex 2 (PRC2) and recruits the complex to repress the expression of p15 (INK4B), a well-known tumor suppressor gene. Moreover, the depletion of ANRIL increases the expression of p15(INK4B) and inhibits cellular proliferation. A similar study identified CBX7 (chromobox homolog 7), a subunit of the polycomb repressive complex 1 (PRC1) as molecular interaction partner of ANRIL. This results in the recruitment of PRC1 to the p16(INK4A)/p14(ARF) locus and subsequent silencing of this gene locus by H3K27-trimethylation.76 Both, CBX7 and ANRIL show elevated levels in human prostate cancer corroborating the importance of this interaction for tumor development. In line with these findings, a recent study from Jeannie T. Lee’s lab identified > 9000 PRC2-interacting RNAs via RIP-Seq (RNA immunoprecipitation and sequencing) technology in embryonic stem cells making it very likely that more and more genes will be identified that are regulated by the ncRNA-mediated recruitment of PRC2 and it will be interesting to see how this relates to carcinogenesis.38 However, an unanswered question so far is how ncRNAs can specifically recruit these silencing machineries to only a subset or even only one specific gene.
In addition to these active oncogenic functions of long ncRNAs there are also tumor suppressor functions assigned to them. One very famous example is the ncRNA GAS5 (Growth Arrest-Specific 5). It was originally identified based on its increased levels in growth-arrested mouse NIH3T3 fibroblasts.77 Conversely, GAS5 expression is strongly reduced in actively growing leukemia cells or NIH3T3 cells and in turn increases after density-induced cell cycle arrest.78 The human GAS5 gene is transcribed from chromosome 1q25.1 and is alternatively spliced. Its exons contain a small and poorly conserved open reading frame that does not encode a functional protein.79,80 GAS5 is a host gene for multiple snoRNAs, which are located in the introns and may mediate important biological activities.81 In contrast to SRA, GAS5 functions as a “riborepressor”: the ncRNA interacts with the DNA binding domain of the glucocorticoid receptors, thus competing with the glucocorticoid response elements in the genome for binding to these receptors. This suppresses the induction of several responsive genes including cellular inhibitor of apoptosis 2 (cIAP2) and ultimately sensitizes cells to apoptosis.82 Moreover, GAS5 expression induces growth arrest and apoptosis independently of other stimuli in some prostate and breast cancer cell lines.83 Interestingly, the inhibition of mammalian Target Of Rapamycin (mTOR) pathway via Rapamycin depends on GAS5. The mTOR pathway plays a critical role in control of cell growth and regulates cellular protein synthesis and proliferation.84,85 Downregulation of GAS5 by RNA interference protects both leukemic and primary human T cells from the anti-proliferative effect of Rapamycin, suggesting that GAS5 might–directly or indirectly–be required.86 How do cancer cells cope with this tumor-suppressive ncRNA? Breast cancers show a significantly lower GAS5 expression compared with normal breast epithelial tissues.83 In addition, genetic aberrations of the GAS5 locus have been found in many types of tumors including melanoma, breast and prostate cancers but their functional significance still needs to be established.87-89
However, GAS5 is not the only ncRNA with growth-suppressive functions. John Rinn and colleagues identified several novel ncRNAs by asking how the transcription factor TP53 can do both, activate and repress gene expression.28 One answer to this question was given by the discovery of lincRNA-p21. This ncRNA is a direct p53 target gene residing next to the p21 (Cdkn1a) gene on mouse chromosome 17. Its expression is activated upon DNA damage in different tumor models. LincRNA-p21 associates with hnRNP K, a well-known RNA binding protein and hnRNP K acts as a transcriptional repressor. LincRNA-p21 mediates the binding of hnRNP K to its target genes, which finally leads to gene silencing and the induction of apoptosis. However, this study was conducted in mice and although lincRNA-p21 seems to be conserved and is also induced in human fibroblasts after DNA damage induction, it needs to be determined, whether a similar mechanism can be found in normal human cells. Moreover, it will be interesting to see how human cancers regulate this tumor suppressive ncRNA, especially in TP53-positive cancers that show an intact TP53 response.
Interestingly, the expression of the tumor suppressor p21 is regulated by at least one other non-coding RNA transcript. Kevin Morris and coworkers found that transcriptional activation of p21 gene depends on the post-transcriptional silencing of a p21-specific antisense transcript, which functions in Argonaute 1-mediated transcriptional control of p21 mRNA expression.90 In human cells, this bidirectional transcription could be an endogenous gene regulatory mechanism whereby an antisense RNA directs epigenetic regulatory complexes to a sense promoter, resulting in RNA-directed epigenetic gene regulation. The epigenetic silencing of tumor suppressor genes, such as p21, may be the result of an imbalance in bidirectional transcription levels and this imbalance allows the antisense RNA to direct silent state epigenetic marks to the sense promoter, resulting in stable transcriptional gene silencing.
In summary, these examples clearly demonstrate a role for long ncRNAs in evading growth suppressors and tumor cells must find ways to circumvent both, the activation of protein and non-protein tumor suppressor genes.
Enabling replicative immortality
In line with the first two hallmarks of cancer and closely related to proliferation is the third trait of cancer: unlimited replicative potential. In contrast to normal cells that are able to pass through only a limited number of cell division cycles, tumor cells show nearly unlimited replication. In normal cells, replication potential is limited due to the appearance of either senescence or crisis with the latter one finally ending in cell death. The chromosome ends, the telomeres, are crucial for this replication limit.91,92 In vertebrates, these sequences are composed of multiple repeats of the hexanucleotide “TTAGGG” and protect the ends of chromosomes from end-to-end fusions.93 However, in normal cells, these telomeric repeats shorten after each cell division and therefore the length of telomeric DNA dictates the number of cell division cycles.94 The loss of these protective ends finally leads to crisis. Tumor cells have found two ways to circumvent the loss of telomeres: (I) About 90% of all human cancers express a specialized enzyme, called telomerase, which is able to add telomeric repeats to the end of the chromosomes. (II) The remaining 10% of all tumor cells employs alternative lengthening of telomeres (ALT), a non-conservative telomere lengthening pathway involving the transfer of telomere tandem repeats between sister chromatids.95-98 The exact mechanism of the ALT pathway still needs to be uncovered. However, it could be shown that ALT cells produce abundant t-circles, possible products of intratelomeric recombination and t-loop resolution providing mechanistic insights.99,100
Interestingly, the major pathway involving telomerase critically depends on an ncRNA. The telomerase holoenzyme consists of a protein component, a reverse transcriptase named TERT (Telomerase Reverse Transcriptase) and an RNA primer, also known as TERC (Telomerase RNA Component) or TR (Telomerase RNA).101 The telomeric RNAs are highly divergent between different species, varying in both size and sequence composition, from:150 nt in ciliates and:450 nt in vertebrates up to:930–1300 nt in the budding yeast. Telomere synthesis involves TERT-catalyzed reverse transcription of a small template region within TERC making it essential for immortalization. Not astonishing, the TERC gene is amplified in several human cancers, thus making it a potential target for therapeutic inhibitors of telomerase activity in cancer cells.102-104 For a more detailed discussion on TERC structure and function please see refer to Theimer and Feigon.105 For a more comprehensive summary on telomerase structure and function the interested reader is referred to Wyatt, West and Beattie.106
TERC is not the only RNA associated with telomeres. Recently, a group of long ncRNAs was discovered and named TERRA (telomeric repeat-containing RNA).107-109 TERRA transcripts are derived from several subtelomeric loci. Telomere transcription is an evolutionarily conserved phenomenon in eukaryotic cells suggesting functional importance.110 TERRA localizes to telomeres and is involved in telomeric heterochromatin formation.111 TERRA is thought to be a negative regulator of telomerase, which may act globally or at individual telomeres as a direct inhibitor of the telomerase enzyme.112 HnRNP A1 is a protein interaction partner of TERRA and together with POT1 (protection of telomeres 1), they act in concert to displace RPA (replication protein A) from telomeric ssDNA after DNA replication to promote telomere capping and preserve genomic integrity.113 Reduction of TERRA transcription is necessary for telomerase-mediated telomere lengthening which may link TERRA to cancer. Telomerase-positive cancer cells with high levels of subtelomeric methylation display low levels of TERRA compared with matched ALT-positive cancer cells or normal cells.114 Moreover, when cell extracts are incubated with an excess of synthetic RNA oligonucleotides mimicking TERRA, telomerase activity is inhibited.109 However, a deregulation, silencing or mutation of TERRA in human cancer remains to be discovered. For a more detailed overview about TERRA the interested reader is referred to Caslini.110
Taken together, both introduced ncRNAs, TERC together with TERT forming a functional ribozyme and TERRA, which comprises a group of ncRNA transcripts, demonstrate once more the broad biological importance of long ncRNAs–here as regulators of genome stability and replication.
