Abstract
Besides their well known functions in storage and translation of information nucleic acids have emerged as a target of pattern recognition receptors that drive activation of innate immunity. Due to the paucity of building block monomers used in nucleic acids, discrimination of host and microbial nucleic acids as a means of self/foreign discrimination is a complicated task. Pattern recognition receptors rely on discrimination by sequence, structural features and spatial compartmentalization to differentiate microbial derived nucleic acids from host ones. Microbial nucleic acid detection is important for the sensing of infectious danger and initiating an immune response to microbial attack. Failures in the underlying recognitions systems can have severe consequences: thus, inefficient recognition of microbial nucleic acids may increase susceptibility to infectious diseases. On the other hand, excessive immune responses as a result of failed self/foreign discrimination are associated with autoimmune diseases. This review gives a general overview over the underlying concepts of nucleic acid sensing by Toll-like receptors. Within this general framework, we focus on bacterial RNA and synthetic RNA oligomers.
Keywords: Nucleic acids, autoimmunity, infections, innate immunity, modifications, toll-like receptor
Introduction
Although nucleic acids are mainly recognized as important for storage and translation of genetic information, cutting-edge research has continuously revealed new and exciting functions, including e.g., the myriad roles of regulatory noncoding RNAs, as well as the recognition by the immune system. Interestingly, some of those non-classical functions are interlinked: before the discovery and development of small interfering RNAs (siRNA),1 the use of long double-stranded RNA (dsRNA) in attempts to elicit RNA interference (RNAi) in mammalian cells was unsuccessful, as a consequence of an interferon response by the innate immune system. This feature of the innate immune system has recently received immense attention, as its implications for health and disease became clear. For example, bifunctional RNA based therapeutics displaying immunostimulating and gene silencing activities alike2 emerged as a new concept. At the heart of these developments is the recognition of nucleic acids in general, and of RNA in particular, by receptors of the innate immune system, which are charged with the task to differentiate between RNA from the mammalian host (“self”) and from invading pathogens (“non-self”).
This review gives an overview of these highly interesting interactions. After an introduction to the topic of nucleic acids as pathogen-associated molecular patterns (PAMPs), we will focus on recognition of nucleic acids by Toll-like receptors (TLR). Particularly, this review will highlight recognition of bacterial and synthetic RNA because this topic has received less attention so far. However, we will also discuss recognition of bacterial DNA to support our ideas of three main principles by which the immune system differentiates self- and microbial-derived nucleic acids. We will finish with an analysis of the importance of nucleic acid recognition during infections as well as mislead activation in autoimmune diseases.
Innate immune sensing of microbes relies on recognition of conserved patterns
With last year’s Nobel Prize of Medicine being awarded to Jules A. Hoffmann, Bruce A. Beutler and Ralph M. Steinman for their discoveries in the field of innate immunity,3-5one of the most exciting developments in immunological research over the last two decades has now been honored. In fact, the rediscovery of innate immunity and its role for early defense of infections but also for inducing and shaping adaptive immunity has definitively changed our way of understanding of how the immune system works on the systemic level. Central to this field has been the seminal concept proposed by the late Charles Janeway in 1989.6 He suggested that the co-stimulatory signal delivered by innate antigen presenting cells was inducible and regulated itself through the recognition of conserved microbial products. Thus, adaptive immunity was placed under the control of innate immunity.
Moreover, for the first time, a conceptual framework of how innate immunity is activated was proposed: Conserved microbial products, termed pathogen-associated molecular patterns (PAMPs), stimulate pattern recognition receptors (PRR), thus driving innate immunity’s activation. Innate immunity can therefore function with a limited set of target structures, without the need to produce a large repertoire of different receptor specificities by somatic recombination and clonal selection. Since this seminal proposal of Janeway, the immunological community has made considerable progress in defining “pattern recognition” at the molecular basis.7
By now, pattern recognition receptors expressed by professional innate immune cells comprise several major groups8: Toll-like receptors (TLRs),4,9-11 Nucleotide-oligomerization domain protein (Nod)-like receptors (NLRs),12-14 C-type lectins (CLRs),15 Retinoic acid inducible gene I (RIG-I) and Melanoma differentiation-associated gene-5 (Mda5),16,17 the NLR family, pyrin domain containing proteins (Nlrp)18 and Absent in melanoma 2 (AIM2) inflammasomes19-21
For most of those receptors, microbial ligands have been identified that fit into the category of molecular patterns: they are expressed by a variety of different microbes and differ in their fine structure, but nevertheless are recognized by a common receptor. Additionally, molecules have been identified that are of self-origin but still drive pattern recognition receptors, especially under non-physiological conditions.22 Conceptually, this fits to a guard theory7,23 whereby innate immune receptors also survey integrity of cellular processes that are often targeted by pathogenic microbes.
Sensing of a microbial infection, once effected by a PRR, is relayed into a downstream cascade of signaling events reviewed elsewhere.24 The first line of relay factors, e.g., the central adaptor molecule Myeloid differentiation factor 88 (MyD88) as well as Interleukin-1 receptor associated kinases (IRAKs) and Tumor necrosis factor (TNF) receptor associated factors (TRAFs) are integral parts of innate immunity, while downstream elements funnel into more general pathways, such as e.g., nuclear factor κB (NFκB). Studies on signal transduction have led to the discovery of important interconnections among several signaling cascades, and provided insights into a complex signaling network (reviewed in25). Clearly, the recognition of the various PAMPs by PRRs of varying specificity for a given microbe is a highly orchestrated event. Its multiple facets may include degrees of redundancy26 to ensure broad and unspecific recognition on one hand, as well as features aimed at high specificity such as cooperativity among PRRs.27
Furthermore, several recent studies on PRRs and relay proteins have promoted the notion that nucleic acid sensing by players of innate immunity significantly impacts adaptive immunity.28-30 This connection extends to Systemic lupus erythematosus (SLE), a prominent autoimmune disease involving the erroneous recognition of self-nucleic acids by the adaptive immune system.
General Aspects of Innate Immune Sensing of Microbial Nucleic Acids
Among the various PAMPs, nucleic acids stand out for several reasons, the most obvious being the chemical and structural resemblances between nucleic acids of the host (“self”) and those of a potential pathogen (“non-self”). Consequently, faithful discrimination cannot exclusively rely on differences in the structure, and erroneous recognition occurs more readily than e.g., in the recognition of such distinctively bacterial molecules as flagellin or lipopolysaccharide. Correspondingly, many nucleic acid-sensing PRRs including e.g., TLRs3, 7–9, RIG-I, Mda5, AIM2, and DExD/H helicases, are known to also respond to nucleic acids of self-origin under pathological conditions.31
Nucleic acids of either provenance form very similar structures, including in particular the unsophisticated double-helix. Typical cellular RNA populations are mostly composed of ribosomal and transfer RNA, who’s ubiquitously conserved three-dimensional structures account for 90% of total RNA. Hence, leverage for discrimination may be found in sequence elements or on the atomic level, i.e. in post-transcriptional modifications or differentially processed 5′- or 3′-extremities.
Indeed, from the published results modes of pattern discrimination emerge that may be classified according to the hierarchy of nucleic acid structure: (i) primary structure, including nucleotide composition, chemical modification, and sequence, (ii) higher order structures including elements of secondary and tertiary structure, and (iii) discrimination by localization. Those modes of recognition appear to be realized in each of the nucleic acid recognizing PRRs.
Here, we focus on recognition by TLRs and will only briefly touch other PRRs. We will start with a short introduction on individual TLRs that will highlight each of the three recognition principles, including also TLR9 as a DNA-sensing PRR. This will be followed by a more detailed analysis of bacterial and synthetic RNA by TLRs with respect to the named modes of self/foreign differentiation.
Structural recognition of RNA through TLR3
TLR3 has first been recognized to be stimulated by double-stranded RNA, a molecular pattern that will occur during replication of RNA viruses.32 TLR333-35 as well as the other nucleic acids recognizing TLRs (TLR7,8,9) resides in the endosomes, thus being accessible to dsRNA taken up from extracellular (mode iii). Of the TLRs sensing nucleic acids, crystal structures are only available for the TLR3 ectodomain (TLR3-ECD). A structure of TLR3-ECD complexed to a 46 basepair (bp) dsRNA helix36 reveals the typical horseshoe shape of the ectodomain. TLR3-ECD is dimerized (Fig. 1) for activation of the cytosolic signal transduction domain and ensuing signal transmission as a consequence of RNA binding. In its general shape, the TLR3 structure resembles X-ray based structures of TLR4,37 and TLR1-TLR2.38 TLR3-ECD binds dsRNA of at least 40 bps36,39 at two sites located at opposite ends of the TLR3 horseshoe. An intermolecular contact between the two TLR3-ECD C-terminal domains coordinates and stabilizes the dimer. This critical length is above that of a typical helix occurring in normal cellular RNA such as e.g., tRNA, and, in particular, miRNA and siRNA. Indeed, before the discovery of siRNA as duplexes of some 20 bps, attempts to induce RNAi in mammals with larger dsRNA was unsuccessful, presumably also as a consequence of TLR3 action.1 Two RNA binding sites in each horseshoe function at pH below 6.5, which is a hallmark of the endosomal compartment,36,40,41 likely due to protonation of histidine residues, whose positive charge then undergoes ionic interaction with the negatively charged RNA backbone.
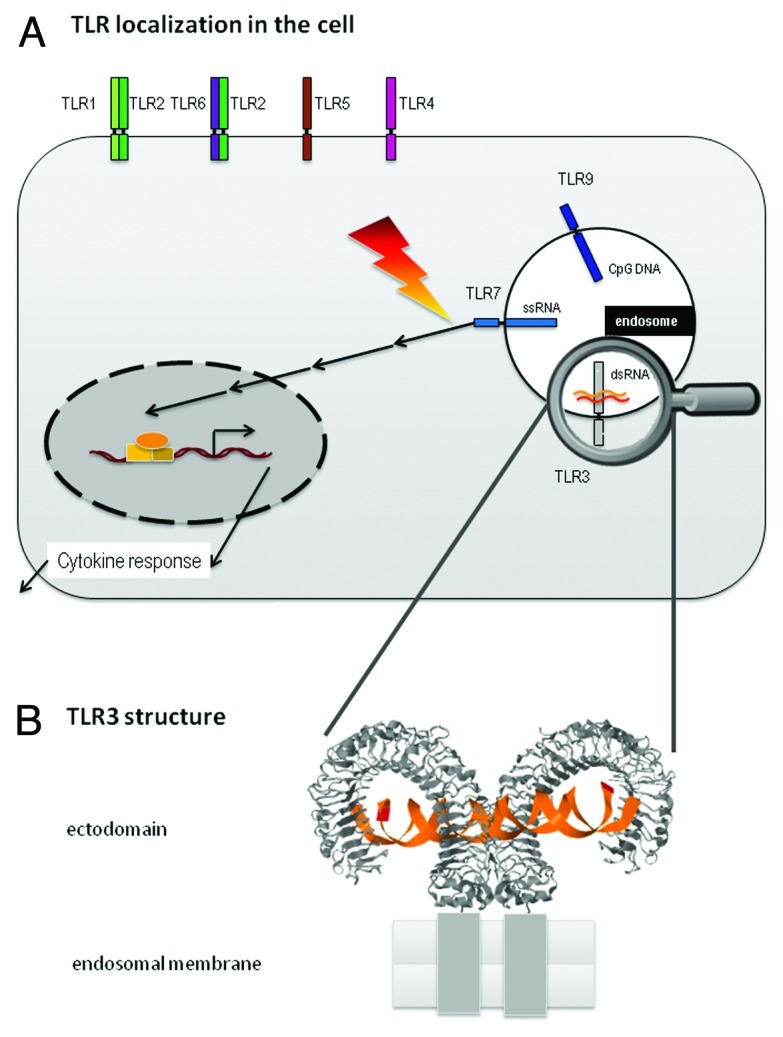
Figure 1. (A) Localization of TLRs. One set of TLRs is situated at the cell membrane. The set of TLRs which is responsible for sensing nucleic acids is located in the membrane of late endosomes/lysosomes. Upon activation by non-self nucleic acid, an inflammation signal is triggered through a signal cascade including MyD88, IRAK1/2/4, TRAF3/6, which ultimately leads to a cytokine response. (B) Horseshoe structure of TLR-ectodomain complexed to a 46mer dsRNA.36
Recognition of a ds helix by TLR3 is a perfect example of pattern recognition by secondary structure alone (mode ii, structural feature), which evidently occurs independently of any specific sequence motif. It involves recognition of relatively long stretches of dsRNA, a pattern that is common during RNA virus infection and replication but largely absent (at least within the endosome where TLR3 is expressed) in normal cell physiology. Indeed, recognition of dsRNA by TLR3 has been shown to be sequence independent.32 Many studies involve the use of poly I:C as a surrogate for natural dsRNA in the stimulation of for TLR3. Although certainly helpful, such results need to be interpreted with some caution as different commercially available poly I:C compounds exert somewhat differential effects, [e.g., interleukin (IL)-12p70 induction in DCs], probably dependent on differences in molecular weight and the fact that it is not a physiologically or even naturally occurring substance. Also, stimulation of additional receptors including cytosolic Mda5 and RIG-I has been observed.42 Of note, mammalian mRNA, i.e., self-RNA has also been postulated to be a TLR-3 ligand,43 which might signal the presence of necrotic debris from neighboring cells. Possibly, structured elements, especially in the UTRs of mRNA might form secondary structures that could induce TLR-3 signaling.
