Abstract
Single nucleotide polymorphisms (SNPs) in microRNAs (miRNAs) or their target sites (miR-SNPs) within the 3′-UTR of mRNAs are increasingly thought to play a major role in pathological dysregulation of gene expression. Here, we studied the functional role of miR-SNPs on miRNA-mediated post-transcriptional regulation of gene expression. First, analyses were performed on a SNP located in the miR-155 target site within the 3′-UTR of the Angiotensin II type 1 receptor (AGTR1; rs5186, A > C) mRNA. Second, a SNP in the 3′-UTR of the muscle RAS oncogene homolog (MRAS; rs9818870, C > T) mRNA was studied which is located outside of binding sites of miR-195 and miR-135. Using these SNPs we investigated their effects on local RNA structure, on local structural accessibility and on functional miRNA binding, respectively. Systematic computational RNA folding analyses of the allelic mRNAs in either case predicted significant changes of local RNA structure in the vicinity of the cognate miRNA binding sites. Consistently, experimental in vitro probing of RNA showing differential cleavage patterns and reporter gene-based assays indicated functional differences of miRNA-mediated regulation of the two AGTR1 and MRAS alleles. In conclusion, we describe a novel model explaining the functional influence of 3′-UTR-located SNPs on miRNA-mediated control of gene expression via SNP-related changes of local RNA structure in non-coding regions of mRNA. This concept substantially extends the meaning of disease-related SNPs identified in non protein-coding transcribed sequences within or close to miRNA binding sites.
Keywords: AGTR1, MRAS, RNA secondary structure analysis, SNP, cardiovascular disease, miR-SNP, miRNA
Introduction
MicroRNAs (miRNAs) are evolutionarily conserved, endogenous, single-stranded RNAs between 18 and 25 nt in length1 and play a major role in the post-transcriptional regulation of gene expression. Binding to the 3′-UTR of their target mRNAs recruits the RNA induced silencing complex (RISC), mostly leading to inhibition of translation, either via interfering with initiation or elongation or via degradation of the mRNA. Recognition of the target site is mainly mediated by the seed region, i.e., the first 6–8 nt from the 5′ end of the mature miRNA.2 However, underlying mechanisms which crucially involve RNA-RNA interactions are not yet understood and subject to further analyses.3,4 Since their discovery in C. elegans in 1993,5 more than 100 miRNAs have been identified in invertebrates and between 500–1000 miRNAs can be found in mammals and plants (miRBase).6 Each miRNA can potentially regulate a large number of mRNAs7 and, on the other hand, one mRNA can be regulated by several miRNAs. Thus there are estimates ranging between 30% and 92% of all protein-coding genes being under miRNA control.8,9 In addition to their essential role in development, differentiation, proliferation and apoptosis, there is increasing evidence for their involvement in the pathogenesis of human diseases including cancer and metabolic diseases.10,11 Several studies have shown that miRNA expression is strictly controlled but differs markedly between normal tissue and tumor tissue and thus can be used as a prognostic and predictive biomarker. Besides, it is well documented that miRNAs can serve either as tumor suppressors or as oncogenes and therefore represent novel therapeutic target molecules.12,13
A correlation between pathogenesis and the occurrence of SNPs in miRNAs or miRNA target sites (miR-SNPs) was predicted in 2007 based on bioinformatic analyses14 and was experimentally validated by Wu et al.15 for different types of human cancer. Further, it was shown that SNPs are less frequent in miRNAs or their target sites than in other portions of the genome.14,16,17 This negative selection of sequence variations in miRNAs emphasizes their importance for crucial cellular processes including the regulation of gene expression. Different scenarios explaining the impact of miR-SNPs are conceivable. On the one hand, SNPs in a miRNA-coding sequence might affect the expression of a multitude of different genes, for example due to an impaired processing or maturation process of the miRNA. SNPs in target sites, on the other hand, can either modulate existing binding sites or create novel binding sites exerting their influence on one or only a few specific target molecules. Meanwhile, miR-SNPs are recognized as a unique class of functional polymorphisms in the human genome. Their patho-mechanistic role is progressively acknowledged and their biological relevance is beginning to be examined.18-20
Recently, an increasing number of studies on the association between miR-SNPs and the risk of common diseases are published.21 Different approaches are used to predict and to identify functional polymorphisms. For example, Landi et al.22 screened putative miRNA binding sites in candidate genes of colorectal cancer for the occurrence of SNPs that affect the binding of the miRNA to its target. Together with data from a case-control association study, two miR-SNPs were found to be significantly associated with the risk for colorectal cancer. Similar analyses based on association studies provided further evidence for genetic variants that may be associated with different types of cancer.23-25 Another rare sequence variant has been suggested to be associated with Tourette’s syndrome and affects the binding site of miR-189 in SLITRK1.26
An example for SNPs that create novel miRNA binding sites was described by Clop et al.27 They demonstrated that a G to A transition in Texel sheep creates target sites for two miRNAs that are highly expressed in skeletal muscle. This additional regulation via translational inhibition of the myostatin gene is related to muscular hypertrophy. A SNP (rs10065172) that has been correlated with Crohn’s disease and is located in the coding region of IRGM within a miRNA binding site was recently studied by Brest et al.28 Their analyses showed that the SNP alters the regulation of IRGM expression by miR-196 which affects the efficacy of autophagy and thus has an impact on the outcome of intestinal inflammation.