Activating invasion and metastasis
The fourth hallmark capability, the ability to invade and form distant metastases, is a highly challenging one and underlies a plethora of complex interactions and regulatory mechanisms. It was noted decades ago that epithelial carcinomas of higher grade show a more invasive phenotype and distant metastases and most patients die from these metastases and not from the primary tumor. Often, cancer cells undergo morphological alterations and change their cell-cell or cell-matrix interactions. All this enables them to successfully pass through the first steps of the multistep process of invasion and metastasis. This invasion-metastasis cascade comprises multiple biological changes that allow cancer cells to invade into healthy tissues, followed by intravasation into blood and lymphatic vessels.115,116 During their transit through the lymphatic system and blood circulation, cancer cells must escape immune surveillance and show anchorage-independent growth and survival. Next, cancer cells must extravasate from the vessels into their target tissues to form micrometastases and eventually lateron a secondary tumor. Much progress was made over the last years and interesting concepts, e.g., the regulation of invasion by the developmental regulatory program known as “epithelial-mesenchymal transition” (EMT) were put forward.117-119 Also, several important factors of this cascade had been identified. For example, E-cadherin (CDH1) is a key cell-to-cell adhesion molecule and helps to assemble epithelial cell layers. An increased E-cadherin expression therefore inhibits invasion and metastasis formation. E-cadherin expression itself is regulated in multiple ways and involves also a natural antisense transcript (NAT) that regulates expression of Zeb2, a transcriptional repressor of E-cadherin.42 Not surprisingly, E-cadherin is frequently downregulated or inactivated in human carcinomas.120,121 In addition to E-cadherin, several other adhesion molecules that either mediate cell-to-cell or cell-to-matrix interactions are altered in some aggressive carcinomas. On the other hand, N-Cadherin (CDH2), which is found in migrating neurons and mesenchymal cells, is often upregulated in many invasive tumors. Besides these alterations within cancer cells, it becomes more and more obvious that also crosstalk between cancer cells and tumor stroma cells is required for this hallmark capability.122-125 Also, cells of the immune system contribute to this capability by supplying, e.g., matrix-degrading enzymes and in that way support invasive tumor cell behavior.123,126,127 One of the major tasks for the future is to identify a “metastatic signature” of genes that support the invasion of tumor cells and facilitate the formation of distant metastases in specific tissues.
As depicted earlier, some protein factors with important functions could already be identified. In addition, early work by Ji and Diederichs et al.128 established a role for long ncRNAs in metastasis formation. They identified MALAT1 (Metastasis-Associated Lung Adenocarcinoma Transcript 1, MALAT-1), also later referred to as NEAT2 (Nuclear-Enriched Abundant Transcript 2) as a prognostic marker for metastasis and patient survival in non-small cell lung cancer (NSCLC). This ncRNA is extremely abundant in many human cell types and is highly conserved across several species underscoring its functional importance.44,129 Moreover, the 8 kb long MALAT1 transcript can be processed into a highly conserved tRNA-like small cytoplasmic RNA of 61 nucleotides that is broadly expressed in human tissues.130 However, the function of this so-called mascRNA is so far unknown.
MALAT1 is retained in the nucleus and specifically localizes to nuclear speckles.29 These structures play a role in pre-mRNA processing and recently MALAT1 has been shown to regulate alternative splicing of pre-mRNA by modulating the levels of active serine/arginine splicing factors.44 These factors regulate tissue- or cell-type specific alternative splicing in a concentration- and phosphorylation-dependent manner. Depletion of MALAT1 alters the processing of a subset of pre-mRNAs, which play important roles in cancer biology, e.g., Tissue Factor or Endoglin.131 This supports the hypothesis that MALAT1 could be a regulator of post-transcriptional RNA processing or modification. However, a recent study from the Rosenfeld lab indicates additional functions for MALAT1 in the nucleus.132 Here, MALAT1 was shown to interact with the unmethylated form of CBX4 and this controls relocation of growth-control genes between polycomb bodies and interchromatin granules, places of silent or active gene expression respectively. Therefore, the exact mechanism of MALAT1 function is still a mystery and it seems not unlikely that this lncRNA might fulfill cell type- or tissue-specific functions.
MALAT1 expression can be found in many healthy organs with the highest levels of expression in pancreas and lung.128 In several human cancers including lung cancer, uterine endometrial stromal sarcoma, cervical cancer and hepatocellular carcinoma (HCC), MALAT1 is upregulated.128,133-135 In addition, it is significantly associated with metastasis in NSCLC patients. This association with metastasis is stage- and histology-specific. Therefore, MALAT1 can serve as an independent prognostic parameter for patient survival in early stage lung adenocarcinoma.128 A recent study shed light onto the role of the metastasis marker MALAT1 as a potentially active player in the metastatic process. MALAT1 promotes cell motility of lung cancer cells through transcriptional or post-transcriptional regulation of motility-related genes.136 Additionally, MALAT1 supports proliferation and invasion of cervical cancer cells and knockdown of MALAT1 in CaSki cells led to an upregulation of caspase-8 and -3 and Bax and the downregulation of Bcl-2 and Bcl-xL.133 Thus, both studies find a plethora of potential MALAT1 functions linked to proliferation, apoptosis, migration or gene regulation and future studies will have to unravel the specificity of these effects. Since both studies were performed with individual siRNAs only and lack rescue experiments, further investigations are necessary to ensure the specificity of the observed effects and to corroborate the functional importance of MALAT1 in carcinogenesis or metastasis. For this reason, we have recently developed a novel gene knockout strategy based on the stable bi-allelic integration of RNA destabilizing elements into the human genome with the help of so called Zinc Finger Nucleases.129 This method yielded a highly specific and more than 1000-fold reduction of MALAT1 expression in human A549 lung cancer cells and will allow deeper analysis of MALAT1 function in lung cancer and its role in migration and metastasis in a clean loss-of-function model.
A second long ncRNA involved in cancer metastasis is known as HOTAIR (HOX Antisense Intergenic RNA). It was discovered by the lab of Howard Chang as a 2.2 kb long ncRNA transcribed in antisense direction from the HOXC gene cluster.23 The same study also revealed that HOTAIR functions in trans by interacting and recruiting the polycomb repressive complex 2 (PRC2) to the HOXD locus which leads to transcriptional silencing across 40 kb. Later, HOTAIR was found to interact with a second histone modification complex, the LSD1/CoREST/REST complex, which coordinates targeting of PRC2 and LSD1 to chromatin for coupled histone H3K27 methylation and K4 demethylation.37 Given its important role in the epigenetic regulation of gene expression, it is not surprising that HOTAIR is deregulated in different types of cancer.36,137-139 In human breast cancer, HOTAIR expression is increased in primary tumors and metastases and its expression level in primary tumors positively correlates with metastasis and poor outcome. Overexpression of HOTAIR in epithelial cancer cells alters H3K27 methylation via PRC2 and therefore alters target gene expression. This leads to increased cancer invasiveness and metastases. On the other hand, HOTAIR depletion inhibits cancer invasiveness.36 In HCC, HOTAIR levels are increased compared with non-cancerous tissues and for those HCC patients, who received a liver transplantation, high HOTAIR expression levels are an independent prognostic marker for HCC recurrence and shorter survival.137 Similar to breast cancer, HOTAIR depletion in liver cancer cells reduces cell invasion and cell viability. Moreover, HOTAIR might be a potential biomarker for the existence of lymph node metastasis in HCC.140 In addition, HOTAIR suppression sensitizes cancer cells to tumor necrosis factor α induced apoptosis and renders them more sensitive to the chemotherapeutic agents cisplatin and doxorubicin.137
These two examples indicate already the functional importance of long ncRNAs for the activation of invasion and metastasis formation. Hence, future studies should foster the identification of such transcripts, e.g., in the context of epithelial-to-mesenchymal transition. LncRNAs could be important regulated genes during EMT or act as possible regulators for other EMT-relevant genes. In addition, ncRNAs could be involved in the complex interplay of tumor cells with the tumor stroma or might also influence inflammatory cell behavior. HOTAIR and MALAT1 could regulate a broad subset of genes involved in cancer cell invasion and metastasis. So, these lncRNAs are on the one hand part of the “metastatic signature” of gene expression and on the other hand they contribute to shaping it.
Inducing angiogenesis
When cancer cells grow and proliferate, tumor mass and size increases. This process would be limited by the natural diffusion limit of oxygen and nutrients, if tumor cells would not acquire the fifth trait: the ability to induce angiogenesis. The formation of new blood vessels, induced by tumor cells, secures not only a supply with nutrients and oxygen, but allows tumors to dispose their metabolic (toxic) wastes and enter the hematogenous metastatic process, as well. The processes of vasculogenesis and angiogenesis are usually restricted to embryonic development, but can become re-activated under specific conditions in adults. For example, angiogenesis is an important process in wound healing and female reproductive cycling. An “angiogenic switch” is also turned on during tumor progression, which leads to continuous sprouting of new vessels that help to sustain tumor growth.141 To achieve this, tumor cells induce pro-angiogenic factors or block antiangiogenic signals.142 Some of these angiogenic regulators are signaling proteins that bind to activating or inhibiting receptors present on the surface of endothelial cells.143 One well-known activator of angiogenesis is VEGF-A (vascular endothelial growth factor-A), a secreted protein whose expression can be induced by hypoxia or oncogenic signals.144 Its effect could be counterbalanced by Thrombospondin-1, which also binds to transmembrane receptors displayed by endothelial cells.145 Thus, the expression of these factors needs to be tightly controlled and it is noteworthy that oncogenes like Ras and Myc contribute to proliferative signaling and angiogenesis. Therefore, it will be interesting to see, whether long ncRNAs with a role in cellular proliferation, as discussed above, will also play a role in tumor angiogenesis.