Sensing of RNA through TLR7
TLR7 and TLR8, the latter of unclear functional competence in mice, have first been identified to be activated by single-stranded RNA (ssRNA) and were suggested to participate in virus recognition, e.g., influenza virus.44,45 TLR7 (as well as TLR3, 8, 9, 13) is expressed intracellularly within endosomes,35 a compartment where RNA can be sensed during e.g., uptake of viral particles45 but which has only sporadic contact with host-derived RNA, e.g., after endocytic uptake of debris from necrotic tissue.46,47
Specifically, TLR7 exerts an important function to trigger IFNα release from plasmacytoid dendritic cells (pDCs).44,45 Self/foreign discrimination involves a certain sequence dependency, and nucleotide modifications modulate recognition48-50: TLR7 is activated by single stranded RNA ONs with an apparent preference for G and U rich sequences.44,45,51 Although length dependence of single stranded RNA ONs has been controversially discussed, it appears that efficient activation requires a length of at least 21 nucleotides.44,52,53 This discussion is complicated by the fact that contradictory studies were often conducted with various read-out systems on ONs of different sequence, composition, and modification state.
It has been suggested that the nucleotide preference might be related to a similar nucleotide composition of viral sequences in certain RNA viruses, including e.g., Influenza A and human immunodeficiency virus.44,54,55 Synthetic RNA ONs encoding such sequences stimulate sequence-dependent cytokine responses via TLR7 and TLR8. It has therefore been speculated that these GU rich sequences have evolved as a PAMP of invariant nucleotide composition for the recognition of viruses.44,54
However, TLR7 is also a major player in the recognition of bacterial RNA: in pDCs bacterial RNA stimulated TLR7 thus leading to secretion of Interferon α (IFNα)56 but in myeloid DCs TLR7 was dispensable. In contrast to the findings of TLR7 independency of bRNA immunostimulation in myeloid DCs, another group showed that IFNβ production in response to phagosomal but not cytosolic bacteria used TLR7. Immunostimulation was achieved through lysosomal recognition of bacterial RNA and activated TLR7, MyD88 and interferon regulatory factor-1 (IRF1).57 It might be that induction of IFNα occurs through a different pathway as compared with typical NFκB dependent innate immune genes. The latter report suggested that bacterial mRNA was specifically active whereas others showed that rRNA (rRNA) is equally potent.56
By conjecture of the known TLR horseshoe structures, it may be assumed hat TLR7, 8 form a similar double horseshoe structure upon activation.58 However, the binding modes of their ligands are still ill understood. While typical approaches to the definition of key features have focused on either two of the parameters length, sequence, modification state, or secondary structure, the solution, as in the case of TLR3, is of three-dimensional nature and therefore most likely must include three-dimensional features of the RNA ligand. Our current understanding of TLR7 ligands thus cannot be consolidated by a simple model such as dsRNA, as shown in the TLR3 structure.36 In simplified presentations, TLR7 is often portrayed as the ssRNA-sensing TLR with a structural specificity that is seemingly complementary to the dsRNA-sensing TLR3. However higher structural features are also of importance, since TLR7 was recently shown to recognized bacterial tRNAs.49,50
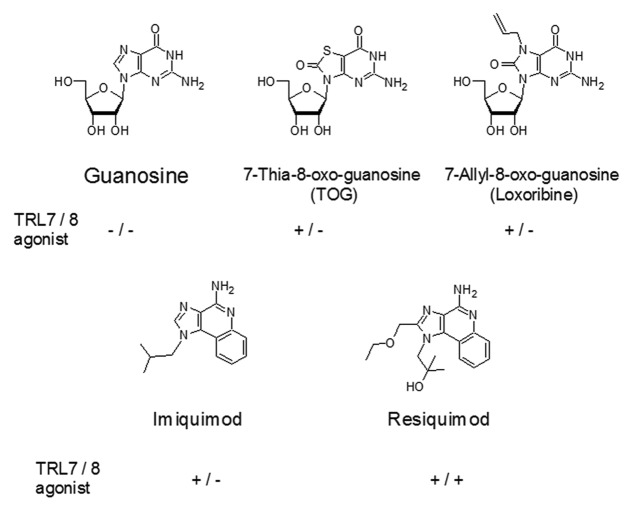
Even more puzzling than the recognition of such divergent structures as ssRNA and tRNA, but not of dsRNA, is the existence of a number of of small molecular compounds that activate TLR7, TLR8, or both.59-61 Among these small molecules, Imiquimod and Resiquimod are shown in Figure 2 because of their clinical relevance. Both may be viewed as nucleoside analogs, although it is unclear if the recognition mode by TLR7 bears any resemblance to RNA recognition. More striking however, is the TLR recognition of several guanosine derivatives including Loxoribine, while, guanosine itself shows no interaction.61 While it is out of the scope of this review to elucidate structure-function relationships of such compounds in depth, it is duly noted that the efficiency of such small molecules implies a mode of action that appears fundamentally different from that of RNA ON recognition by TLRs. Yet, the recognition of guanosine derivatives provokes a comparison with the guanosine-rich RNA ONs mentioned above.52 In the absence of any conclusive in-depth study, we speculate that small molecules as well as ssRNA might aggregate to form higher ordered structures.62 Beyond the discussed implications from the structure of Loxoribine, this hypothesis is based on the fact that many highly active ssRNA ONs do not span a distance comparable to the 46mer in the TLR3 structure. It has indeed been suggested, that TLR7 activating RNA ONs that are presumed single stranded, actually form secondary structures,48,63 possibly including hybridization of several RNA ONs to a larger complex, which then might be able to bridge the TLR subunits for dimerization. This concept has also been discussed based on stimulation of TLR7/8 using phosphorothioate RNA ONs.63
Figure 2. Small molecule purine agonists of TLR7 and/or TLR8. The respective names are indicted under the chemical structures, and triggering of TLR7 and/or TLR861 is indicated below the name.
Although there is so far no experimental structural basis for recognition by TLR7, the recognition of tRNA structures by TLR749,50 strongly suggests an involvement of larger structures.
DNA sensing by TLR9
Although TLR9 itself is known for DNA recognition, it is discussed here as a relative of the RNA-sensing TLRs. Indeed many clues in the studies of TLRs 3 and 7 have been taken from pioneering work on TLR9. TLR9 has been shown to recognize hypomethylated bacterial DNA that is rich in CG dinucleotides.64 Whereas hypomethylated CG dinucleotides are frequent in bacterial DNA, CG motifs are suppressed in eukaryotic DNA and furthermore are highly methylated.65 As bacterial DNA mediated TLR9 stimulation can be mimicked by synthetic CpG ONs66 and is abolished by GC inversion, this receptor provides a clear example for a (albeit minimal) sequence-dependent pattern recognition (mode i). However, a recent report claims that CG sequence dependency can be observed mainly when using synthetic, phosphorothioate modified DNA ONs but might be less important for natural backbones.67 Despite the bias of CG sequences in bacterial vs. mammalian genomes, this dinucleotide element alone contains too little information for efficient discrimination. Indeed, further control is achieved through recognition of cytosine methylation which acts as a negative determinant, i.e., its presence in mammalian DNA prevents triggering of TLR9.
In addition to these structural features (mode i), the endosomal expression of TLR968 adds an additional level of control (compartmentalization, mode iii). Interestingly, forced expression of TLR9 at the cellular surface promotes recognition of self-DNA, thus abolishing self/foreign discrimination.69 Also, increasing the amount of self-DNA either by interference with lysosomal DNA degradation in DNase I knockouts70,71or through increased delivery by anti-DNA antibodies72,73, results in increased stimulation through self-derived DNA. Using a combined approach of mutational analysis and homology modeling, it was suggested that TLR9 recognizes bacterial DNA in a manner similar to TLR3 with two binding sites in the extracellular domain that possibly interact through charges with the nucleic acid.74 TLR9 has to be cleaved in the extracellular domain75,76 which appears to be necessary to bind the double stranded helix in a curvature-dependent manner.77
Excursion: Cytoplasmic nucleic acid recognition also relies on sequence, spatial and structural features
Besides endosomal TLRs a variety of cytoplasmic PRRs rely on similar principles for the recognition of foreign nucleic acids, adapted to the parameters of a different cellular compartment. Although we focus on TLRs, this paragraph will give an abbreviated overview to avoid the impression that sensing is taking place exclusively in the endosome. Interestingly, cytoplasmic PRRs rely on similar structural recognition principles, although they differ in fine details. For example RIG-I and Mda5 are cytosolic receptors that recognize viral RNA. The mode of pattern recognition is best characterized for RIG-I as crystal structures with and without RNA are now reported.78,79 RIG-I has first been shown to interact with uncapped, 5′-tri-phosphorylated RNA, a pattern that would be produced during viral replication processes but not in eukaryotic RNA biology as capping and some nucleotide modifications occur during eukaryotic posttranscriptional RNA processing in the nucleus, before export of mRNAs into the cytosol.80 Yet, double-stranded RNA longer than 100bp can stimulate RIG-I without the need for 5′-tri-phosphate ends.81 The crystal structure of RIG-I with RNA shows that ligand-free RIG-I has an open conformation in which the signaling domain is sequestered, but closes upon dsRNA binding. Blunt end 5′ppp-dsRNA is bound by the helicase and the C-terminal domain thus enabling signal induction and inducing further cooperative RIG-I binding on dsRNA. Thus, RIG-I combines the sensing of 5′-trisphosphates and dsRNA with compartmentalization.
Cytosolic DNA recognition also relies on localization as a major discriminatory principle, because lack of cytosolic DNA is typical of normal cell physiology. AIM2 was reported to recognize cytosolic DNA and induce maturation of IL-1β and IL-18 through a new type of inflammasome.19-21,82 IFI16 is a cytoplasmic receptor, which was recently identified as being also stimulated by intracellular DNA.83 AT-rich stem loop DNA motifs in the genome of plasmodia are recognized by still another unknown DNA receptor.84
Aspects of endosomal recognition of nucleic acids
As has been outlined in the separate presentations of the nucleic acid receptors TLR3, 7 and 9, all these PRRs recognize their ligands in intracellular endolysosomes, thus adding a layer of control to pattern recognition by means of compartmentalization.27 The similarities among these TLRs, which are probably rooted in their evolutionary relationship, do not only include recognition modes, but extend to signaling cascades and intracellular trafficking.85 It is unclear how many components of these networks are yet to be discovered, but probably the present picture is still incomplete.