In the human system, Angiotensin II plays important roles in normal physiology as well as in the progression of cardiac diseases. Most of the responses to Angiotensin II are mediated by the Angiotensin II type 1 receptor (AGTR1), which belongs to the family of G-protein coupled receptors.29 The AGTR1 gene has been found to be highly variable as more than 600 mostly rare genetic variants30 and at least 50 SNPs have been described.29,31 Of these, SNP rs5186:A > C located in the 3′-UTR, is one of the most-studied and best-evaluated polymorphisms. In particular, the C allele of this A/C transversion has been associated with essential hypertension and other related cardiovascular diseases.29,32 Moreover, this SNP is located in a target site for hsa-miR-155 and two groups33-35 have shown that expression of the A allele is downregulated by this miRNA. Strikingly, this regulation by miR-155 is abolished for the C allele and leads to elevated levels of AGTR1, suggesting a functional association of this miR-SNP with the onset of hypertension. In a cohort of hypertensive patients, Ceolotto et al. analyzed the role of the interaction of miR-155 and rs5186 in conjunction with AGTR1 mRNA and protein levels in the regulation of blood pressure.36 Thus, the implication of this and other miRNAs in cardiovascular disease makes them promising therapeutic targets.37
On the mechanistic level of miRNA-target recognition, key features include imperfect base pairing between miRNA and target mRNA with increased matching toward the 5′-end of the miRNA guide strand. More recently, it has been shown that target sites with decreased 5′-complementarity also exist, but require additional compensatory or supplementary base pairing at the 3′-end.7 In addition, the position and sequence context of miRNA target sites within the 3′-UTR are also important.38 Meanwhile, these and other features are the basis for most target prediction algorithms. However, additional factors might affect miRNA function, for example RNA secondary structure and RNA binding proteins. It has already been convincingly shown that target site accessibility, i.e., global as well as local folding characteristics of RNA secondary structure, are important determinants for the efficacy of functional RNA-RNA interactions including antisense nucleic acids, ribozymes as well as siRNAs.39,40 Recently, several groups studied whether target accessibility could be involved in miRNA-mediated target suppression. On the basis of free energy (ΔG) predictions Zhao et al.41 showed that the 5′ or 3′ flanking regions of miRNA binding sites are less stable than random sequences, suggesting a locally linear RNA structure around the target site. Kertesz et al.42 also performed free energy predictions, additionally taking into account the difference between the free energy gained by the binding of the miRNA to the target and the free energy lost by mismatching target site nucleotides (ΔΔG). On the basis of this model, they experimentally showed that target site accessibility is as important as sequence match in the seed region and therefore, miRNA target sites are preferentially positioned in mRNA regions with high accessibility. Two other groups included local RNA secondary structures in their target prediction algorithms and validated their results in C.elegans43 and Drosophila,44 respectively.