That long ncRNAs can also have functions in regulating the angiogenic process is becoming more and more evident. Nearly a decade ago, it was shown that aHIF, a natural antisense transcript (NAT) complementary to the 3′ untranslated region of the hypoxia inducible factor α (HIF1α), negatively regulates the expression of HIF1α, a critical regulator of angiogenesis.146,147 Overexpression of aHIF triggers HIF1α mRNA decay and HIF-1a and aHIF constitute a negative feedback loop.148 Interestingly, aHIF transcripts can be detected in several human tissues and cancers and αHIF is a marker for poor prognosis in breast cancer.146,147,149
However, aHIF is not the only antisense RNA associated with angiogenesis. Another NAT was discovered when studying the transcriptional unit of the human endothelial nitric-oxide synthase (eNOS). The transcript was termed sONE or NOS3AS and it regulates the expression of eNOS in a post-transcriptional manner under normoxia and hypoxic conditions.150,151 In contrast to eNOS, sONE expression is detectable in a variety of cell types except endothelial cells. In tumors, protumorigenic agents, such as estrogen, induce eNOS expression and several cancer treatment methods influence eNOS expression and activity.152 For a more detailed overview about eNOS function in cancer biology, please refer to Ying and Hofseth.153 At the moment it is a matter of debate, whether sONE really acts as RNA, because also a protein product derived from this RNA has recently been described.154
Finally, a very recent study identified an NAT for tyrosine kinase containing immunoglobulin and epidermal growth factor homology domain-1 (tie-1), tie-1AS.155 This long ncRNA is conserved in zebrafish, mouse and humans where it selectively binds to tie-1 mRNA in vivo and regulates tie-1 transcript levels, resulting in specific defects in endothelial cell contact junctions in vivo and in vitro. In addition, the ratio of tie-1 vs. tie-1AS is altered in human vascular-related disease states and it would be interesting to know if and how cancer cells can manipulate this ratio to achieve proper blood vessel formation.
The presented examples directly implicate long ncRNA-mediated post-transcriptional regulation of gene expression as a fundamental control mechanism for physiological processes, such as vascular development and show their importance for cancer development. Future work should focus on the identification of novel long ncRNAs regulated by or being involved in the “angiogenic switch” of cancer cells. Most important questions are: (1) Can HIF1α, the major transcriptional regulator under hypoxic conditions, actively regulate long ncRNA expression? (2) Are lncRNAs involved in recruiting this transcription factor to its target genes? (3) Which signaling cascades can regulate lncRNA expression in endothelial or tumor cells, e.g., is VEGF signaling involved? (4) Can lncRNAs act as second messengers, i.e., can they function in a cytokine-like way to activate or repress endothelial cells or other cells involved in tumor angiogenesis like immune cells? (5) Are long ncRNAs involved in signal transduction pathway relevant for blood vessel formation?
Finding answers to these and similar questions will broaden our view on the complex interplay between tumor cells and supporting tumor stroma and could provide new anti-angiogenic treatment options.
Resisting cell death
The dream of immortality–for cancer cells it can come true, if they acquire the last hallmark capability: resisting cell death. Three major pathways can lead to cell death and their activation must be carefully controlled by tumor cells. The first mechanism leading to controlled cell death is apoptosis. Apoptosis can be induced by various external as well as internal stimuli and several studies have shown how highly malignant cancers can attenuate apoptosis and become therapy resistant.156,157 Induction of DNA damage, e.g., by chemotherapeutic agents (cisplatinum, etoposide, etc.) is one way to trigger apoptosis via the TP53 (p53) pathway. TP53 induces the expression of pro-apoptotic proteins Noxa and Puma (p53-upregulated modulator of apoptosis) leading to cell death. As mentioned earlier, 50% of all human cancers have either lost the TP53 gene or show mutations.68 This makes them more resistant to such cellular stresses. Alternatively, tumors show an increased expression of survival factors or anti-apoptotic regulators like Bcl-2 and Bcl-xL. A second mechanism leading to controlled cell death is autophagy. This mechanism usually operates at low levels in cells, but it can be activated by certain kinds of cellular stress, e.g., nutrient deficiency.158 Autophagy can be seen as a recycling program that allows cells to break down their organelles and to use the degradation products to fuel biosynthesis pathways or use for energy production. Several regulatory components and effector proteins are known and the interested reader might refer to Levine and Kroemer and Mizushima.158,159 Interestingly, autophagy can have both, beneficial effects for cancer cells or block carcinogenesis. For example, mice lacking the Beclin-1 gene, a critical factor for autophagy induction, show increased susceptibility to cancer. The same holds true for other components of the autophagy machinery.158,160 On the other hand, severely stressed cells shrink via autophagy to a state of reversible dormancy and this survival response might be the reason for cancer recurrence after therapy.160 The last mode of cell death is constituted by necrosis. Although often referred to as “uncontrolled” cell death, more and more evidence is accumulating that also necrosis is a controlled process rather than an undirected way to die.161,162 Similar to autophagy, necrosis can do both–eliminate cancer cells or promote their expansion. Necrotic cells can attract proinflammatory cells, which in turn can activate angiogenesis, cancer cell proliferation and invasiveness.161,163 Thus, more studies are necessary to fully understand the double-edged nature of this process and how it can be manipulated to achieve a beneficial effect for the patient.
Maybe long ncRNAs can provide some insights here, as they were already shown to influence cell death decisions. Over a decade ago, PCGEM1 (Prostate-specific transcript 1) was identified as a prostate tissue-specific and prostate cancer-associated long ncRNA.164 Later studies provided some insights into its function. They ruled out a role for this ncRNA in cell proliferation and colony formation and established a function for this ncRNA in apoptosis inhibition after doxorubicin treatment of prostate cancer cells.33,165 The anti-apoptotic effect might result from a delayed induction of p53 and p21(Waf1/Cip1) after PCGEM1 overexpression in LNCaP cells. In addition, cleaved caspase 7 and cleaved PARP levels were strongly reduced after doxorubicin treatment in PCGEM1 transfected cells compared with normal cells. This PCGEM1-associated delay in apoptotic responses seems to be androgen-dependent, as androgen-independent variants of LNCaP cells did not exhibit this delay.
However, PCGEM1 is not the only long ncRNA with anti-apoptotic functions. A more global approach using differential display identified genes conveying drug resistance to cancer cells.166 This led to the discovery of CUDR (cancer upregulated drug resistant), an ncRNA that confers resistance to doxorubicin and etoposide as well as drug-induced apoptosis in squamous carcinoma cells A431. One possible explanation for this function might be the observed downregulation of effector caspase 3 after CUDR overexpression.
A study from George Calin and Carlo Croce indicates that more long ncRNAs can be discovered that play a role in cell death control and that this might be an evolutionary conserved function.31 Their study identified several hundred transcripts derived from ultraconserved regions (T-UCRs), which are consistently altered at the genomic level in human leukemias and carcinomas. More importantly, they examined the biologic effect of the ncRNA uc.73A(P), a significant upregulated T-UCR in colon cancer. Depletion of this ncRNA resulted in reduced cellular proliferation of COLO-320 cells and an increase in sub-G1 cells, suggesting higher apoptosis rates. Thus, uc.73A(P) could act as an oncogene by increasing the number of malignant cells as a consequence of reduced apoptosis.
Another study shows that long ncRNAs have a plethora of functions in tumorigenesis including apoptosis prevention.167 Here, several long ncRNA, which are differentially expressed in melanoma cell lines in comparison to melanocytes and keratinocytes had been discovered. One of these long ncRNAs, SPRY4-IT1, is derived from an intron of the SPRY4 gene. SPRY4-IT1 is predominantly localized in the cytoplasm of melanoma cells, and SPRY4-IT1 knockdown results in defects in cell growth, differentiation, and higher rates of apoptosis in melanoma cell lines.
Finally, DNA damage can induce five long ncRNAs from the p21 promoter, and one of these was named PANDA (P21 associated ncRNA DNA damage activated).67 Further experiments showed that PANDA acts in trans via interaction with the transcription factor NF-YA and limits the expression of pro-apoptotic genes. Consequently, PANDA depletion markedly sensitized human fibroblasts to apoptosis by doxorubicin. In contrast, DNA damage can also lead to the activation of cis-acting ncRNAs. Wang et al. showed that ncRNAs induced by DNA damage derive from regulatory regions of the human CCND1 promoter and bind to TLS (translocated in liposarcoma) protein.41 TLS in turn inhibits cyclin D1 expression via interaction with and inhibition of CBP (CREB-binding protein) and p300.
Future studies, conducted to understand cell death regulation, will have to include long ncRNAs into the analyses to achieve a complete overview about activators and inhibitors of this important cancer hallmark. Especially, the growing fields of autophagy and necrosis research are promising areas to unravel further functions for long ncRNAs in healthy and malignant states.