The recognition of nucleic acids itself may actually be aided by accessory proteins, as suggested by the fact that high-mobility group box (HMGB) proteins 1, 2 and 3 were found to bind to immunogenic nucleic acids and to contribute to activation of TLR3, 7, 9 by their cognate ligands. HMGBs increased nucleic acid uptake, thus acting as universal sentinels for nucleic acids.86 Moreover, HMGB1 was suggested to affect DNA curvature thus contributing to DNA-TLR9 interaction.77
An interesting aspect of intracellular trafficking was discovered in a mutagenesis screen, which identified a mutant named 3d85 that abolished immunostimulation by the three endosomal TLRs. The underlying mutation was found to reside in Unc93b1, a conserved protein of the endoplasmic reticulum that turned out to be crucial for delivering TLR3, 7 and 9 to the endosome87 (as well as TLR13 in mice). The Unc93b1 mutant D34A upregulated ligand-induced trafficking of TLR7 but downregulated delivery of TLR9, from which it was inferred that wild-type Unc93b1 might have evolved to naturally bias responses to nucleic acids toward DNA- but against RNA-sensing.88 Importantly, Unc93b1 was found to be involved in resistance to infections with Toxoplasma gondii89 and streptococci90; human Unc93b1 deficiency resulted in susceptibility to Herpes simplex virus encephalitis.91
Evolutionary relations between TLR7, 8 and 9 are also observed when modeling the extracellular receptor domains.58 All nucleic acid recognizing TLRs are cleaved by proteolysis in a stepwise manner,92,93 however it remains controversial to what extent this cleavage represents simple degradation, or a processing step that contributes to recognition.74-76
Co-receptor usage might be an additional way to shape nucleic acid responses. CD14, a well-known co-receptor for TLR4 in lipopolysaccharide (LPS) recognition recently was shown to be involved in TLR7 and TLR9 recognition as well.94 CD14 was shown to contribute to nucleic acid uptake as well as promoting endosomal TLR activation in response to vesicular stomatitis virus.
Posttranscriptional processing and modifications that discriminate self from foreign RNA
Microbial foreign RNA comes from many sources and in various compositions, therefore distinction by biased nucleotide composition alone is insufficient. However, bacterial and eukaryotic RNA processing differs significantly and thus offers leverage for efficient identification of prokaryotic RNA. Features that may be used to identify eukaryotic RNA include the cap structure of mRNA (which will also pass inspection by cytoplasmic RIG-I as discussed above), polyadenylation, and post-transcriptional nucleotide modification, the latter being generally more pronounced in eukaryotic RNA. Of note, since viral RNA is processed in eukaryotic cells, its recognition by these features may be less efficient than that of bacterial RNA (bRNA).
An effect of polyadenlation was discovered by comparison of IL-12 secretion from human monocyte-derived DCs, which was high upon stimulation with bRNA, but low with eukaryotic RNA.95 In-vitro transcribed mRNA that lacked a poly(A) tail was also immunostimulatory and this property was lost by enzymatic 3′-polyadenylation. DCs activated by bRNA to secrete high amounts of IL-12 in turn induced T-helper cell differentiation to the T-helper-1 subset which was discussed to be a biological meaningful shift to defend against intracellular microbes.
A fundamental work also based on the comparison of eukaryotic RNA vs. bRNA was published in 2005.46 Here, activation of TLR3, 7 and 8 was specifically addressed and found to be inefficient with mammalian cytosolic RNA, but efficient with bRNA and, most interestingly, with mitochondrial RNA. Of note, the presence of stimulatory mitochondrial RNA in eukaryotic cells is a further example of the contribution of compartmentalization in RNA sensing. As mitochondria are presumed descendents from bacterial symbionts, mitochondrial transcription and translation carry distinct bacterial traits, which also extend to RNA processing and thus mitochondrial RNA. This includes a distinctly lower content of modified nucleotides, which was subsequently revealed to be a major contribution to the observed effect.
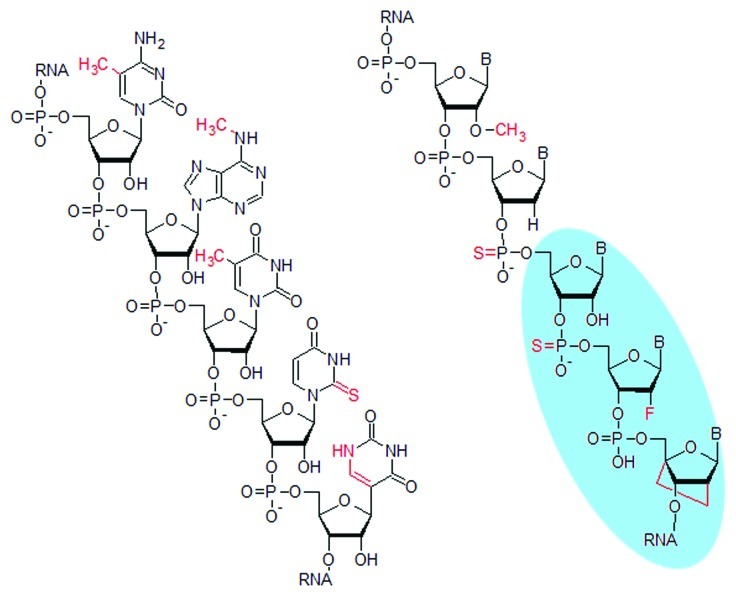
It was observed that in vitro transcribed RNA containing randomly incorporated m6A or s2U lacked TLR3 stimulation and that m6A, m5C, m5U, s2U and Ψ modifications suppressed activation of TLR7 and 8 (see Figure 3 for structures). Primary DCs on the other hand, could be stimulated by m6A and m5C modified RNA and were inhibited when Ψ was introduced in parallel. In further studies, these modifications were also shown to improve translation of the respective mRNA by diminishing activation of cytosolic components of innate immunity.97,98
Figure 3. Selected chemical modifications (red) at the nucleobase shown on the left strand include the naturally occurring m5C, m6A, m5U, s2U and Ψ, (see 96 for nomenclature). Modifications at the ribose shown on the right strand include the naturally occurring 2’-O-Me, and desoxyphosphorothioate, as well as the synthetic ribophosphorothioate, 2’-fluoro and locked nucleic acid (LNA) modifications highlighted in blue, which have not (yet) been discovered in nature.
This concept of a “relative hypomodification” of bRNA vs. eukaryotic RNA as a basis for pattern recognition by TLRs was subsequently taken up in many studies, from which 2’-O-methylation of the ribose emerged as a recurrent feature of eukaryotic nature, which efficiently prevents triggering of TLR response. Several studies based on synthetic 2’-O-methylated RNA observed suppressing effects for TLR7 in human as well as murine plasmacytoid DCs and monocytes.48,99,100 Those recent findings fit well to old observations that within dsRNA-homopolymers 2’-O-methylation (poly I:Cm, poly A:Um) induced less type I IFN.101 2’-O-methylation is naturally occurring at significantly higher abundance in eukaryotic as compared with prokaryotic (and mitochondrial) RNA. This assessment is valid for various RNA species including mRNA, rRNA, tRNA and several other noncoding RNAs.102-104
Eukaryotes (and archaea) have evolved a sophisticated system that allows variable targeting of 2’-O-methylation by guide RNAs, most of which direct abundant modification to rRNA. By virtue of their abundance, rRNA, which actually presents 80–90% of a cell’s RNA population, and tRNA (tRNA, ~10%) should in principle account for most of the RNA mediated TLR stimulation, and the frequent occurrence of 2’-O-methylated residues in rRNA may have a function in TLR mediated RNA sensing. However, since rRNA exists mostly in ribosomes and is thus tightly covered by proteins, tRNA is probably the most abundant free RNA species being sampled in endosomes.
Two recent studies show that most bacterial tRNA isoacceptor species as well as all unmodified in vitro transcribed tRNAs elicit a strong interferon response from pDCs via TLR7.49,50 In contrast, mammalian tRNA isoacceptor species and certain bacterial tRNAs were non-stimulatory. A detailed analysis identified 2’-O-methylated guanosines at two different positions, Gm18 and Gm34, as efficient in suppressing a TLR7 response. Of note, Gm18 is present in certain bacteria and tRNAs containing Gm18 act as TLR7 antagonists, suppressing a response to other bacterial tRNA molecules that would otherwise elicit a response. More antagonistic modifications remain to be identified in mammalian tRNAs,49 which, by containing up to 25% modified nucleotides, are the most heavily modified RNAs known so far.105
In summary, the recent dynamic development of this field has identified post-transcriptional processing and modification features as a leverage for the definition of PAMPs in RNA. This has major implications for potential therapeutic applications, including RNAi based strategies, as well as vaccination strategies106 that employ mRNA.107 Both, avoiding recognition or deliberate activation of innate immunity may be desirable, depending on the therapeutic strategy. For either case, the use of naturally occurring or non-natural synthetic modifications is being actively investigated.97,98,107
Modulation of TLR activation by synthetic RNA
Following the analysis of principles that allow discrimination of natural, microbial RNA from host RNA we will now center on synthetic RNA in a therapeutic perspective. Aside from mRNA based vaccination,106 synthetic RNA is being develop with three main features: (1) antisense, ribozyme, or RNAi based gene silencing,108,109 or activation,110,111 (2) target recognition of aptamer structures109 and (3) deliberate immunostimulation or avoidance thereof.2
Therapeutic antisense approaches have been in development for decades, and unwanted activation of the immune system is a long standing problem. Academia and industry alike have responded with the development of ever new building blocks (known as phosphoramidites) for incorporation into synthetic oligonucleotides by solid phase synthesis.112 In addition to modulation of immunostimulation, desirable properties of artificial modifications include resistance against nucleolytic degradation, and optimized base-pairing which in turn should result in maximized antisense or RNAi effects. Phosphoramidite building blocks corresponding to chemical alterations of almost every atom of RNA nucleosides have been explored,112 and may be roughly divided into modifications of the nucleobase and modifications of the ribose-phosphate backbone; some examples are given in Figure 3. This concerns in particular the 2’-OH position113 that, for one, marks the border between RNA and DNA, second, is critical for resistance against nucleases, and third, appears to be an important feature in TLR7 recognition, as is evident from the role of 2’-O-methlyation. Modern oligonucleotide therapeutics typically contain a mix of modified nucleotides, some of which are naturally occurring, e.g., 2’-O-methlyated nucleosides, and some of which are clearly artificial, such as 2’-fluorinated nucleosides, which defy definitions of DNA and RNA. Phosphorothioates (Fig. 3) present a very special case: originally conceived as a human invention by Eckstein, DNA phosphorothioates were recently revealed to be naturally occurring.114-116 The phosphorothioate modification in both DNA and RNA enhances stability, improves cellular uptake both via active transport117 and by increased membrane permeability, and importantly, increases activation of the immune system.118
Oligonucleotides for targeted TLR stimulation
Many nucleic acid preparations elicit an ill-defined response of the innate immune system because of pleiotropic effects: for example, synthetic poly I:C can be used to stimulate TLR3, however this is not specific as cytosolic Mda5 and RIG-I might be triggered as well.119 Because of differential expression of the various TLRs in different professional immune cells,120 activation of multiple PRRs may seriously complicate clinical development. Development of ONs with sequence elements or chemical modifications that allow selective targeting of a given TLR or immune cell is therefore of high interest. Early implementations of the idea to use ONs for deliberate activation of the innate immune system were targeting TLR9 because its recognition of CpG DNA ONs was best understood among the TLRs early on.66,121,122 The emergence of ligand profiles for TLR332 and TLR7,44,45 went hand in hand with the development of motifs for selective TLR targeting: such motifs include 5′-GUCCUUCAA-3′,53 5′-UGUGU-3′,123 as well as a 5′-CUGAAUU-3′ motif, which stimulated TLR7 and pDCs when contained in duplex-forming ONs.53 Forsbach et al. report sequences that specifically trigger a specific set of cytokines via TLR8, while avoiding the type I IFNs response that is typical of TLR7-mediated pDC activation.124 Along the same line, it was shown that by manipulation of molecular structure and mode of delivery, either selective TLR7 stimulation and IFNα secretion from pDC, or TLR8 stimulation and IL12p70 secretion from monocytes can be achieved.125