The aim of our study was to analyze the mechanisms underlying the influence of miR-SNPs in miRNA target sites on the interaction between a miRNA and its target mRNA leading to dysregulated gene expression. On the molecular level, such a mismatch could either cause a destabilization of the interaction due to changes in the free binding energy or a change in target accessibility due to alterations in the RNA secondary/tertiary structure. To this end, we established a model system, which allowed performing thermodynamic studies on the level of linear RNA sequences as well as structural analyses on the level of secondary structure accompanied by functional assays using different miRNA tools to study miRNA-mediated regulation. We chose a common SNP (rs5186:A > C) in the AGTR1 gene as a model miR-SNP and our data suggest that the observed differences in miRNA-mediated regulation of the two alleles are correlated with altered RNA secondary structure. Additional experimental evidence for the functional role of SNP-affected local RNA structure was derived from analyses of rs9818870, a SNP located in the muscle RAS oncogene homolog (MRAS) 3′-UTR which indicates that SNPs even outside of miRNA target sites can influence miRNA-mediated regulation. In summary, this study provides strong experimental evidence supporting the functional importance of SNPs even outside coding sequences, i.e., potentially all miR-SNPs within the 3′-UTR.
Results
A SNP in the miR-155 target site in AGTR1 is correlated with dysregulated reporter gene expression
The relevant AGTR1 nucleotide sequences including miR-155 and its target sites are depicted in Figure 1 which also shows the AGTR1 mRNA transcript variant 4 (NCBI Reference Sequence: NM_031850.2) with its 5′-UTR (position 1 - 446), coding sequence (CDS, position 447 – 1523) and 3′-UTR (position 1524 – 2412). With regard to this numbering the position of rs5186 in the AGTR1 3′-UTR is 1612 and the SNP will therefore be referred to as A1612C throughout this study. In contrast, many other publications call the same polymorphic site A1166C which is the position in relation to the start of the coding sequence. Figure 1 further shows the two different reporter transcripts used in this study that either contain the complete AGTR1 3′-UTR (AGTR1 “long,” position 1527 - 2407, Figure 1B) or the miR-155 target site only (AGTR1 “short,” position 1590 - 1620, Figure 1C) in the firefly luciferase 3′-UTR of pMIR-REPORT.

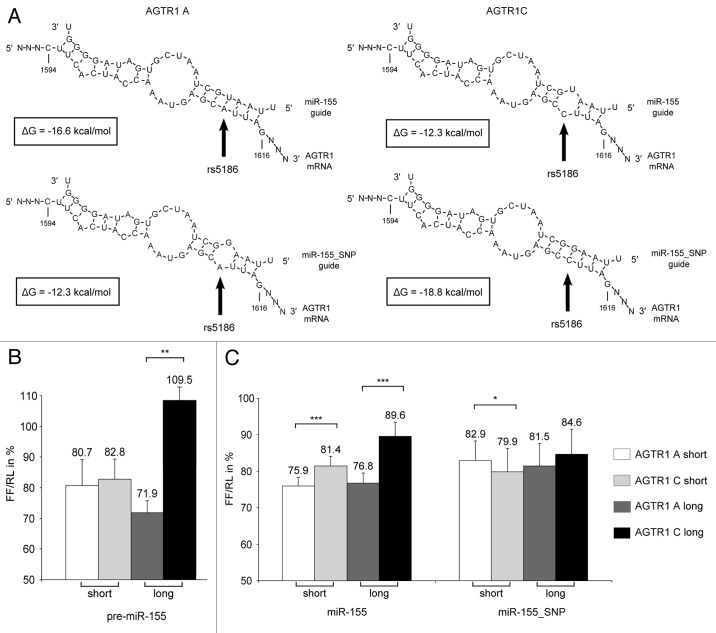
Figure 1. Schematic depiction of AGTR1 transcripts and miRNAs. (A) Structural and functional portions of AGTR1 mRNA. The numbering is according to NCBI Reference Sequence: NM_031850.2. Thus, the position of rs5186 within the miR-155 target site is designated as 1612 (which equates to position 1166 relative to the start of the coding sequence) and the miR-1 target site is located appr. Thirty nt upstream of the SNP. (B) Schematic depiction of the recombinant firefly luciferase-AGTR1 3′-UTR in vitro transcript (AGTR1 IVT “long”). (C) Schematic depiction of the recombinant firefly luciferase-AGTR1 miR-155 target site in vitro transcript (AGTR1 IVT “short”). (D) Nucleotide sequences of miRNA-tools used here (miR-155 and miR-155_SNP) to analyze the target-miR-SNP interaction.