All in all, the presented studies strongly emphasize the functional importance of long ncRNAs and provide first mechanistic insights how long ncRNAs can contribute to the hallmark capacities of cancer cells. A complete list with the herein described lncRNAs and additional examples can be found in Table 1. An extensive list of lncRNAs with a connection to cancer is provided in an overview article from Spizzo et al.168
Table 1. LncRNAs and the hallmarks of cancer.
| Cancer Hallmark | LncRNA | Mode of action | Reference |
|---|---|---|---|
| I. sustaining proliferative signaling |
SRA PCAT-1 RN7SK ncRNAs derived from cell cycle gene promoters KRASP1 PR antisense |
Transcriptional co-activator Regulating gene expression Regulating transcription Unknown miRNA sponge Regulating gene expression |
53, 54, 58 22 62–64 67 203 204 |
| II. Evading growth suppressors |
PSF-interacting RNA ANRIL GAS5 lincRNA-p21 E2F4 antisense |
Modulating protein activity Chromatin remodeling Competitor Transcriptional co-repressor Regulating gene expression |
71 75, 76 77–83, 86–89 28 205 |
| III. Enabling replicative immortality |
TERC TERRA |
RNA primer Enzymatic inhibitor |
101–103, 105 107–114 |
| IV. Activating invasion and metastasis |
MALAT1 HOTAIR HULC BC200 |
Modulating protein activity; sensor; scaffold Chromatin remodeling miRNA sponge Translational modulator |
29, 44, 128–136 23, 36, 137 173, 174, 200 206, 207 |
| V. Inducing angiogenesis |
aHIF sONE/NOS3AS tie-1AS ncR-uPAR |
RNA decay RNA decay RNA decay Regulating gene expression |
146–149 150, 151, 154 155 208 |
| VI. Resisting cell death |
PCGEM1 CUDR uc.73A(P) SPRY4-IT1 PANDA LUST PINC |
Regulating gene expression Regulating gene expression Unknown Unknown Modulating protein activity RNA-Splicing unknown |
33, 164, 165 166 31 167 67 209 210 |
Next, we want to provide a brief overview on what is needed for a better understanding of long ncRNA biology, including their discovery and functional analysis.
Long ncRNAs: Future challenges
Long ncRNAs have proven to be important regulators in health and disease. However, only individual examples have been functionally studied in detail so far and many important questions remain to be addressed. Biochemical analyses are needed to explore the functions and mechanisms of action of this novel class of biomolecules. This is where long ncRNAs raise a lot of challenges due to their mechanistic heterogeneity that is just beginning to emerge from the first discoveries in this field. However, a good way to start with are classical loss-of-function or gain-of-function studies using RNA interference to knockdown long ncRNAs, genetic loss-of-function models or overexpression. Subsequent analyses of tumor-relevant phenotypes such as proliferation, migration, cell viability or apoptosis could provide first insights into ncRNA functions. However, RNA interference-mediated loss-of-function studies should always be scrutinized to exclude frequent off-target effects and a validation of the phenotype specificity in rescue experiments reversing the phenotype by overexpression of the targeted gene should always be included. Furthermore, studying the localization of lncRNA can also provide valuable insights into its function. Establishing the cellular fraction, in which the lncRNA is normally present, can guide further analyses on the molecular function.
To unravel ncRNA mechanisms at the molecular level, identification of protein interaction partners of individual lncRNAs is informative and necessary to fully understand the mechanistic process accomplished by the ncRNA–based on the assumption that most ncRNAs will function in ribonucleoprotein complexes. A direct way to identify these proteins would be the application of RNA affinity purification, which uses the ncRNA as bait to fish out all putative important protein interaction partners. This could also be expanded to identifying ncRNA-interacting RNA or DNA molecules.169 These methods are of special interest as lncRNAs have been shown to play important regulatory roles and can function at the level of chromatin. To determine where lncRNAs bind to chromatin, two methods, called ChIRP and CHART, have been developed recently using complementary oligonucleotides to pull down lncRNAs associated with chromatin.170,171 Alternatively, RNA immunoprecipitation-sequencing (RIP-Seq) or PAR-CLiP (Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation) represent complementary approaches to study RNA-protein interactions.172 Here, all RNAs that bind to a specific protein of interest are pulled down to identify substrate, target, or regulator RNAs–either coding or non-coding. Expanding RIP-based analysis to long ncRNAs will help to create an experimentally documented RNA-protein interactome atlas, including both coding and non-coding transcripts. Such atlas can be a helpful guide for in-depth studies on the functions of each long ncRNA.11
Besides their molecular analysis, long ncRNA expression patterns should be further analyzed with genome-wide approaches, especially to discover these molecules in a wide variety of human diseases, e.g., cancers. This could be achieved by microarray-based profiling, deep sequencing or RNA-Seq followed by careful and detailed validation of the expression and sequencing data by qRT-PCR (quantitative polymerase chain reaction), Northern Blotting and Rapid amplification of cDNA ends techniques.
Long ncRNAs specifically expressed or silenced in human cancers could play an important role in these cancer entities and therefore might represent novel therapeutic target genes. Thus, in the last paragraph we will introduce several novel approaches in cancer therapy that target RNA molecules or make use of their tumor-specific expression. In addition, we will outline promising concepts to interfere with ncRNA function or expression, which could become beneficial in cancer therapy.
Fighting cancer: Are ncRNAs a new answer?
Currently, cancer therapy is greatly hampered by many difficulties, e.g., specific targeting of cancer cells without interfering with normal tissue function, specific delivery of antitumor drugs, and, in case of carcinoma of unknown primary, exact characterization of the malignancy. Here, long ncRNAs could offer a number of advantages, both as diagnostic and prognostic markers but also as novel specific therapeutic targets. The latter, of course, first requires detailed knowledge about the tumor-specific ncRNA function and its requirement for essential cancer cell properties.
For instance, the long ncRNA HULC (highly upregulated in liver cancer) is a liver-specific RNA that is highly expressed in primary liver tumors and hepatic metastases of colorectal carcinoma, but is not found in primary colon cancers or in non-liver metastases.173,174 PCGEM1, PCA3 (Prostate cancer gene 3) or PRNCR1 (prostate cancer non-coding RNA 1) are three long ncRNAs exclusively associated with prostate cancer.175-177 This makes them interesting candidates for tumor- and tissue-specific treatments and their expression can be used to identify unknown primary tumors. Moreover, long ncRNAs can also be used to differentiate between subtypes of the same cancer.22,36 As ncRNAs can be easily detected from minute amounts, e.g., in biological fluids like blood and urine, using qRT-PCR amplification, they are of great diagnostic value. For example, HULC can be easily detected in the blood of HCC patients using qRT-PCR.174 In addition, a quantitative PCA3 urine test with the potential for general use in clinical settings was developed; the Progensa™ PCA3 urine test. This specific test can help patients who had a first negative prostate biopsy to avoid unnecessary repeated biopsies.178 Furthermore, lncRNA expression can correlate with patient response to chemotherapy and therefore can be seen as predictive marker. For example, the expression of the long ncRNA Xist strongly associates with the disease-free survival of Taxol-treated cancer patients.179 LncRNAs are also powerful predictors of patient outcome, e.g., MALAT1 can serve as an independent prognostic parameter for patient survival in early-stage lung adenocarcinoma.128 The same is true for HOTAIR whose expression positively correlates with metastasis and poor outcome in primary breast tumors, gastrointestinal, hepatocellular and colorectal cancers.36,137,139,140
These few examples of long ncRNAs show already the great value of these newly discovered transcripts and it is highly likely that many other long ncRNA markers will be discovered in the near future. Hence, currently generated cancer genome data can only be fully exploited if also the non-coding content of the human cancer genome is studied in great detail - after all, it constitutes the large majority of the genomic information! The more we learn about long ncRNA expression patterns in different types of cancer–as well as in healthy cells–the higher the chances for an improved diagnosis and better prognosis will be.
Finally, long ncRNAs are interesting targets in cancer therapy and especially their cancer- and tissue-specific expression could be a major advantage over other therapeutic options. In fact, antitumor strategies that focus on RNA as target molecule are currently under development.