Phosphorothioate modifications in the backbone as well as cationic lipid formulation were shown to be effective to use RNA as DC stimulus.126 Intra-tumor injection of stabilized single stranded RNA could be used to trigger anti-tumor immunity and thus opens the field for the development of synthetic TLR7 agonists.127 Potential therapeutic fields include adjuvant activity for new vaccines106 as well as treatment of viral infections, cancer and allergies.128,129 In another approach RNA segments were coupled through their 3′ ends resulting in enhanced nuclease resistance and increased immunostimulation of TLR8 without lipid carriers.130 Substitution with 7-deazaguanosine for guanosine also resulted in an immunomodulatory compound with increased stimulation of TLR7 and 8. Another RNA ON that additionally contained a CpG motif was reported which induced IL-12 in peripheral blood mononuclear cells (PBMCs) without special delivery agents when phosphorothioate modified.131 Overcoming the need for cellular delivery might be achieved by presence of a poly(G) motif that leads to higher order structures and increased nucleic acid uptake.118,132
Immune activation by siRNA
Based on the hallmark observation that RNA interference can be induced in mammalian cells with 21-nucleotide duplexes, which avoid the typical powerful immune response of longer dsRNA,1 siRNA has become a universal molecular tool and potential therapeutic drug.108 An early report claiming that siRNA does not induce type I IFN133 has been convincingly contradicted in several studies.134-137 One such case reports an IFN response to shRNA vectors.134 In a genome wide expression analysis, 21 bp siRNAs induced a considerable number of genes of the IFN response.135 This response involved the dsRNA-activated serine-threonine kinase PKR. However, as found by analysis of cells deficient in IFN signaling, the gene silencing effect was independent of an IFN response, showing that both functions – gene knockdown and immunostimulation – rely on pathways that are clearly different.135
Careful recent analysis strongly suggests that immune response to siRNA in a TLR3, 7 or 8 dependent manner is sequence-dependent, thus reconciling several of the above reports.53,138,139 Sequence specific TLR7 activation depended upon delivery by cationic lipoplexes and endosomal maturation.53,139 There is recurrent emergence of GU-rich or U-rich regions which are particularly efficient in TLR7 stimulation53,123,140 by “entire” siRNAs, i.e., duplexes composed of sense and antisense strand. These nucleotide combinations appear to be particularly effective in ss siRNA components (sense or antisense),48,139 possibly because they have a higher propensity for forming secondary structures through mismatched G:U basepairs.141
Another report claimed that siRNA and short hairpin RNA (shRNA) induced IFNα and TNFα secretion in professional innate immune cells. The secretion could be inhibited by blocking Toll/Interleukin-1 receptor domain-containing adaptor inducing IFN-β (TRIF) or interferon regulatory factor 3 signaling and could be increased by TLR3 overexpression, thus identifying sequence independent TLR3 activation as an additional mode of action.138 Understanding the molecular details and requirements of RNA/TLR stimulation will in turn allow the development of more specific siRNAs that avoid immunostimulation, as well as to predict the potential of any given RNA sequence in terms of innate immune activation.142
Besides stimulation of endosomal TLRs, it was shown that 27-mer siRNAs with blunt ends but not with 2-base 3′overhangs triggered cytoplasmic RIG-I to induce type I IFN143 thus providing an explanation why endogenous miRNAs with a Dicer signature avoid immunorecognition. Importantly, as many cell lines have impaired IFN response pathways, use of primary cells will be necessary to assess the stimulatory potential of synthetic RNAs. An independent study confirmed that the expression of 90% of genes that were regulated by the TLR7 ligand R848 (Fig. 3) was also modified by ss siRNA. However, TLR-independent genes were activated as well confirming the existence of multiple siRNA recognition systems.144
While activation of the innate immune system is a serious concern in the development of siRNA therapeutics, where it must typically be avoided as an aspect of siRNA toxicity,112,113 alternative strategies include immunostimulation as a beneficial therapeutic feature, e.g., in antiviral or anticancer strategies.2,145-148
Using modifications to decrease immunostimulation by siRNA
In contrast to the deliberate immunostimulation mentioned above, avoidance of an interferon response is a necessity in most therapeutic applications of RNA. Beyond sequence optimization, chemical modification is the method of choice, which, though occasionally cumbersome, allows optimization of other parameters as well, including nuclease resistance etc. First generation siRNAs in clinical trials typical contain a mix of phosphorothioates, 2’-O methyl- and 2’-desoxynucleotide modifications.108,149 The advantages of 2’-O methylation include immunosilencing, increased nuclease resistance, low cost, and the fact that its degradation products occur naturally in the cell. Indeed, siRNAs containing 2’-O-methyl modifications at every other nucleotide,150 are now in clinical trials.151 The 2’-hydroxyl group does not play a critical role at most positions in siRNA135 and thus is an attractive candidate to dissect immunostimulation from RNAi . Indeed, substitution of 2'-hydroxyl uridines with either 2'-fluorouridines, 2'-deoxyuridines (dU) or 2'-O-methyluridines (Um) decreased immune activation of siRNAs within PBMCs.140 Among those modifications, 2’-O-methylation not only abolished immunostimulation (in PBMCs including pDCs) but also suppressed activity of an unmodified, stimulatory strand, thus acting as an antagonist. Um-containing ONs inhibited stimulation by unmodified siRNAs at nanomolar concentrations. Using global gene expression analysis, Um-siRNA abrogated nearly all 270 genes that were induced by ss siRNA in human monocytes. Thus, Um acts as dominant negative, suppressing modification and indeed, it also suppressed TLR7/8 stimulation in PBMCs by bRNA.100 In contrast, dU lacked immunostimulation but did not act in a suppressive manner, an observation also confirmed by others.48 Likewise, incorporation of 2’-O-methyl residues into siRNA was sufficient to suppress immunostimulation even if the modification was not incorporated into the immunostimulatory strand, confirming a role as antagonistic modification.99 Of note in this report the nucleoside modification did not alter RNA interference. In contrast, others observed that 2’-O-methyl modifications also decreased efficacy of gene knockdown.48 It was reported that modifications have to be restricted to certain positions in a siRNA152 thus possibly explaining the slightly varying findings.
In general, modifications in the middle of the siRNA duplex are not well tolerated, as this part is important for activity of the RNA induced silencing complex (RISC),153 an important fact in attempts to use modifications to decrease siRNA side effects. Also, incorporation of such modifications in the passenger strand might be recommended as this should have less effect on RNAi.154
Incorporation of dU or dT into siRNA allowed for dissection of immunostimulation and RNA interference.48 Whereas IFNα secretion from PBMCs was decreased (or abolished at concentration necessary for RNA interference), gene knockdown was as efficient as with unmodified siRNA. For maximal effects, both strands of the siRNA had to be modified as those modifications behaved as “silent” but not “suppressive” modifications. Interestingly, 2’-deoxyriboses with nucleobases other than uridine were not effective, arguing for a specific recognition of uridine by TLR7. Also, neither 5-methylcytidine nor 7-deazaguanosine, both modifications affecting the surface of the major groove of a duplex, modified IFNα secretion. Abrogation of immunostimulation by incorporation of Um or Gm into one strand of the siRNA duplex were confirmed to be effective and such modified siRNA were efficient and specific in systemic gene knockdown.144,155 From those results it was suggested to use alternating 2’-O-ribose methylation as a general strategy to generate siRNA.150 In contrast to suppressive effects of Am, Um and Gm, we observed that Cm was not sufficient to suppress immunostimulation48 arguing for a lack of recognition of cytidine residues.
RNA immunostimulation in infections
The role of nucleic acid recognizing TLRs in infections is largely defined by pDCs, for which a large body of evidence156 documents the requirement for TLR9 to respond to DNA viruses157-159 and for TLR7 mediated type I IFN induction in response to RNA viruses, including influenza virus, respiratory syncytial virus, Sendai virus or vesicular stomatitis virus.45,55 MyD88 dependent but TLR9 independent recognition of murine cytomegalovirus might indicate that further receptors exist.160
TLR-related innate immunity affects induction of adaptive immunity29 and therefore defects in pattern recognition receptors can affect both limbs of the immune system. Some valuable insight comes from recent reports on human patients with genetic deficiencies of components of the innate immune system, which frequently result in predispositions to bacterial or viral infections. For example, children with a dominant-negative allele of TLR3 were found to be susceptible to Herpes simplex-virus-1 encephalitis.161 Patients suffering from primary deficiency of IRAK4, an important signaling molecule for most TLRs, display a loss of PBMC responsiveness to TLR ligands including R848 (also known as Resiquimod, a TLR 7, 8 ligand displayed in Figure 2) and CpG ON (TLR9 ligand).162,163 As an apparent consequence, patients were prone to invasive pneumococcal diseases in young childhood and half of them died. With increasing age the infection susceptibility vanished. IFN responses to most viruses tested were not severely affected and no specific viral infections in IRAK-4 deficient patients have been reported arguing for a redundant role of IFN inducing TLRs, especially the nucleic acid receptors, in viral defense.
Genetic predispositions to viral infections have been reported for autosomal recessive deficiency in Unc93b1, a protein necessary for trafficking of endosomal, nucleic acid detecting TLRs164,165). This resulted in impaired antiviral IFN responses and Herpes simplex virus-1 encephalitis in children.91 Unc93b1 defect that abrogates TLR3, 7 and 9 functions was also reported to decrease resistance to infection with the parasite Toxoplasma gondii.89 Furthermore sensing of group B streptococci by macrophages and cellular activation was dependent on bacterial ssRNA and involved MyD88 and Unc93b1, although the established RNA sensors TLR3 and TLR7 were dispensable.90 Further studies in a mouse model of group A streptococci-induced lethal subcutaneous cellulitis analyzed type I IFN induction of macrophages and pDCs.166 Type I IFN induction in macrophages was dependent on endosomal recognition of streptococcal DNA. In contrast myeloid DCs recognized streptococcal RNA in a MyD88 but TLR7 independent manner. The results suggest that nucleic acid detection is important for defense of Streptococcus pyogenes which evades immune recognition by other TLRs.
A role of TLR7 in signaling toward fungal infections was reported for IFNβ induction in DCs in response to Candida infection. IFNβ induction was dependent on TLR7, MyD88 and IRF1 and occurred after completed phagocytosis.167 A similar report also showed that IFNβ induction in myeloid DCs to Candida infection required MyD88 and partly TLR7 or TLR9.168 Moreover it was shown that Candida DNA as well as RNA is recognized. Type I IFN induction was necessary to control fungal growth. Although yeast RNA has been shown to bear nucleoside modifications similar to other eukaryotes and was not overly stimulatory in vitro,56 it is possible that enrichment within the endolysosome during severe infections overcomes classical self/foreign discrimination principles. This would be compatible with a model in which intrinsic structural features (nucleoside modifications) in cooperation with spatial control (endolysosome) would account for overall discrimination of self and foreign nucleic acids.