Three different double-stranded miR-155 mimics were used for analyzing the miRNA-mediated regulation of AGTR1 rs5186 variants A and C (further termed AGTR1 A and AGTR1 C, respectively). First, Pre-miR™, a commercially available miRNA precursor molecule (Ambion, further termed pre-miR-155) which - according to the manufacturer - is a small, partially double-stranded, chemically modified RNA. However, its sequence as well as the nature and position of the chemical modifications are not disclosed. Second, we synthesized miR-155 guide and passenger strands (further named miR-155) according to sequence information from miRBase.6 Double-stranded miR-155 (Fig. 1D, top) is characterized by a fully matching seed sequence to the A allele of the AGTR1 target site (Fig. 1D, seed region indicated by line, arrow indicates position opposite the polymorphic site) but one C-U mismatch to the C allele of AGTR1 at position 5 of the mature miRNA. For analyzing a possible rescue of the miRNA-mediated regulation of AGTR1 C we restored base-pairing at the seed region by producing a miR-155 guide strand with a U > G substitution (further named miR-155_SNP, Figure 1D, bottom) at position 5. Please note that this modification was introduced in the guide strand only, i.e., we did not modify the corresponding passenger strand position.
To study the thermodynamics of binding, minimal free energies were calculated for each of the possible pairs of miRNA guide strand and AGTR1 miR-155 target site (Fig. 2A) using the program RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/45). The upper panel in Figure 2A displays hybridization of the miR-155 guide strand with the two miR-155 target site variants (A on the left vs C on the right) whereas the lower panel shows hybrids with the miR-155_SNP guide strand. Minimal free energies for the binding at the two target sites (A vs. C allele) with their matching miRNA guide strands differ slightly (-16.6 vs -18.8 kcal/mol) which can be explained by the occurrence of a different Watson-Crick base pair at the SNP position, i.e., an A-U base pair for the A allele compared with a C-G base pair for the C allele. However, the minimal free energy of both hybrids, each harboring a mismatch at position 5 of the seed, is the same (-12.3 kcal/mol). For each allele of the AGTR1 mRNA miR-155 target site, the loss of a complete match in the seed region caused by the mismatch in the miRNA guide strand results in a reduction of the minimal free energy (for A + 4.3 kcal/mol and for C + 6.5 kcal/mol). This suggests that the presence of the C allele is accompanied by a weaker interaction with the authentic miR-155 affecting miRNA-mediated regulation of AGTR1 gene expression.
Figure 2. Structures and thermodynamic stability of complexes of miR-155 and AGTR1 mRNA, and miR-155-mediated effects on reporter gene expression in HeLa cells. (A) Secondary structures and stability of the target-miR-SNP interaction. Minimal free energies of AGTR1 A and C miR-155 target site and miR-155 (upper panel) and miR-155_SNP guide strand (lower panel), respectively, were calculated using the program RNAhybrid. The loss of seed complementarity leads to a reduction in ΔG values for both AGTR1 miR-155 target sites. Effects of pre-miR-155 (B) and of miR-155 and miR-155_SNP (C) on reporter gene expression in HeLa cells. Cells were co-transfected with 25 ng pMIR constructs, 0.25 ng pRL-CMV as transfection control and 25–150 nM miRNA. Values are means of the lowest relative firefly luciferase activity (FF/RL) in percent normalized to cells that were transfected with plasmids but no miR-155 tool of at least four independent experiments. P values were determined by analysis of variance (ANOVA) and error bars represent the SEM, *p ≤ 0.05, **p ≤ 0.001, ***p ≤ 0.0001.
In order to set up a reporter system for miRNA-mediated regulation of gene expression we cloned minimal miR-155 target sites into the 3′-UTR of the firefly luciferase of the pMIR-REPORT vector using DNA oligonucleotides (Fig. 1C). These constructs are further termed AGTR1 “short” A and C, respectively. Next, we performed co-transfection experiments in HeLa cells of these constructs together with a control plasmid coding for Renilla luciferase and different amounts of pre-miR-155. HeLa cells were chosen because they do not express miR-155 according to the database microRNA.org.46 Thus it is reasonable to assume that reporter gene expression is not influenced by endogenous miR-155.