For example, ribonucleases, small proteins that can enter cells by endocytosis, translocate to the cyctoplasm where they evade mammalian protein nuclease inhibitors and degrade RNA. This can lead to cell death and can be used as anticancer therapy. One of these ribonucleases called Onconase, an amphibian nuclease, even reached clinical trials.180,181 Onconase is very stable, less catalytically efficient but more cytotoxic than most RNase A homologs. It targets tRNA, rRNA, mRNA as well as miRNAs and therefore shows cytostatic and cytotoxic effects.182-186 Treatment of cancer cell lines with Onconase leads to suppression of cell cycle progression, predominantly through G1, followed by apoptosis or cell senescence and Onconase even shows anticancer properties in animal models.187-189 Onconase sensitizes cells to a variety of anticancer modalities, suggesting its application as an adjunct to chemotherapy or radiotherapy.190-193
In addition to this general approach that targets all cellular RNAs, there are currently new strategies under development that make use of the cancer- or tissue-specific expression of long ncRNAs to reduce the risk of affecting normal tissues during genetic treatment. For example, the expression of the lncRNA H19 is increased in a wide range of human cancers. A plasmid, BC-819 (DTA-H19), has been developed to make use of this tumor-specific expression of H19. The plasmid carries the gene for the A subunit of diphtheria toxin under the regulation of the H19 promoter. Intratumoral injections of this plasmid induce the expression of high levels of diphtheria toxin specifically in the tumor resulting in a reduction of tumor size in human trials. Recent studies have yielded encouraging results in a broad range of carcinomas including NSCLC, colon, bladder, pancreatic and ovarian cancers.194-198
Alternatively, one could also think about manipulating the expression of specific tumor-suppressive long ncRNAs that were discussed earlier:
GAS5 expression induces growth arrest and apoptosis independently of other stimuli in some prostate and breast cancer cell lines.83 Therefore, finding a way to induce GAS5 expression in tumors or designing a vector that would induce the expression of GAS5 when injected into the tumor might provide an attractive therapeutic approach. The same holds true for TERRA, a negative regulator of telomerase.114 A potential therapeutic strategy in telomerase-positive cancer cells would be to enhance TERRA expression or to administer synthetic TERRA mimics.
A supplementary approach would target oncogenic long ncRNAs that show an increased expression in cancer. Here, one could block their activity via multiple ways: (1) One could try to decrease their expression. This is not a trivial task and silencing of long ncRNA expression via siRNAs can be complicated, possibly because of the extensive secondary structure or intracellular localization. Therefore, our lab has developed a specific approach that highly efficiently silences lncRNA expression via genomic integration of RNA destabilizing elements.129 This approach could be of special interest for blood-borne diseases where one could modulate the patient’s genome in hematopoietic stem cells and use these “corrected” cells for gene therapy. (2) One could block molecular interactions and therefore functionally silence lncRNAs. This could be achieved via application of either small molecule inhibitors that mask the binding site in protein interaction partners or antagonistic oligonucleotides that bind to the ncRNA and therefore hinder proteins from binding. (3) As long ncRNA function is most likely attributed to their secondary structure, one could develop methods to efficiently disrupt it. Again, small molecule inhibitors could be developed, e.g., via systematic evolution of ligands by exponential enrichment, which bind to lncRNAs and change their secondary structure or mimic their secondary structure and compete for their binding partners. Also, antagonistic oligonucleotides could target a specific region of the ncRNA and block its correct folding. (4) Once we have understood how ncRNAs can specifically recruit chromatin-remodeling complexes to specific genes, we could make use of this by creating artificial ncRNAs with pre-designed targeting specificities. With this, one could silence driving oncogenes, e.g., Ras or Myc at the genomic level in cancer cells.
However, before we can make use of these new therapeutic options many more functional and structural studies are necessary to fully understand long ncRNA biology - even for individual examples. At the moment, we are just taking our first steps on the road to understanding the role of long ncRNAs in cancer. But as we move forward, we will discover new ncRNAs and find out more about their importance in cancer, which will inevitably help us to design better therapeutic agents. Given the exponentially growing number of newly discovered long ncRNAs, many great discoveries may be expected in this field strongly encouraging basic science as well as clinical research in this exciting field.
Acknowledgments
We apologize to all scientists whose important work could not be cited in this review due to space constraints. Our research is supported by the German Research Foundation (DFG Transregio TRR77, TP B03), the Marie Curie Program of the European Commission, the Helmholtz Society (VH-NG-504), the Virtual Helmholtz Institute for Resistance in Leukemia, the German Cancer Research Center (DKFZ), and the Institute of Pathology, University of Heidelberg. T.G. is supported by a DKFZ PhD Fellowship.
Glossary
Abbreviations:
- ALT
alternative lengthening of telomeres
- AR
androgen receptor
- ANRIL
antisense non-coding RNA in the INK4 locus
- Bax
BCL2-associated X protein
- Bcl-2
B cell CLL/Lymphoma 2
- Bcl-xL
B cell Lymphoma-extra large
- CBX
chromobox homolog
- cIAP2
cellular inhibitor of apoptosis
- CUDR
cancer up-regulated drug resistant
- EMT
epithelial-mesenchymal-transition
- eNOS
endothelial nitric-oxide synthase
- ER
estrogen receptor
- GAGE6
G antigene 6
- GAS5
growth-arrest specific 5
- GR
glucocorticoid receptor
- HCC
Hepatocellular carcinoma
- HIF
hypoxia-inducible factor
- HMGA1
high mobility group AT-hook 1
- hnRNP
heterogenous ribonucleoprotein
- HOTAIR
HOX Antisense intergenic RNA
- HOX
homeobox
- HULC
highly up-regulated in liver cancer
- lncRNA
long non-coding RNA
- lincRNA
long intergenic RNA
- MALAT1
Metastasis associated long adenocarcinoma transcript 1
- Mdm2
murine double minute 2
- miRNA
microRNA
- mRNA
messenger RNA
- mTOR
mammalian target of rapamycin
- Myc
myelocytomatosis oncogene
- NAT
natural antisense transcript
- ncRNA
non-coding RNA
- NEAT2
Nuclear-Enriched Abundant Transcript 2
- NSCLC
non-small cell lung cancer
- PANDA
P21 associated ncRNA DNA damage activated
- PAR-CLiP
photoactivatable-ribonucleoside.enhanced crosslinking and immunoprecipitation
- PCA3
prostate cancer gene 3
- PCAT-1
prostate cancer associated transcript 1
- piRNA
PIWI-interacting RNA
- PCGEM1
prostate-specific transcript 1
- POT1
protection of telomeres 1
- PR
progesterone receptor
- PRC
polycomb repressive complex
- PRNCR1
prostate cancer non-coding RNA 1
- PSF
PTB-associated splicing factor
- P-TEFb
positive transcription elongation factor b
- PTEN
phosphatase and tensin homolog
- Puma
p53-upregulated modulator of apoptosis
- qRT-PCR
quantitative reverse transcription-polymerase chain reaction
- RB
retinoblastoma
- RIP-Seq
RNA immunoprecipitation and sequencing
- rRNA
ribosomal RNA
- siRNA
small interfering RNA
- snRNA
small nuclear RNA
- snoRNA
small nucleolar RNA
- SPRY4-IT1
sprouty homolog 4 intronic transcript 1
- SRA
steroid receptor RNA activator
- SUZ12
suppressor of zeste 12 homolog
- TERC
telomerase RNA component
- TERRA
telomeric repeat-containing RNA
- TERT
telomerase reverse transcriptase
- tie-1AS
tyrosine kinase containing immunoglobulin and epidermal growth factor homology domain-1 antisense transcript
- TP53
tumor protein 53
- tRNA
transfer RNA
- T-UCR
Transcribed ultra-conserved genomic region
- VEGF-A
vascular endothelial growth factor-A
- Xist
X inactive-specific transcript
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/20481
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–6. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 3.Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, et al. ENCODE Project Consortium. NISC Comparative Sequencing Program. Baylor College of Medicine Human Genome Sequencing Center. Washington University Genome Sequencing Center. Broad Institute. Children’s Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. FANTOM Consortium. RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 5.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–54. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 6.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 8.Trang P, Weidhaas JB, Slack FJ. MicroRNAs as potential cancer therapeutics. Oncogene. 2008;27(Suppl 2):S52–7. doi: 10.1038/onc.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 10.Chen LL, Carmichael GG. Long noncoding RNAs in mammalian cells: what, where, and why? Wiley Interdiscip Rev RNA. 2010;1:2–21. doi: 10.1002/wrna.5. [DOI] [PubMed] [Google Scholar]