Manipulating the course of natural infections through RNA-mediated TLR stimulation
As nucleic acid recognizing TLRs contribute to resistance against virus infections it was speculated that TLR7 agonists might be used to increase anti-viral responses especially when chronic infections are established. In this line it was shown that TLR7 stimulation results in increased anti-Hepatitis C (HCV) immunity by means involving IFN dependent as well as independent effects.169 In a cell culture system HCV levels were reduced by treatment with a TLR7 agonist. As TLR7 is expressed in hepatocytes it was speculated that TLR7 stimulation could be used to reinforce HCV immunity.
Usage of antagonists for nucleic acid sensing TLRs might be a strategy to avoid overshooting reactions toward such pathogens for which nucleic acid detection is of importance during natural immune responses. It was reported that cerebral malaria in an experimental model with Plasmodium berghei is caused by excessive immunostimulation with TLR9 and possibly other nucleic acid recognizing TLRs.170 Using E6446, a synthetic antagonist composed of benzoxazole with two-sided pyrrolidine rings that blocks TLR9, and TLR8 at higher concentrations, severe symptoms of cerebral malaria could be successfully inhibited.
Mislead self RNA recognition and autoimmune diseases
Whereas under healthy, physiological conditions discrimination of self- and foreign-derived nucleic acids is achieved through sequence and structure as well as spatial control (outlined above), recent publications indicate that self-tolerance of nucleic acids can be broken in certain pathologies. Thus, recognition of self-nucleic acids through TLRs is not entirely prevented. An important finding was first published for recognition of self-DNA by TLR9. B cells prone to produce IgG autoantibodies known as rheumatoid factors were shown to be stimulated by dual engagement of the antigen receptor as well as TLR9.72 Chromatin-antibody immune complexes activated autoreactive B cells by triggering an endosomal TLR. Increased uptake and delivery of self-DNA within those chromatin complexes contributed to the breakdown of TLR-mediated self/foreign discrimination. Similarly, dsDNA-specific antibodies triggered autoreactive B cells through antigen receptor/TLR9 coengagement.171 A two-stage model for autoimmunity in systemic lupus erythematosus was proposed that discriminates TLR-independent and TLR-dependent processes.172 TLR-independent dendritic cell uptake of self-derived cell debris is followed by amplification through TLR recognition of nucleic acids in respective complexes. Both phases are dependent on type I IFN. These observation fit well to an experimental observation which shows that ectopic expression of TLR9 at the cell surface results in reactivity with mammalian DNA, emphasizing the need for control by localization.69 In turn, conditions in which spatial control is overcome, e.g., increased occurrence and uptake of self-DNA, defects in vesicle trafficking or membrane integrity, autoimmunity might develop through unphysiological triggering of TLRs in conjunction with self-antigen presentation.
Similar to the findings of synergistic activation of the B-cell receptor and TLR9 it was subsequently shown that this paradigm also holds true for RNA autoantigens. RNA containing immune complexes triggered the B-cell receptor together with TLR7.173 This response was markedly enhanced by IFNα thus resembling a situation of disease progression in patients with autoimmune systemic lupus erythematosus. Autoimmune-prone mice lacking the nucleic acid response through MyD88 deficiency had reduced chromatin autoantibody titers. Of note, pDC activation and IFNα secretion can occur with self-nucleic acids through increased endolysosomal delivery, thus overcoming spatial control. It was shown that nucleic acid/immunoglobulin complexes can be taken up into pDCs through the low-affinity Fc receptor (FcγRIIA). Internalization is followed by TLR9 activation overcoming the physiological spatial control.174 Another way to deliver self-DNA is by interaction with the antimicrobial peptide LL37.175 LL37 increased delivery of aggregated DNA into the early endosome where it triggered TLR9. Thus, LL37 converts self-DNA to stimulatory DNA and, interestingly, LL37 plays a role in pDC activation in psoriasis.176 It can be speculated that similar findings hold true for RNA delivery and TLR7 stimulation.
It has also been shown that high mobility group box protein 1 is a general nucleic acid sensor that delivers nucleic acids into TLR-bearing vesicles and synergistically activates the cell through its receptor for advanced glycation end products (RAGE).69 HMGB1 is a nuclear-DNA binding protein that can be released by dying cells and interacts with aggregated DNA. Later, it was shown that HMGB1, 2, 3 act as general nucleic acid sensors including DNA and RNA and facilitate stimulation of TLR3, 7, 9 by their cognate ligands.86 HMGB knockout mice were defective in DNA or RNA induced type I IFN secretion and proinflammatory cytokine induction. Thus, HMGBs could be promiscuous nucleic acid sensors that act together with specific TLRs to mediate self/foreign discrimination.
Similar to findings for DNA it was shown that self-RNA rich in uridine and guanosine and RNA in small nuclear ribonucleoprotein can induce type I IFN in plasmacytoid DCs in a TLR7 dependent manner.177 Thus, nucleic acid recognizing TLRs might play a role in pathological activation of plasmacytoid DCs and B-lymphocytes to produce autoantibodies against DNA and RNA as observed in systemic lupus erythematosus.178 In this line TLR7 gene duplication has been found to promote autoimmune diseases, plasmacytoid DC activation, type I IFN production and autoantibody formation in mice.179,180 In turn, TLR7 deficiency results in decreased susceptibility for autoimmune diseases with lowered serum autoantibody levels.181,182
Fc gamma receptor independent, but strictly TLR7 dependent induction of autoimmunity was reported in a model with 2,6,10,14-tetramethylpentadecane induced lupus-like disease with immune complex nephritis and autoantibodies to DNA and ribonucleoproteins.183 TLR7 triggered type I IFN was also shown to upregulate MHC class I expression and induction of autoimmune T-cell responses in a model of pancreatic lymphocytic choriomeningitis virus glycoprotein expression.184 Autoreactive cytotoxic T cells were only activated when an additional TLR stimulus was provided.
TLR3 contribution to autoimmunity has been reported for autoimmune liver damage and rheumatoid arthritis.185,186 Thus, incubation of fibroblasts that bear TLR3 with necrotic synovial fluid cells from rheumatoid arthritis patients resulted in increased chemokine and cytokine secretion. Interestingly, Unc93b1 signaling (implicating TLR3, 7 8, 9 nucleic acid sensing) was necessary to remove developing autoreactive B cells and patients with Unc93b1 deficiency had defective central and peripheral B-cell tolerance with accumulation of autoreactive mature B cells.187
Glucocorticoids are used to treat autoimmune diseases and glucocorticoids decrease proinflammatory NF-κB signaling. However, glucocorticoids lose activity in long-term treatment. Recently, it was shown that pDCs triggered through self-nucleic acids via TLR7 or TLR9 induced NF-κB for cell survival. NF-κB activation in pDCs however was not sensitive to glucocorticoids and in turn pDCs produced IFNα was not decreased.188 TLR7/9 antagonists might therefore be of use to decrease pDC activation and to spare glucocorticoid. In a model of collagen-induced arthritis reduction of TLR7 expression by means of lentiviral transfer resulted in decreased inflammation and increased clinical outcome.189
The common topic in all reports that claim breakdown of self-tolerance for nucleic acid recognizing TLRs is that the underlying patterns that mediate self/foreign discrimination are not entirely specific for microbes. Perhaps this is the reason why spatial restriction occurs for TLR3, 7, 8, 9. This allows for an additional level of control that under healthy conditions excludes self-reactivity.
Closing Remarks
Recognition of nucleic acids should be, as such, a simple task on the molecular level, because of the few building blocks present. However, discrimination of nucleic acids originating from the host organism against that of an invading pathogen is even more daunting, probably for the very same reasons. As we have outlined here, mammals have evolved multiple sophisticated sensors in different cellular compartments. More discrimination principles unravel as newer and more realistic assays are being developed. In all likelihood, more receptors for nucleic acid recognition await detection, and after completion of this task, new challenges appear. Not only do all parameters for recognition and discrimination have to be clarified, if therapeutic use is to emerge from this research; a much more challenging task may be to define the interplay between the various receptors, whose signaling, in its sum, must be what alarms our body to an imminent infection. Although single receptors may make dominating contributions in the detection of e.g., cytosolic viral RNA, the variety of different receptors and the use of common molecular relays in the signaling pathways strongly suggests the possibility of cooperative effects in pathogen sensing, where the full-fledged alarm mode might be most efficiently stimulated upon detection of multiple PAMPs by different receptors.
Acknowledgments
We thank Steffen Kaiser for technical assistance and graphical concepts. This work has been supported by a grant from SAMT (M.H.).
Glossary
Abbreviations:
- PBMCs
peripheral blood mononuclear cells
- pDCs
plasmacytoid dendritic cells
- DC
dendritic cells
- CLRs
C-type lectin receptors
- siRNA
small interfering RNAs
- dsRNA
double stranded RNA
- RNAi
RNA interference
- bRNA
bacterial RNA
- tRNA
transfer RNA
- rRNA
ribosomal RNA
- TLR
Toll-like receptors
- NLR
nucleotide-oligomerization domain protein (Nod)-like receptors
- RIG-I
retinoic acid inducible gene I
- Mda5
melanoma differentiation-associated gene-5
- Nlrp
NLR family, pyrin domain containing
- AIM2
absent in melanoma 2
- PAMP
pathogen-associated molecular pattern
- PRR
pattern recognition receptor
- MyD88
myeloid differentiation factor 88
- IRAK
interleukin-1 receptor-associated kinase
- TRAF
tumor necrosis factor (TNF) receptor associated factor
- SLE
systemic lupus erythematosus
- HIV
human immunodeficiency virus
- TLR3-ECD
TLR3 ectodomain
- IFNα
interferon alpha
- IFN
interferon
- IRF1
interferon regulatory factor-1
- NFκB
nuclear factor κB
- IL
interleukin
- HMGB
high-mobility group box
- ON
oligonucleotide
- Bp
basepair
- shRNA
short hairpin RNA
- RISC
RNA-induced silencing complex
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/20206
References
- 1.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.Schlee M, Hornung V, Hartmann G. siRNA and isRNA: two edges of one sword. Mol Ther. 2006;14:463–70. doi: 10.1016/j.ymthe.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–83. doi: 10.1016/S0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 4.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–8. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 5.Steinman RM, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. I. Morphology, quantitation, tissue distribution. J Exp Med. 1973;137:1142–62. doi: 10.1084/jem.137.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janeway CA., Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54:1–13. doi: 10.1101/SQB.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Medzhitov R. Approaching the asymptote: 20 years later. Immunity. 2009;30:766–75. doi: 10.1016/j.immuni.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–84. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 9.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–7. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 10.Yang RB, Mark MR, Gray A, Huang A, Xie MH, Zhang M, et al. Toll-like receptor-2 mediates lipopolysaccharide-induced cellular signalling. Nature. 1998;395:284–8. doi: 10.1038/26239. [DOI] [PubMed] [Google Scholar]
- 11.Kirschning CJ, Wesche H, Merrill Ayres T, Rothe M. Human toll-like receptor 2 confers responsiveness to bacterial lipopolysaccharide. J Exp Med. 1998;188:2091–7. doi: 10.1084/jem.188.11.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inohara N, Koseki T, del Peso L, Hu Y, Yee C, Chen S, et al. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J Biol Chem. 1999;274:14560–7. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- 13.Bertin J, Nir WJ, Fischer CM, Tayber OV, Errada PR, Grant JR, et al. Human CARD4 protein is a novel CED-4/Apaf-1 cell death family member that activates NF-kappaB. J Biol Chem. 1999;274:12955–8. doi: 10.1074/jbc.274.19.12955. [DOI] [PubMed] [Google Scholar]
- 14.Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–72. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 15.Brown GD, Gordon S. Immune recognition. A new receptor for beta-glucans. Nature. 2001;413:36–7. doi: 10.1038/35092620. [DOI] [PubMed] [Google Scholar]
- 16.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–7. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 17.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–5. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 18.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–26. doi: 10.1016/S1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 19.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–8. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–13. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bürckstümmer T, Baumann C, Blüml S, Dixit E, Dürnberger G, Jahn H, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–72. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 22.Ogura Y, Sutterwala FS, Flavell RA. The inflammasome: first line of the immune response to cell stress. Cell. 2006;126:659–62. doi: 10.1016/j.cell.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–33. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 24.O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–64. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 25.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–50. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Nish S, Medzhitov R. Host defense pathways: role of redundancy and compensation in infectious disease phenotypes. Immunity. 2011;34:629–36. doi: 10.1016/j.immuni.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol. 2009;9:535–42. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasare C, Medzhitov R. Control of B-cell responses by Toll-like receptors. Nature. 2005;438:364–8. doi: 10.1038/nature04267. [DOI] [PubMed] [Google Scholar]
- 29.Schnare M, Barton GM, Holt AC, Takeda K, Akira S, Medzhitov R. Toll-like receptors control activation of adaptive immune responses. Nat Immunol. 2001;2:947–50. doi: 10.1038/ni712. [DOI] [PubMed] [Google Scholar]
- 30.Kasturi SP, Skountzou I, Albrecht RA, Koutsonanos D, Hua T, Nakaya HI, et al. Programming the magnitude and persistence of antibody responses with innate immunity. Nature. 2011;470:543–7. doi: 10.1038/nature09737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deane JA, Bolland S. Nucleic acid-sensing TLRs as modifiers of autoimmunity. J Immunol. 2006;177:6573–8. doi: 10.4049/jimmunol.177.10.6573. [DOI] [PubMed] [Google Scholar]
- 32.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–8. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 33.de Bouteiller O, Merck E, Hasan UA, Hubac S, Benguigui B, Trinchieri G, et al. Recognition of double-stranded RNA by human toll-like receptor 3 and downstream receptor signaling requires multimerization and an acidic pH. J Biol Chem. 2005;280:38133–45. doi: 10.1074/jbc.M507163200. [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto M, Funami K, Tanabe M, Oshiumi H, Shingai M, Seto Y, et al. Subcellular localization of Toll-like receptor 3 in human dendritic cells. J Immunol. 2003;171:3154–62. doi: 10.4049/jimmunol.171.6.3154. [DOI] [PubMed] [Google Scholar]
- 35.Nishiya T, DeFranco AL. Ligand-regulated chimeric receptor approach reveals distinctive subcellular localization and signaling properties of the Toll-like receptors. J Biol Chem. 2004;279:19008–17. doi: 10.1074/jbc.M311618200. [DOI] [PubMed] [Google Scholar]
- 36.Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, Segal DM, et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–81. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–5. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 38.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–82. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 39.Leonard JN, Ghirlando R, Askins J, Bell JK, Margulies DH, Davies DR, et al. The TLR3 signaling complex forms by cooperative receptor dimerization. Proc Natl Acad Sci U S A. 2008;105:258–63. doi: 10.1073/pnas.0710779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukuda K, Watanabe T, Tokisue T, Tsujita T, Nishikawa S, Hasegawa T, et al. Modulation of double-stranded RNA recognition by the N-terminal histidine-rich region of the human toll-like receptor 3. J Biol Chem. 2008;283:22787–94. doi: 10.1074/jbc.M802284200. [DOI] [PubMed] [Google Scholar]
- 41.Pirher N, Ivicak K, Pohar J, Bencina M, Jerala R. A second binding site for double-stranded RNA in TLR3 and consequences for interferon activation. Nat Struct Mol Biol. 2008;15:761–3. doi: 10.1038/nsmb.1453. [DOI] [PubMed] [Google Scholar]
- 42.Avril T, de Tayrac M, Leberre C, Quillien V. Not all polyriboinosinic-polyribocytidylic acids (Poly I:C) are equivalent for inducing maturation of dendritic cells: implication for alpha-type-1 polarized DCs. J Immunother. 2009;32:353–62. doi: 10.1097/CJI.0b013e31819d29bf. [DOI] [PubMed] [Google Scholar]
- 43.Karikó K, Ni H, Capodici J, Lamphier M, Weissman D. mRNA is an endogenous ligand for Toll-like receptor 3. J Biol Chem. 2004;279:12542–50. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 44.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–9. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 45.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–31. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 46.Karikó K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–75. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 47.Diken M, Kreiter S, Selmi A, Britten CM, Huber C, Türeci O, et al. Selective uptake of naked vaccine RNA by dendritic cells is driven by macropinocytosis and abrogated upon DC maturation. Gene Ther. 2011;18:702–8. doi: 10.1038/gt.2011.17. [DOI] [PubMed] [Google Scholar]
- 48.Eberle F, Giessler K, Deck C, Heeg K, Peter M, Richert C, et al. Modifications in small interfering RNA that separate immunostimulation from RNA interference. J Immunol. 2008;180:3229–37. doi: 10.4049/jimmunol.180.5.3229. [DOI] [PubMed] [Google Scholar]
- 49.Gehrig S, Eberle ME, Botschen F, Rimbach K, Eberle F, Eigenbrod T, et al. Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J Exp Med. 2012;209:225–33. doi: 10.1084/jem.20111044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jöckel S, Nees G, Sommer R, Zhao Y, Cherkasov D, Hori H, et al. The 2′-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition. J Exp Med. 2012;209:235–41. doi: 10.1084/jem.20111075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gantier MP, Tong S, Behlke MA, Xu D, Phipps S, Foster PS, et al. TLR7 is involved in sequence-specific sensing of single-stranded RNAs in human macrophages. J Immunol. 2008;180:2117–24. doi: 10.4049/jimmunol.180.4.2117. [DOI] [PubMed] [Google Scholar]
- 52.Diebold SS, Massacrier C, Akira S, Paturel C, Morel Y, Reis e Sousa C. Nucleic acid agonists for Toll-like receptor 7 are defined by the presence of uridine ribonucleotides. Eur J Immunol. 2006;36:3256–67. doi: 10.1002/eji.200636617. [DOI] [PubMed] [Google Scholar]
- 53.Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11:263–70. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 54.Forsbach A, Nemorin JG, Völp K, Samulowitz U, Montino C, Müller C, et al. Characterization of conserved viral leader RNA sequences that stimulate innate immunity through TLRs. Oligonucleotides. 2007;17:405–17. doi: 10.1089/oli.2007.0098. [DOI] [PubMed] [Google Scholar]
- 55.Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci U S A. 2004;101:5598–603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eberle F, Sirin M, Binder M, Dalpke AH. Bacterial RNA is recognized by different sets of immunoreceptors. Eur J Immunol. 2009;39:2537–47. doi: 10.1002/eji.200838978. [DOI] [PubMed] [Google Scholar]
- 57.Mancuso G, Gambuzza M, Midiri A, Biondo C, Papasergi S, Akira S, et al. Bacterial recognition by TLR7 in the lysosomes of conventional dendritic cells. Nat Immunol. 2009;10:587–94. doi: 10.1038/ni.1733. [DOI] [PubMed] [Google Scholar]
- 58.Wei T, Gong J, Jamitzky F, Heckl WM, Stark RW, Rössle SC. Homology modeling of human Toll-like receptors TLR7, 8, and 9 ligand-binding domains. Protein Sci. 2009;18:1684–91. doi: 10.1002/pro.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jurk M, Heil F, Vollmer J, Schetter C, Krieg AM, Wagner H, et al. Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat Immunol. 2002;3:499. doi: 10.1038/ni0602-499. [DOI] [PubMed] [Google Scholar]
- 60.Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 61.Lee J, Chuang TH, Redecke V, She L, Pitha PM, Carson DA, et al. Molecular basis for the immunostimulatory activity of guanine nucleoside analogs: activation of Toll-like receptor 7. Proc Natl Acad Sci U S A. 2003;100:6646–51. doi: 10.1073/pnas.0631696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Forsbach A, Samulowitz U, Völp K, Hofmann HP, Noll B, Tluk S, et al. Dual or triple activation of TLR7, TLR8, and/or TLR9 by single-stranded oligoribonucleotides. Nucleic Acid Ther. 2011;21:423–36. doi: 10.1089/nat.2011.0323. [DOI] [PubMed] [Google Scholar]
- 63.Lan T, Putta MR, Wang D, Dai M, Yu D, Kandimalla ER, et al. Synthetic oligoribonucleotides-containing secondary structures act as agonists of Toll-like receptors 7 and 8. Biochem Biophys Res Commun. 2009;386:443–8. doi: 10.1016/j.bbrc.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 64.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–5. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 65.Wagner H. Bacterial CpG DNA activates immune cells to signal infectious danger. Adv Immunol. 1999;73:329–68. doi: 10.1016/S0065-2776(08)60790-7. [DOI] [PubMed] [Google Scholar]