- 11.Lipovich L, Johnson R, Lin CY. MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochim Biophys Acta. 2010;1799:597–615. doi: 10.1016/j.bbagrm.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol. 2008;4:e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, et al. FANTOM Consortium. RIKEN Genome Exploration Research Group Phase I & II Team Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–73. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 15.Kapranov P, Drenkow J, Cheng J, Long J, Helt G, Dike S, et al. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 2005;15:987–97. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 17.Gibb EA, Vucic EA, Enfield KS, Stewart GL, Lonergan KM, Kennett JY, et al. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011;6:e25915. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutzinger R, Mrázek J, Vorwerk S, Hüttenhofer A. NcRNA-microchip analysis: a novel approach to identify differential expression of noncoding RNAs. RNA Biol. 2010;7:586–95. doi: 10.4161/rna.7.5.12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–7. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng X, Gralinski L, Armour CD, Ferris MT, Thomas MJ, Proll S, et al. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio. 2010;1 doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–9. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA. 2010;16:1478–87. doi: 10.1261/rna.1951310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chodroff RA, Goodstadt L, Sirey TM, Oliver PL, Davies KE, Green ED, et al. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010;11:R72. doi: 10.1186/gb-2010-11-7-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–5. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 31.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–29. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 32.Amaral PP, Neyt C, Wilkins SJ, Askarian-Amiri ME, Sunkin SM, Perkins AC, et al. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA. 2009;15:2013–27. doi: 10.1261/rna.1705309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu X, Ravindranath L, Tran N, Petrovics G, Srivastava S. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol. 2006;25:135–41. doi: 10.1089/dna.2006.25.135. [DOI] [PubMed] [Google Scholar]
- 34.Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–9. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–53. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–84. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–70. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–30. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beltran M, Puig I, Peña C, García JM, Alvarez AB, Peña R, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–69. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–30. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–38. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 46.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 47.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 48.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 49.McKenna NJ, Lanz RB, O’Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321–44. doi: 10.1210/er.20.3.321. [DOI] [PubMed] [Google Scholar]
- 50.Lonard DM, O’Malley BW. Expanding functional diversity of the coactivators. Trends Biochem Sci. 2005;30:126–32. doi: 10.1016/j.tibs.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 51.Smith CL, O’Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- 52.Xu J, Qiu Y, DeMayo FJ, Tsai SY, Tsai MJ, O’Malley BW. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science. 1998;279:1922–5. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- 53.Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/S0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 54.Leygue E, Dotzlaw H, Watson PH, Murphy LC. Expression of the steroid receptor RNA activator in human breast tumors. Cancer Res. 1999;59:4190–3. [PubMed] [Google Scholar]
- 55.Chooniedass-Kothari S, Hamedani MK, Troup S, Hubé F, Leygue E. The steroid receptor RNA activator protein is expressed in breast tumor tissues. Int J Cancer. 2006;118:1054–9. doi: 10.1002/ijc.21425. [DOI] [PubMed] [Google Scholar]
- 56.Kawashima H, Takano H, Sugita S, Takahara Y, Sugimura K, Nakatani T. A novel steroid receptor co-activator protein (SRAP) as an alternative form of steroid receptor RNA-activator gene: expression in prostate cancer cells and enhancement of androgen receptor activity. Biochem J. 2003;369:163–71. doi: 10.1042/BJ20020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hube F, Guo J, Chooniedass-Kothari S, Cooper C, Hamedani MK, Dibrov AA, et al. Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol. 2006;25:418–28. doi: 10.1089/dna.2006.25.418. [DOI] [PubMed] [Google Scholar]
- 58.Cooper C, Guo J, Yan Y, Chooniedass-Kothari S, Hube F, Hamedani MK, et al. Increasing the relative expression of endogenous non-coding Steroid Receptor RNA Activator (SRA) in human breast cancer cells using modified oligonucleotides. Nucleic Acids Res. 2009;37:4518–31. doi: 10.1093/nar/gkp441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gajjar M, Candeias MM, Malbert-Colas L, Mazars A, Fujita J, Olivares-Illana V, et al. The p53 mRNA-Mdm2 interaction controls Mdm2 nuclear trafficking and is required for p53 activation following DNA damage. Cancer Cell. 2012;21:25–35. doi: 10.1016/j.ccr.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 60.Lohrum MA, Ashcroft M, Kubbutat MH, Vousden KH. Identification of a cryptic nucleolar-localization signal in MDM2. Nat Cell Biol. 2000;2:179–81. doi: 10.1038/35004057. [DOI] [PubMed] [Google Scholar]
- 61.Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–6. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 62.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–5. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 63.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–22. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 64.Eilebrecht S, Brysbaert G, Wegert T, Urlaub H, Benecke BJ, Benecke A. 7SK small nuclear RNA directly affects HMGA1 function in transcription regulation. Nucleic Acids Res. 2011;39:2057–72. doi: 10.1093/nar/gkq1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chiappetta G, Avantaggiato V, Visconti R, Fedele M, Battista S, Trapasso F, et al. High level expression of the HMGI (Y) gene during embryonic development. Oncogene. 1996;13:2439–46. [PubMed] [Google Scholar]
- 66.Chiappetta G, Manfioletti G, Pentimalli F, Abe N, Di Bonito M, Vento MT, et al. High mobility group HMGI(Y) protein expression in human colorectal hyperplastic and neoplastic diseases. Int J Cancer. 2001;91:147–51. doi: 10.1002/1097-0215(200002)9999:9999<::AID-IJC1033>3.3.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 67.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–9. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 69.Subramanya D, Grivas PD. HPV and cervical cancer: updates on an established relationship. Postgrad Med. 2008;120:7–13. doi: 10.3810/pgm.2008.11.1928. [DOI] [PubMed] [Google Scholar]
- 70.Brooks CL, Gu W. p53 regulation by ubiquitin. FEBS Lett. 2011;585:2803–9. doi: 10.1016/j.febslet.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li L, Feng T, Lian Y, Zhang G, Garen A, Song X. Role of human noncoding RNAs in the control of tumorigenesis. Proc Natl Acad Sci U S A. 2009;106:12956–61. doi: 10.1073/pnas.0906005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- 73.Song X, Sui A, Garen A. Binding of mouse VL30 retrotransposon RNA to PSF protein induces genes repressed by PSF: effects on steroidogenesis and oncogenesis. Proc Natl Acad Sci U S A. 2004;101:621–6. doi: 10.1073/pnas.0307794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Song X, Sun Y, Garen A. Roles of PSF protein and VL30 RNA in reversible gene regulation. Proc Natl Acad Sci U S A. 2005;102:12189–93. doi: 10.1073/pnas.0505179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–62. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–74. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787–93. doi: 10.1016/S0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 78.Coccia EM, Cicala C, Charlesworth A, Ciccarelli C, Rossi GB, Philipson L, et al. Regulation and expression of a growth arrest-specific gene (gas5) during growth, differentiation, and development. Mol Cell Biol. 1992;12:3514–21. doi: 10.1128/mcb.12.8.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muller AJ, Chatterjee S, Teresky A, Levine AJ. The gas5 gene is disrupted by a frameshift mutation within its longest open reading frame in several inbred mouse strains and maps to murine chromosome 1. Mamm Genome. 1998;9:773–4. doi: 10.1007/s003359900862. [DOI] [PubMed] [Google Scholar]
- 80.Raho G, Barone V, Rossi D, Philipson L, Sorrentino V. The gas 5 gene shows four alternative splicing patterns without coding for a protein. Gene. 2000;256:13–7. doi: 10.1016/S0378-1119(00)00363-2. [DOI] [PubMed] [Google Scholar]
- 81.Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18:6897–909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 84.Fingar DC, Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23:3151–71. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- 85.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–45. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 86.Mourtada-Maarabouni M, Hasan AM, Farzaneh F, Williams GT. Inhibition of human T-cell proliferation by mammalian target of rapamycin (mTOR) antagonists requires noncoding RNA growth-arrest-specific transcript 5 (GAS5) Mol Pharmacol. 2010;78:19–28. doi: 10.1124/mol.110.064055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morrison LE, Jewell SS, Usha L, Blondin BA, Rao RD, Tabesh B, et al. Effects of ERBB2 amplicon size and genomic alterations of chromosomes 1, 3, and 10 on patient response to trastuzumab in metastatic breast cancer. Genes Chromosomes Cancer. 2007;46:397–405. doi: 10.1002/gcc.20419. [DOI] [PubMed] [Google Scholar]
- 88.Nupponen NN, Carpten JD. Prostate cancer susceptibility genes: many studies, many results, no answers. Cancer Metastasis Rev. 2001;20:155–64. doi: 10.1023/A:1015557308033. [DOI] [PubMed] [Google Scholar]
- 89.Smedley D, Sidhar S, Birdsall S, Bennett D, Herlyn M, Cooper C, et al. Characterization of chromosome 1 abnormalities in malignant melanomas. Genes Chromosomes Cancer. 2000;28:121–5. doi: 10.1002/(SICI)1098-2264(200005)28:1<121::AID-GCC14>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 90.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat Rev Genet. 2005;6:611–22. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 92.Shay JW, Wright WE. Hayflick, his limit, and cellular ageing. Nat Rev Mol Cell Biol. 2000;1:72–6. doi: 10.1038/35036093. [DOI] [PubMed] [Google Scholar]