- 66.Krieg AM, Yi AK, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–9. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 67.Haas T, Metzger J, Schmitz F, Heit A, Müller T, Latz E, et al. The DNA sugar backbone 2′ deoxyribose determines toll-like receptor 9 activation. Immunity. 2008;28:315–23. doi: 10.1016/j.immuni.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 68.Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF, et al. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5:190–8. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 69.Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat Immunol. 2006;7:49–56. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- 70.Napirei M, Karsunky H, Zevnik B, Stephan H, Mannherz HG, Möröy T. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat Genet. 2000;25:177–81. doi: 10.1038/76032. [DOI] [PubMed] [Google Scholar]
- 71.Yasutomo K, Horiuchi T, Kagami S, Tsukamoto H, Hashimura C, Urushihara M, et al. Mutation of DNASE1 in people with systemic lupus erythematosus. Nat Genet. 2001;28:313–4. doi: 10.1038/91070. [DOI] [PubMed] [Google Scholar]
- 72.Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak-Rothstein A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–7. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 73.Christensen SR, Kashgarian M, Alexopoulou L, Flavell RA, Akira S, Shlomchik MJ. Toll-like receptor 9 controls anti-DNA autoantibody production in murine lupus. J Exp Med. 2005;202:321–31. doi: 10.1084/jem.20050338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peter ME, Kubarenko AV, Weber AN, Dalpke AH. Identification of an N-terminal recognition site in TLR9 that contributes to CpG-DNA-mediated receptor activation. J Immunol. 2009;182:7690–7. doi: 10.4049/jimmunol.0900819. [DOI] [PubMed] [Google Scholar]
- 75.Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi GP, Chapman HA, et al. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456:658–62. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Park B, Brinkmann MM, Spooner E, Lee CC, Kim YM, Ploegh HL. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat Immunol. 2008;9:1407–14. doi: 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y, Berke IC, Modis Y. DNA binding to proteolytically activated TLR9 is sequence-independent and enhanced by DNA curvature. EMBO J. 2011;31:919–31. doi: 10.1038/emboj.2011.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–35. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 79.Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–22. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–7. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 81.Binder M, Eberle F, Seitz S, Mücke N, Hüber CM, Kiani N, et al. Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid-inducible gene-I (RIG-I) J Biol Chem. 2011;286:27278–87. doi: 10.1074/jbc.M111.256974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–60. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 83.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sharma S, DeOliveira RB, Kalantari P, Parroche P, Goutagny N, Jiang Z, et al. Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome. Immunity. 2011;35:194–207. doi: 10.1016/j.immuni.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, et al. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7:156–64. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- 86.Yanai H, Ban T, Wang Z, Choi MK, Kawamura T, Negishi H, et al. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature. 2009;462:99–103. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]
- 87.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452:234–8. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 88.Fukui R, Saitoh S, Matsumoto F, Kozuka-Hata H, Oyama M, Tabeta K, et al. Unc93B1 biases Toll-like receptor responses to nucleic acid in dendritic cells toward DNA- but against RNA-sensing. J Exp Med. 2009;206:1339–50. doi: 10.1084/jem.20082316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Melo MB, Kasperkovitz P, Cerny A, Könen-Waisman S, Kurt-Jones EA, Lien E, et al. UNC93B1 mediates host resistance to infection with Toxoplasma gondii. PLoS Pathog. 2010;6:e1001071. doi: 10.1371/journal.ppat.1001071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Deshmukh SD, Kremer B, Freudenberg M, Bauer S, Golenbock DT, Henneke P. Macrophages recognize streptococci through bacterial single-stranded RNA. EMBO Rep. 2011;12:71–6. doi: 10.1038/embor.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Casrouge A, Zhang SY, Eidenschenk C, Jouanguy E, Puel A, Yang K, et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314:308–12. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- 92.Ewald SE, Engel A, Lee J, Wang M, Bogyo M, Barton GM. Nucleic acid recognition by Toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopeptidase. J Exp Med. 2011;208:643–51. doi: 10.1084/jem.20100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sepulveda FE, Maschalidi S, Colisson R, Heslop L, Ghirelli C, Sakka E, et al. Critical role for asparagine endopeptidase in endocytic Toll-like receptor signaling in dendritic cells. Immunity. 2009;31:737–48. doi: 10.1016/j.immuni.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 94.Baumann CL, Aspalter IM, Sharif O, Pichlmair A, Blüml S, Grebien F, et al. CD14 is a coreceptor of Toll-like receptors 7 and 9. J Exp Med. 2010;207:2689–701. doi: 10.1084/jem.20101111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Koski GK, Karikó K, Xu S, Weissman D, Cohen PA, Czerniecki BJ. Cutting edge: innate immune system discriminates between RNA containing bacterial versus eukaryotic structural features that prime for high-level IL-12 secretion by dendritic cells. J Immunol. 2004;172:3989–93. doi: 10.4049/jimmunol.172.7.3989. [DOI] [PubMed] [Google Scholar]
- 96.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, et al. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39(Database issue):D195–201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Anderson BR, Muramatsu H, Jha BK, Silverman RH, Weissman D, Karikó K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–38. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Anderson BR, Muramatsu H, Nallagatla SR, Bevilacqua PC, Sansing LH, Weissman D, et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–92. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Robbins M, Judge A, Liang L, McClintock K, Yaworski E, MacLachlan I. 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol Ther. 2007;15:1663–9. doi: 10.1038/sj.mt.6300240. [DOI] [PubMed] [Google Scholar]
- 100.Sioud M, Furset G, Cekaite L. Suppression of immunostimulatory siRNA-driven innate immune activation by 2′-modified RNAs. Biochem Biophys Res Commun. 2007;361:122–6. doi: 10.1016/j.bbrc.2007.06.177. [DOI] [PubMed] [Google Scholar]
- 101.De Clercq E, Zmudzka B, Shugar D. Antiviral activity of polynucleotides: role of the 2′-hydroxyl and a pyrimidine 5-methyl. FEBS Lett. 1972;24:137–40. doi: 10.1016/0014-5793(72)80845-7. [DOI] [PubMed] [Google Scholar]
- 102.Lacoux C, Di Marino D, Pilo Boyl P, Zalfa F, Yan B, Ciotti MT, et al. BC1-FMRP interaction is modulated by 2′-O-methylation: RNA-binding activity of the tudor domain and translational regulation at synapses. Nucleic Acids Res. 2012 doi: 10.1093/nar/gkr1254. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA. 2011;2:611–31. doi: 10.1002/wrna.79. [DOI] [PubMed] [Google Scholar]
- 104.Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37(Database issue):D159–62. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, et al. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009;37(Database issue):D118–21. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kreiter S, Diken M, Selmi A, Türeci O, Sahin U. Tumor vaccination using messenger RNA: prospects of a future therapy. Curr Opin Immunol. 2011;23:399–406. doi: 10.1016/j.coi.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 107.Karikó K, Weissman D. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Curr Opin Drug Discov Devel. 2007;10:523–32. [PubMed] [Google Scholar]
- 108.Vaishnaw AK, Gollob J, Gamba-Vitalo C, Hutabarat R, Sah D, Meyers R, et al. A status report on RNAi therapeutics. Silence. 2010;1:14. doi: 10.1186/1758-907X-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chem Biol. 2012;19:60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–7. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 112.Watts JK, Deleavey GF, Damha MJ. Chemically modified siRNA: tools and applications. Drug Discov Today. 2008;13:842–55. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 113.Deleavey GF, Watts JK, Alain T, Robert F, Kalota A, Aishwarya V, et al. Synergistic effects between analogs of DNA and RNA improve the potency of siRNA-mediated gene silencing. Nucleic Acids Res. 2010;38:4547–57. doi: 10.1093/nar/gkq181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhou X, He X, Liang J, Li A, Xu T, Kieser T, et al. A novel DNA modification by sulphur. Mol Microbiol. 2005;57:1428–38. doi: 10.1111/j.1365-2958.2005.04764.x. [DOI] [PubMed] [Google Scholar]
- 115.Eckstein F. Phosphorothioation of DNA in bacteria. Nat Chem Biol. 2007;3:689–90. doi: 10.1038/nchembio1107-689. [DOI] [PubMed] [Google Scholar]
- 116.Wang L, Chen S, Xu T, Taghizadeh K, Wishnok JS, Zhou X, et al. Phosphorothioation of DNA in bacteria by dnd genes. Nat Chem Biol. 2007;3:709–10. doi: 10.1038/nchembio.2007.39. [DOI] [PubMed] [Google Scholar]
- 117.Detzer A, Sczakiel G. Phosphorothioate-stimulated uptake of siRNA by mammalian cells: a novel route for delivery. Curr Top Med Chem. 2009;9:1109–16. doi: 10.2174/156802609789630884. [DOI] [PubMed] [Google Scholar]
- 118.Dalpke AH, Zimmermann S, Albrecht I, Heeg K. Phosphodiester CpG oligonucleotides as adjuvants: polyguanosine runs enhance cellular uptake and improve immunostimulative activity of phosphodiester CpG oligonucleotides in vitro and in vivo. Immunology. 2002;106:102–12. doi: 10.1046/j.1365-2567.2002.01410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lan T, Dai M, Wang D, Zhu FG, Kandimalla ER, Agrawal S. Toll-like receptor 7 selective synthetic oligoribonucleotide agonists: synthesis and structure-activity relationship studies. J Med Chem. 2009;52:6871–9. doi: 10.1021/jm901145s. [DOI] [PubMed] [Google Scholar]
- 120.Hornung V, Rothenfusser S, Britsch S, Krug A, Jahrsdörfer B, Giese T, et al. Quantitative expression of toll-like receptor 1-10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J Immunol. 2002;168:4531–7. doi: 10.4049/jimmunol.168.9.4531. [DOI] [PubMed] [Google Scholar]
- 121.Lipford GB, Bauer M, Blank C, Reiter R, Wagner H, Heeg K. CpG-containing synthetic oligonucleotides promote B and cytotoxic T cell responses to protein antigen: a new class of vaccine adjuvants. Eur J Immunol. 1997;27:2340–4. doi: 10.1002/eji.1830270931. [DOI] [PubMed] [Google Scholar]
- 122.Kerkmann M, Rothenfusser S, Hornung V, Towarowski A, Wagner M, Sarris A, et al. Activation with CpG-A and CpG-B oligonucleotides reveals two distinct regulatory pathways of type I IFN synthesis in human plasmacytoid dendritic cells. J Immunol. 2003;170:4465–74. doi: 10.4049/jimmunol.170.9.4465. [DOI] [PubMed] [Google Scholar]
- 123.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–62. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 124.Forsbach A, Nemorin JG, Montino C, Müller C, Samulowitz U, Vicari AP, et al. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J Immunol. 2008;180:3729–38. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- 125.Ablasser A, Poeck H, Anz D, Berger M, Schlee M, Kim S, et al. Selection of molecular structure and delivery of RNA oligonucleotides to activate TLR7 versus TLR8 and to induce high amounts of IL-12p70 in primary human monocytes. J Immunol. 2009;182:6824–33. doi: 10.4049/jimmunol.0803001. [DOI] [PubMed] [Google Scholar]
- 126.Scheel B, Braedel S, Probst J, Carralot JP, Wagner H, Schild H, et al. Immunostimulating capacities of stabilized RNA molecules. Eur J Immunol. 2004;34:537–47. doi: 10.1002/eji.200324198. [DOI] [PubMed] [Google Scholar]
- 127.Scheel B, Aulwurm S, Probst J, Stitz L, Hoerr I, Rammensee HG, et al. Therapeutic anti-tumor immunity triggered by injections of immunostimulating single-stranded RNA. Eur J Immunol. 2006;36:2807–16. doi: 10.1002/eji.200635910. [DOI] [PubMed] [Google Scholar]
- 128.Kanzler H, Barrat FJ, Hessel EM, Coffman RL. Therapeutic targeting of innate immunity with Toll-like receptor agonists and antagonists. Nat Med. 2007;13:552–9. doi: 10.1038/nm1589. [DOI] [PubMed] [Google Scholar]
- 129.Barchet W, Wimmenauer V, Schlee M, Hartmann G. Accessing the therapeutic potential of immunostimulatory nucleic acids. Curr Opin Immunol. 2008;20:389–95. doi: 10.1016/j.coi.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 130.Lan T, Kandimalla ER, Yu D, Bhagat L, Li Y, Wang D, et al. Stabilized immune modulatory RNA compounds as agonists of Toll-like receptors 7 and 8. Proc Natl Acad Sci U S A. 2007;104:13750–5. doi: 10.1073/pnas.0706059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sugiyama T, Gursel M, Takeshita F, Coban C, Conover J, Kaisho T, et al. CpG RNA: identification of novel single-stranded RNA that stimulates human CD14+CD11c+ monocytes. J Immunol. 2005;174:2273–9. doi: 10.4049/jimmunol.174.4.2273. [DOI] [PubMed] [Google Scholar]