- 93.Meyne J, Ratliff RL, Moyzis RK. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci U S A. 1989;86:7049–53. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, et al. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–9. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–4. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 96.Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AY, Pickett HA, et al. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol. 2009;27:1181–5. doi: 10.1038/nbt.1587. [DOI] [PubMed] [Google Scholar]
- 97.Molenaar C, Wiesmeijer K, Verwoerd NP, Khazen S, Eils R, Tanke HJ, et al. Visualizing telomere dynamics in living mammalian cells using PNA probes. EMBO J. 2003;22:6631–41. doi: 10.1093/emboj/cdg633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–91. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 99.Tomaska L, Nosek J, Kramara J, Griffith JD. Telomeric circles: universal players in telomere maintenance? Nat Struct Mol Biol. 2009;16:1010–5. doi: 10.1038/nsmb.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tomaska L, Nosek J, Makhov AM, Pastorakova A, Griffith JD. Extragenomic double-stranded DNA circles in yeast with linear mitochondrial genomes: potential involvement in telomere maintenance. Nucleic Acids Res. 2000;28:4479–87. doi: 10.1093/nar/28.22.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, et al. The RNA component of human telomerase. Science. 1995;269:1236–41. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 102.Andersson S, Wallin KL, Hellström AC, Morrison LE, Hjerpe A, Auer G, et al. Frequent gain of the human telomerase gene TERC at 3q26 in cervical adenocarcinomas. Br J Cancer. 2006;95:331–8. doi: 10.1038/sj.bjc.6603253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nowak T, Januszkiewicz D, Zawada M, Pernak M, Lewandowski K, Rembowska J, et al. Amplification of hTERT and hTERC genes in leukemic cells with high expression and activity of telomerase. Oncol Rep. 2006;16:301–5. [PubMed] [Google Scholar]
- 104.Phatak P, Burger AM. Telomerase and its potential for therapeutic intervention. Br J Pharmacol. 2007;152:1003–11. doi: 10.1038/sj.bjp.0707374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr Opin Struct Biol. 2006;16:307–18. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 106.Wyatt HD, West SC, Beattie TL. InTERTpreting telomerase structure and function. Nucleic Acids Res. 2010;38:5609–22. doi: 10.1093/nar/gkq370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 108.Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. EMBO J. 2009;28:2503–10. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–36. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 110.Caslini C. Transcriptional regulation of telomeric non-coding RNA: implications on telomere biology, replicative senescence and cancer. RNA Biol. 2010;7:18–22. doi: 10.4161/rna.7.1.10257. [DOI] [PubMed] [Google Scholar]
- 111.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell. 2009;35:403–13. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38:5797–806. doi: 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Flynn RL, Centore RC, O’Sullivan RJ, Rai R, Tse A, Songyang Z, et al. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature. 2011;471:532–6. doi: 10.1038/nature09772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ng LJ, Cropley JE, Pickett HA, Reddel RR, Suter CM. Telomerase activity is associated with an increase in DNA methylation at the proximal subtelomere and a reduction in telomeric transcription. Nucleic Acids Res. 2009;37:1152–9. doi: 10.1093/nar/gkn1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–8. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 116.Talmadge JE, Fidler IJ. AACR centennial series: the biology of cancer metastasis: historical perspective. Cancer Res. 2010;70:5649–69. doi: 10.1158/0008-5472.CAN-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Klymkowsky MW, Savagner P. Epithelial-mesenchymal transition: a cancer researcher’s conceptual friend and foe. Am J Pathol. 2009;174:1588–93. doi: 10.2353/ajpath.2009.080545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–73. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 119.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 120.Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol. 2009;1:a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat Rev Cancer. 2004;4:118–32. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- 122.Egeblad M, Rasch MG, Weaver VM. Dynamic interplay between the collagen scaffold and tumor evolution. Curr Opin Cell Biol. 2010;22:697–706. doi: 10.1016/j.ceb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–52. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 125.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mohamed MM, Sloane BF. Cysteine cathepsins: multifunctional enzymes in cancer. Nat Rev Cancer. 2006;6:764–75. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 128.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–41. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 129.Gutschner T, Baas M, Diederichs S. Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res. 2011;21:1944–54. doi: 10.1101/gr.122358.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–32. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lin R, Roychowdhury-Saha M, Black C, Watt AT, Marcusson EG, Freier SM, et al. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett. 2011;585:671–6. doi: 10.1016/j.febslet.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–88. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Guo F, Li Y, Liu Y, Wang J, Li Y, Li G. Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai) 2010;42:224–9. doi: 10.1093/abbs/gmq008. [DOI] [PubMed] [Google Scholar]
- 134.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–8. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 135.Yamada K, Kano J, Tsunoda H, Yoshikawa H, Okubo C, Ishiyama T, et al. Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Sci. 2006;97:106–12. doi: 10.1111/j.1349-7006.2006.00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, et al. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584:4575–80. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 137.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–50. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 138.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 139.Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H, et al. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 2012;72:1126–36. doi: 10.1158/0008-5472.CAN-11-1803. [DOI] [PubMed] [Google Scholar]
- 140.Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39:2119–28. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 141.Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86:353–64. doi: 10.1016/S0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 142.Baeriswyl V, Christofori G. The angiogenic switch in carcinogenesis. Semin Cancer Biol. 2009;19:329–37. doi: 10.1016/j.semcancer.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 143.Augustin HG, Koh GY, Thurston G, Alitalo K. Control of vascular morphogenesis and homeostasis through the angiopoietin-Tie system. Nat Rev Mol Cell Biol. 2009;10:165–77. doi: 10.1038/nrm2639. [DOI] [PubMed] [Google Scholar]
- 144.Ferrara N. Vascular endothelial growth factor. Arterioscler Thromb Vasc Biol. 2009;29:789–91. doi: 10.1161/ATVBAHA.108.179663. [DOI] [PubMed] [Google Scholar]
- 145.Kazerounian S, Yee KO, Lawler J. Thrombospondins in cancer. Cell Mol Life Sci. 2008;65:700–12. doi: 10.1007/s00018-007-7486-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rossignol F, Vaché C, Clottes E. Natural antisense transcripts of hypoxia-inducible factor 1alpha are detected in different normal and tumour human tissues. Gene. 2002;299:135–40. doi: 10.1016/S0378-1119(02)01049-1. [DOI] [PubMed] [Google Scholar]
- 147.Thrash-Bingham CA, Tartof KD. aHIF: a natural antisense transcript overexpressed in human renal cancer and during hypoxia. J Natl Cancer Inst. 1999;91:143–51. doi: 10.1093/jnci/91.2.143. [DOI] [PubMed] [Google Scholar]
- 148.Uchida T, Rossignol F, Matthay MA, Mounier R, Couette S, Clottes E, et al. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression in lung epithelial cells: implication of natural antisense HIF-1alpha. J Biol Chem. 2004;279:14871–8. doi: 10.1074/jbc.M400461200. [DOI] [PubMed] [Google Scholar]
- 149.Cayre A, Rossignol F, Clottes E, Penault-Llorca F. aHIF but not HIF-1alpha transcript is a poor prognostic marker in human breast cancer. Breast Cancer Res. 2003;5:R223–30. doi: 10.1186/bcr652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fish JE, Matouk CC, Yeboah E, Bevan SC, Khan M, Patil K, et al. Hypoxia-inducible expression of a natural cis-antisense transcript inhibits endothelial nitric-oxide synthase. J Biol Chem. 2007;282:15652–66. doi: 10.1074/jbc.M608318200. [DOI] [PubMed] [Google Scholar]
- 151.Robb GB, Carson AR, Tai SC, Fish JE, Singh S, Yamada T, et al. Post-transcriptional regulation of endothelial nitric-oxide synthase by an overlapping antisense mRNA transcript. J Biol Chem. 2004;279:37982–96. doi: 10.1074/jbc.M400271200. [DOI] [PubMed] [Google Scholar]
- 152.Xia Y, Krukoff TL. Estrogen induces nitric oxide production via activation of constitutive nitric oxide synthases in human neuroblastoma cells. Endocrinology. 2004;145:4550–7. doi: 10.1210/en.2004-0327. [DOI] [PubMed] [Google Scholar]
- 153.Ying L, Hofseth LJ. An emerging role for endothelial nitric oxide synthase in chronic inflammation and cancer. Cancer Res. 2007;67:1407–10. doi: 10.1158/0008-5472.CAN-06-2149. [DOI] [PubMed] [Google Scholar]
- 154.Yamada T, Carson AR, Caniggia I, Umebayashi K, Yoshimori T, Nakabayashi K, et al. Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J Biol Chem. 2005;280:18283–90. doi: 10.1074/jbc.M413957200. [DOI] [PubMed] [Google Scholar]