- 132.Kerkmann M, Costa LT, Richter C, Rothenfusser S, Battiany J, Hornung V, et al. Spontaneous formation of nucleic acid-based nanoparticles is responsible for high interferon-alpha induction by CpG-A in plasmacytoid dendritic cells. J Biol Chem. 2005;280:8086–93. doi: 10.1074/jbc.M410868200. [DOI] [PubMed] [Google Scholar]
- 133.Heidel JD, Hu S, Liu XF, Triche TJ, Davis ME. Lack of interferon response in animals to naked siRNAs. Nat Biotechnol. 2004;22:1579–82. doi: 10.1038/nbt1038. [DOI] [PubMed] [Google Scholar]
- 134.Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34:263–4. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 135.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5:834–9. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 136.Marques JT, Williams BR. Activation of the mammalian immune system by siRNAs. Nat Biotechnol. 2005;23:1399–405. doi: 10.1038/nbt1161. [DOI] [PubMed] [Google Scholar]
- 137.Persengiev SP, Zhu X, Green MR. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10:12–8. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Karikó K, Bhuyan P, Capodici J, Weissman D. Small interfering RNAs mediate sequence-independent gene suppression and induce immune activation by signaling through toll-like receptor 3. J Immunol. 2004;172:6545–9. doi: 10.4049/jimmunol.172.11.6545. [DOI] [PubMed] [Google Scholar]
- 139.Sioud M. Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J Mol Biol. 2005;348:1079–90. doi: 10.1016/j.jmb.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 140.Sioud M. Single-stranded small interfering RNA are more immunostimulatory than their double-stranded counterparts: a central role for 2′-hydroxyl uridines in immune responses. Eur J Immunol. 2006;36:1222–30. doi: 10.1002/eji.200535708. [DOI] [PubMed] [Google Scholar]
- 141.Masquida B, Westhof E. On the wobble GoU and related pairs. RNA. 2000;6:9–15. doi: 10.1017/S1355838200992082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sioud M. Innate sensing of self and non-self RNAs by Toll-like receptors. Trends Mol Med. 2006;12:167–76. doi: 10.1016/j.molmed.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 143.Marques JT, Devosse T, Wang D, Zamanian-Daryoush M, Serbinowski P, Hartmann R, et al. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol. 2006;24:559–65. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 144.Cekaite L, Furset G, Hovig E, Sioud M. Gene expression analysis in blood cells in response to unmodified and 2′-modified siRNAs reveals TLR-dependent and independent effects. J Mol Biol. 2007;365:90–108. doi: 10.1016/j.jmb.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 145.Poeck H, Besch R, Maihoefer C, Renn M, Tormo D, Morskaya SS, et al. 5′-Triphosphate-siRNA: turning gene silencing and Rig-I activation against melanoma. Nat Med. 2008;14:1256–63. doi: 10.1038/nm.1887. [DOI] [PubMed] [Google Scholar]
- 146.Khairuddin N, Gantier MP, Blake SJ, Wu SY, Behlke MA, Williams BR, et al. siRNA-induced immunostimulation through TLR7 promotes antitumoral activity against HPV-driven tumors in vivo. Immunol Cell Biol. 2012;90:187–96. doi: 10.1038/icb.2011.19. [DOI] [PubMed] [Google Scholar]
- 147.Furset G, Sioud M. Design of bifunctional siRNAs: combining immunostimulation and gene-silencing in one single siRNA molecule. Biochem Biophys Res Commun. 2007;352:642–9. doi: 10.1016/j.bbrc.2006.11.059. [DOI] [PubMed] [Google Scholar]
- 148.Gantier MP, Tong S, Behlke MA, Irving AT, Lappas M, Nilsson UW, et al. Rational design of immunostimulatory siRNAs. Mol Ther. 2010;18:785–95. doi: 10.1038/mt.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–4. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 150.Hamm S, Latz E, Hangel D, Müller T, Yu P, Golenbock D, et al. Alternating 2′-O-ribose methylation is a universal approach for generating non-stimulatory siRNA by acting as TLR7 antagonist. Immunobiology. 2010;215:559–69. doi: 10.1016/j.imbio.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 151.Strumberg D, Schultheis B, Traugott U, Vank C, Santel A, Keil O, et al. Phase I clinical development of Atu027, a siRNA formulation targeting PKN3 in patients with advanced solid tumors. Int J Clin Pharmacol Ther. 2012;50:76–8. doi: 10.5414/cpp50076. [DOI] [PubMed] [Google Scholar]
- 152.Amarzguioui M, Holen T, Babaie E, Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–95. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Braasch DA, Jensen S, Liu Y, Kaur K, Arar K, White MA, et al. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967–75. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 154.Sioud M. Does the understanding of immune activation by RNA predict the design of safe siRNAs? Front Biosci. 2008;13:4379–92. doi: 10.2741/3011. [DOI] [PubMed] [Google Scholar]
- 155.Judge AD, Bola G, Lee AC, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 156.Gilliet M, Cao W, Liu YJ. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat Rev Immunol. 2008;8:594–606. doi: 10.1038/nri2358. [DOI] [PubMed] [Google Scholar]
- 157.Krug A, Luker GD, Barchet W, Leib DA, Akira S, Colonna M. Herpes simplex virus type 1 activates murine natural interferon-producing cells through toll-like receptor 9. Blood. 2004;103:1433–7. doi: 10.1182/blood-2003-08-2674. [DOI] [PubMed] [Google Scholar]
- 158.Krug A, French AR, Barchet W, Fischer JA, Dzionek A, Pingel JT, et al. TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity. 2004;21:107–19. doi: 10.1016/j.immuni.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 159.Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J Exp Med. 2003;198:513–20. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hokeness-Antonelli KL, Crane MJ, Dragoi AM, Chu WM, Salazar-Mather TP. IFN-alphabeta-mediated inflammatory responses and antiviral defense in liver is TLR9-independent but MyD88-dependent during murine cytomegalovirus infection. J Immunol. 2007;179:6176–83. doi: 10.4049/jimmunol.179.9.6176. [DOI] [PubMed] [Google Scholar]
- 161.Zhang SY, Jouanguy E, Ugolini S, Smahi A, Elain G, Romero P, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317:1522–7. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 162.Yang K, Puel A, Zhang S, Eidenschenk C, Ku CL, Casrouge A, et al. Human TLR-7-, -8-, and -9-mediated induction of IFN-alpha/beta and -lambda Is IRAK-4 dependent and redundant for protective immunity to viruses. Immunity. 2005;23:465–78. doi: 10.1016/j.immuni.2005.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ku CL, von Bernuth H, Picard C, Zhang SY, Chang HH, Yang K, et al. Selective predisposition to bacterial infections in IRAK-4-deficient children: IRAK-4-dependent TLRs are otherwise redundant in protective immunity. J Exp Med. 2007;204:2407–22. doi: 10.1084/jem.20070628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim YM. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol. 2007;177:265–75. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fukui R, Saitoh S, Kanno A, Onji M, Shibata T, Ito A, et al. Unc93B1 restricts systemic lethal inflammation by orchestrating Toll-like receptor 7 and 9 trafficking. Immunity. 2011;35:69–81. doi: 10.1016/j.immuni.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 166.Gratz N, Hartweger H, Matt U, Kratochvill F, Janos M, Sigel S, et al. Type I interferon production induced by Streptococcus pyogenes-derived nucleic acids is required for host protection. PLoS Pathog. 2011;7:e1001345. doi: 10.1371/journal.ppat.1001345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bourgeois C, Majer O, Frohner IE, Lesiak-Markowicz I, Hildering KS, Glaser W, et al. Conventional dendritic cells mount a type I IFN response against Candida spp. requiring novel phagosomal TLR7-mediated IFN-β signaling. J Immunol. 2011;186:3104–12. doi: 10.4049/jimmunol.1002599. [DOI] [PubMed] [Google Scholar]
- 168.Biondo C, Signorino G, Costa A, Midiri A, Gerace E, Galbo R, et al. Recognition of yeast nucleic acids triggers a host-protective type I interferon response. Eur J Immunol. 2011;41:1969–79. doi: 10.1002/eji.201141490. [DOI] [PubMed] [Google Scholar]
- 169.Lee J, Wu CC, Lee KJ, Chuang TH, Katakura K, Liu YT, et al. Activation of anti-hepatitis C virus responses via Toll-like receptor 7. Proc Natl Acad Sci U S A. 2006;103:1828–33. doi: 10.1073/pnas.0510801103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Franklin BS, Ishizaka ST, Lamphier M, Gusovsky F, Hansen H, Rose J, et al. Therapeutical targeting of nucleic acid-sensing Toll-like receptors prevents experimental cerebral malaria. Proc Natl Acad Sci U S A. 2011;108:3689–94. doi: 10.1073/pnas.1015406108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Viglianti GA, Lau CM, Hanley TM, Miko BA, Shlomchik MJ, Marshak-Rothstein A. Activation of autoreactive B cells by CpG dsDNA. Immunity. 2003;19:837–47. doi: 10.1016/S1074-7613(03)00323-6. [DOI] [PubMed] [Google Scholar]
- 172.Baccala R, Hoebe K, Kono DH, Beutler B, Theofilopoulos AN. TLR-dependent and TLR-independent pathways of type I interferon induction in systemic autoimmunity. Nat Med. 2007;13:543–51. doi: 10.1038/nm1590. [DOI] [PubMed] [Google Scholar]
- 173.Lau CM, Broughton C, Tabor AS, Akira S, Flavell RA, Mamula MJ, et al. RNA-associated autoantigens activate B cells by combined B cell antigen receptor/Toll-like receptor 7 engagement. J Exp Med. 2005;202:1171–7. doi: 10.1084/jem.20050630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lövgren T, Eloranta ML, Båve U, Alm GV, Rönnblom L. Induction of interferon-alpha production in plasmacytoid dendritic cells by immune complexes containing nucleic acid released by necrotic or late apoptotic cells and lupus IgG. Arthritis Rheum. 2004;50:1861–72. doi: 10.1002/art.20254. [DOI] [PubMed] [Google Scholar]
- 175.Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang YH, Homey B, et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–9. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 176.Nestle FO, Conrad C, Tun-Kyi A, Homey B, Gombert M, Boyman O, et al. Plasmacytoid predendritic cells initiate psoriasis through interferon-alpha production. J Exp Med. 2005;202:135–43. doi: 10.1084/jem.20050500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Vollmer J, Tluk S, Schmitz C, Hamm S, Jurk M, Forsbach A, et al. Immune stimulation mediated by autoantigen binding sites within small nuclear RNAs involves Toll-like receptors 7 and 8. J Exp Med. 2005;202:1575–85. doi: 10.1084/jem.20051696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006;6:823–35. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pisitkun P, Deane JA, Difilippantonio MJ, Tarasenko T, Satterthwaite AB, Bolland S. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–72. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- 180.Deane JA, Pisitkun P, Barrett RS, Feigenbaum L, Town T, Ward JM, et al. Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity. 2007;27:801–10. doi: 10.1016/j.immuni.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Berland R, Fernandez L, Kari E, Han JH, Lomakin I, Akira S, et al. Toll-like receptor 7-dependent loss of B cell tolerance in pathogenic autoantibody knockin mice. Immunity. 2006;25:429–40. doi: 10.1016/j.immuni.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 182.Christensen SR, Shupe J, Nickerson K, Kashgarian M, Flavell RA, Shlomchik MJ. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25:417–28. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 183.Lee PY, Kumagai Y, Li Y, Takeuchi O, Yoshida H, Weinstein J, et al. TLR7-dependent and FcgammaR-independent production of type I interferon in experimental mouse lupus. J Exp Med. 2008;205:2995–3006. doi: 10.1084/jem.20080462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lang KS, Recher M, Junt T, Navarini AA, Harris NL, Freigang S, et al. Toll-like receptor engagement converts T-cell autoreactivity into overt autoimmune disease. Nat Med. 2005;11:138–45. doi: 10.1038/nm1176. [DOI] [PubMed] [Google Scholar]
- 185.Lang KS, Georgiev P, Recher M, Navarini AA, Bergthaler A, Heikenwalder M, et al. Immunoprivileged status of the liver is controlled by Toll-like receptor 3 signaling. J Clin Invest. 2006;116:2456–63. doi: 10.1172/JCI28349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Brentano F, Schorr O, Gay RE, Gay S, Kyburz D. RNA released from necrotic synovial fluid cells activates rheumatoid arthritis synovial fibroblasts via Toll-like receptor 3. Arthritis Rheum. 2005;52:2656–65. doi: 10.1002/art.21273. [DOI] [PubMed] [Google Scholar]
- 187.Isnardi I, Ng YS, Srdanovic I, Motaghedi R, Rudchenko S, von Bernuth H, et al. IRAK-4- and MyD88-dependent pathways are essential for the removal of developing autoreactive B cells in humans. Immunity. 2008;29:746–57. doi: 10.1016/j.immuni.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Guiducci C, Gong M, Xu Z, Gill M, Chaussabel D, Meeker T, et al. TLR recognition of self nucleic acids hampers glucocorticoid activity in lupus. Nature. 2010;465:937–41. doi: 10.1038/nature09102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Chen SY, Shiau AL, Li YT, Lin YS, Lee CH, Wu CL, et al. Suppression of collagen-induced arthritis by intra-articular lentiviral vector-mediated delivery of Toll-like receptor 7 short hairpin RNA gene. Gene Ther. 2011 doi: 10.1038/gt.2011.173. [DOI] [PubMed] [Google Scholar]