- 155.Li K, Blum Y, Verma A, Liu Z, Pramanik K, Leigh NR, et al. A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood. 2010;115:133–9. doi: 10.1182/blood-2009-09-242180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–37. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–15. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 158.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–73. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 160.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308–16. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Galluzzi L, Kroemer G. Necroptosis: a specialized pathway of programmed necrosis. Cell. 2008;135:1161–3. doi: 10.1016/j.cell.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 162.Zong WX, Thompson CB. Necrotic death as a cell fate. Genes Dev. 2006;20:1–15. doi: 10.1101/gad.1376506. [DOI] [PubMed] [Google Scholar]
- 163.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Srikantan V, Zou Z, Petrovics G, Xu L, Augustus M, Davis L, et al. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci U S A. 2000;97:12216–21. doi: 10.1073/pnas.97.22.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Petrovics G, Zhang W, Makarem M, Street JP, Connelly R, Sun L, et al. Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene. 2004;23:605–11. doi: 10.1038/sj.onc.1207069. [DOI] [PubMed] [Google Scholar]
- 166.Tsang WP, Wong TW, Cheung AH, Co CN, Kwok TT. Induction of drug resistance and transformation in human cancer cells by the noncoding RNA CUDR. RNA. 2007;13:890–8. doi: 10.1261/rna.359007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, et al. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–62. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- 168.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012 doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–9. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, et al. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci U S A. 2011;108:20497–502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Matouk IJ, Abbasi I, Hochberg A, Galun E, Dweik H, Akkawi M. Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur J Gastroenterol Hepatol. 2009;21:688–92. doi: 10.1097/MEG.0b013e328306a3a2. [DOI] [PubMed] [Google Scholar]
- 174.Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–42. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 175.Chung S, Nakagawa H, Uemura M, Piao L, Ashikawa K, Hosono N, et al. Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci. 2011;102:245–52. doi: 10.1111/j.1349-7006.2010.01737.x. [DOI] [PubMed] [Google Scholar]
- 176.de Kok JB, Verhaegh GW, Roelofs RW, Hessels D, Kiemeney LA, Aalders TW, et al. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002;62:2695–8. [PubMed] [Google Scholar]
- 177.Ifere GO, Ananaba GA. Prostate cancer gene expression marker 1 (PCGEM1): a patented prostate- specific non-coding gene and regulator of prostate cancer progression. Recent Pat DNA Gene Seq. 2009;3:151–63. doi: 10.2174/187221509789318360. [DOI] [PubMed] [Google Scholar]
- 178.Durand X, Moutereau S, Xylinas E, de la Taille A. Progensa™ PCA3 test for prostate cancer. Expert Rev Mol Diagn. 2011;11:137–44. doi: 10.1586/erm.10.122. [DOI] [PubMed] [Google Scholar]
- 179.Huang KC, Rao PH, Lau CC, Heard E, Ng SK, Brown C, et al. Relationship of XIST expression and responses of ovarian cancer to chemotherapy. Mol Cancer Ther. 2002;1:769–76. [PubMed] [Google Scholar]
- 180.Beck AK, Pass HI, Carbone M, Yang H. Ranpirnase as a potential antitumor ribonuclease treatment for mesothelioma and other malignancies. Future Oncol. 2008;4:341–9. doi: 10.2217/14796694.4.3.341. [DOI] [PubMed] [Google Scholar]
- 181.Mikulski SM, Costanzi JJ, Vogelzang NJ, McCachren S, Taub RN, Chun H, et al. Phase II trial of a single weekly intravenous dose of ranpirnase in patients with unresectable malignant mesothelioma. J Clin Oncol. 2002;20:274–81. doi: 10.1200/JCO.20.1.274. [DOI] [PubMed] [Google Scholar]
- 182.Ardelt B, Ardelt W, Darzynkiewicz Z. Cytotoxic ribonucleases and RNA interference (RNAi) Cell Cycle. 2003;2:22–4. doi: 10.4161/cc.2.1.232. [DOI] [PubMed] [Google Scholar]
- 183.Ardelt W, Mikulski SM, Shogen K. Amino acid sequence of an anti-tumor protein from Rana pipiens oocytes and early embryos. Homology to pancreatic ribonucleases. J Biol Chem. 1991;266:245–51. [PubMed] [Google Scholar]
- 184.Darzynkiewicz Z, Carter SP, Mikulski SM, Ardelt WJ, Shogen K. Cytostatic and cytotoxic effects of Pannon (P-30 Protein), a novel anticancer agent. Cell Tissue Kinet. 1988;21:169–82. doi: 10.1111/j.1365-2184.1988.tb00855.x. [DOI] [PubMed] [Google Scholar]
- 185.Saxena A, Saxena SK, Shogen K. Effect of Onconase on double-stranded RNA in vitro. Anticancer Res. 2009;29:1067–71. [PubMed] [Google Scholar]
- 186.Suhasini AN, Sirdeshmukh R. Transfer RNA cleavages by onconase reveal unusual cleavage sites. J Biol Chem. 2006;281:12201–9. doi: 10.1074/jbc.M504488200. [DOI] [PubMed] [Google Scholar]
- 187.Lee I, Lee YH, Mikulski SM, Shogen K. Effect of ONCONASE +/- tamoxifen on ASPC-1 human pancreatic tumors in nude mice. Adv Exp Med Biol. 2003;530:187–96. doi: 10.1007/978-1-4615-0075-9_18. [DOI] [PubMed] [Google Scholar]
- 188.Mikulski SM, Ardelt W, Shogen K, Bernstein EH, Menduke H. Striking increase of survival of mice bearing M109 Madison carcinoma treated with a novel protein from amphibian embryos. J Natl Cancer Inst. 1990;82:151–3. doi: 10.1093/jnci/82.2.151-a. [DOI] [PubMed] [Google Scholar]
- 189.Juan G, Ardelt B, Li X, Mikulski SM, Shogen K, Ardelt W, et al. G1 arrest of U937 cells by onconase is associated with suppression of cyclin D3 expression, induction of p16INK4A, p21WAF1/CIP1 and p27KIP and decreased pRb phosphorylation. Leukemia. 1998;12:1241–8. doi: 10.1038/sj.leu.2401100. [DOI] [PubMed] [Google Scholar]
- 190.Ardelt W, Ardelt B, Darzynkiewicz Z. Ribonucleases as potential modalities in anticancer therapy. Eur J Pharmacol. 2009;625:181–9. doi: 10.1016/j.ejphar.2009.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Mikulski S, Viera A, Shogen K. Invitro synergism between a novel amphibian oocytic ribonuclease (onconase(r)) and tamoxifen, lovastatin and Cisplatin, in human ovcar-3 ovarian-carcinoma cell-line. Int J Oncol. 1992;1:779–85. [PubMed] [Google Scholar]
- 192.Mikulski SM, Viera A, Ardelt W, Menduke H, Shogen K. Tamoxifen and trifluoroperazine (Stelazine) potentiate cytostatic/cytotoxic effects of P-30 protein, a novel protein possessing anti-tumor activity. Cell Tissue Kinet. 1990;23:237–46. doi: 10.1111/j.1365-2184.1990.tb01119.x. [DOI] [PubMed] [Google Scholar]
- 193.Mikulski SM, Viera A, Darzynkiewicz Z, Shogen K. Synergism between a novel amphibian oocyte ribonuclease and lovastatin in inducing cytostatic and cytotoxic effects in human lung and pancreatic carcinoma cell lines. Br J Cancer. 1992;66:304–10. doi: 10.1038/bjc.1992.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Amit D, Hochberg A. Development of targeted therapy for bladder cancer mediated by a double promoter plasmid expressing diphtheria toxin under the control of H19 and IGF2-P4 regulatory sequences. J Transl Med. 2010;8:134. doi: 10.1186/1479-5876-8-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hasenpusch G, Pfeifer C, Aneja MK, Wagner K, Reinhardt D, Gilon M, et al. Aerosolized BC-819 inhibits primary but not secondary lung cancer growth. PLoS One. 2011;6:e20760. doi: 10.1371/journal.pone.0020760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mizrahi A, Czerniak A, Levy T, Amiur S, Gallula J, Matouk I, et al. Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J Transl Med. 2009;7:69. doi: 10.1186/1479-5876-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sidi AA, Ohana P, Benjamin S, Shalev M, Ransom JH, Lamm D, et al. Phase I/II marker lesion study of intravesical BC-819 DNA plasmid in H19 over expressing superficial bladder cancer refractory to bacillus Calmette-Guerin. J Urol. 2008;180:2379–83. doi: 10.1016/j.juro.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 198.Smaldone MC, Davies BJ. BC-819, a plasmid comprising the H19 gene regulatory sequences and diphtheria toxin A, for the potential targeted therapy of cancers. Curr Opin Mol Ther. 2010;12:607–16. [PubMed] [Google Scholar]
- 199.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–26. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–83. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–3. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 202.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–8. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chu Y, Yue X, Younger ST, Janowski BA, Corey DR. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010;38:7736–48. doi: 10.1093/nar/gkq648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yochum GS, Cleland R, McWeeney S, Goodman RH. An antisense transcript induced by Wnt/beta-catenin signaling decreases E2F4. J Biol Chem. 2007;282:871–8. doi: 10.1074/jbc.M609391200. [DOI] [PubMed] [Google Scholar]
- 206.Chen W, Böcker W, Brosius J, Tiedge H. Expression of neural BC200 RNA in human tumours. J Pathol. 1997;183:345–51. doi: 10.1002/(SICI)1096-9896(199711)183:3<345::AID-PATH930>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 207.Iacoangeli A, Lin Y, Morley EJ, Muslimov IA, Bianchi R, Reilly J, et al. BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis. 2004;25:2125–33. doi: 10.1093/carcin/bgh228. [DOI] [PubMed] [Google Scholar]
- 208.Madamanchi NR, Hu ZY, Li F, Horaist C, Moon SK, Patterson C, et al. A noncoding RNA regulates human protease-activated receptor-1 gene during embryogenesis. Biochim Biophys Acta. 2002;1576:237–45. doi: 10.1016/s0167-4781(02)00308-1. [DOI] [PubMed] [Google Scholar]
- 209.Rintala-Maki ND, Sutherland LC. Identification and characterisation of a novel antisense non-coding RNA from the RBM5 gene locus. Gene. 2009;445:7–16. doi: 10.1016/j.gene.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 210.Ginger MR, Shore AN, Contreras A, Rijnkels M, Miller J, Gonzalez-Rimbau MF, et al. A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc Natl Acad Sci U S A. 2006;103:5781–6. doi: 10.1073/pnas.0600745103. [DOI] [PMC free article] [PubMed] [Google Scholar]