Unexpectedly, luciferase activities were decreased for both of the short target sites to a comparable amount (Fig. 2B). Moreover, the observed reduction of reporter gene expression was not correlated with the concentration of pre-miR-155 (data not shown). At a first glance, these results were not in accordance with published data.33 However, in their study the complete 3′-UTR sequence was used rather than a minimal target site construct. Therefore, we also generated reporter plasmids containing the authentic AGTR1 3′-UTR, either according to the wild type sequence, i.e., the A allele, or to the SNP sequence, i.e., the C allele, introduced by site directed mutagenesis (see methods section and Fig. 1B). The resulting recombinant constructs were termed AGTR1 “long” A or C. Co-transfection experiments with these constructs showed a different kind of regulation of reporter gene expression depending on the polymorphic variant (Fig. 2B), i.e., only the wild type target sequence harboring the A variant was strongly repressed by almost 30% whereas no repression was observed for the 3′-UTR construct with the target site mismatch. This is consistent with published results33 and confirms the selective regulation by the miRNA precursor. The inconsistent behavior observed for the short target site constructs could be related to unknown characteristics of the commercially available pre-miR-155 which is a precursor mimic with unknown sequence and proprietary chemical modifications optimized for enhanced binding to the miR-155 target site. These features might exert a very strong influence on miRNA-target site interactions such that even one mismatch within the seed region has no measurable effects.
Furthermore, one of the major differences of the indicator systems is their length and their sequence context in the vicinity of the target site. It seems to be reasonable to assume that longer stretches of authentic 3′-UTR sequences within recombinant constructs fold into local structures that are more similar to local folding of the wild type AGTR1 mRNA than the shorter transcripts AGTR1 A and AGTR1 C. Thus, constructs containing short (i.e., miR-155 target site only) and long stretches of the AGTR1 3′-UTR were further tested in parallel.
Regarding the use of miRNA-derived tools, a number of alternative constructs and approaches have been published so far. These include synthetic single-stranded and double-stranded miRNAs containing various chemical modifications, miRNA precursors (pre-miRNAs), conjugates, as well as endogenously expressed hairpin transcripts. However, there is uncertainty of their validity in specific cases and it does not seem that there is general agreement on their standardized use. Therefore, in addition to pre miR-155, we included two more miRNA tools in our studies, miR-155 and miR-155_SNP, both with known sequences and without chemical modifications (Fig. 1D). Results from transfection experiments with all possible combinations of target sites and miRNAs are shown in Figure 2C. Application of miR-155 leads to strong repression of both target sites harboring the A allele (Fig. 2C, left panel; appr. 25%). Again, for the C allele, there is also a significant decrease in luciferase activity of the short target site (appr. 20%) but only weak repression of the long target site (appr. 10%). In conclusion, in the context of the short target site the SNP has a moderate effect on the miRNA mediated regulation due to the difference in the thermodynamics of binding (Fig. 2A) but in the context of the long target site the SNP has a much stronger effect probably caused by additional differences in the RNA secondary structure of the 3′-UTR. MiR-155_SNP compensates the A1612C SNP with a complementary G residue in the guide strand sequence (Fig. 1D) which should lead to a rescue of miR-155 mediated regulation of the C allele. This phenotype can be observed in case of co-transfection with the short target site constructs (Fig. 2C, right). Here, the C variant is slightly more repressed than the A variant, albeit with very low significance. In case of the long target site constructs such a reversion is not observed and, in general, the decrease in luciferase activity is not very strong (< 15%).
The different results for the short and long reporter constructs suggest that besides the thermodynamics of the miRNA-target site interaction, the location of the polymorphic target site in relation to the secondary structure of the 3′-UTR may play an important role for miR-155 mediated regulation of AGTR1.
The miR-SNP is correlated with changes of the secondary structure of the AGTR1 3'-UTR
If the interaction between a given miRNA and a target site is determined mainly by thermodynamic properties of RNA-RNA recognition, then the corresponding ΔG values (Fig. 2A) should be closely related to the phenotypes observed in transfection experiments. However, comparing miR-155 and miR-155_SNP-mediated regulation, this is not the case. According to the prediction of the RNA binding interactions, miR-155_SNP should show the highest repression of the short AGTR1 C target site due to the lowest ΔG value (-18,8 kcal/mol). However, miR-155 results in a superior suppression of the short AGTR1 A target site (ca. 25% vs < 20%) despite a slightly reduced free energy value (-16.6 kcal/mol). Additionally, both seed-mismatching interactions (miR-155 with AGTR1 C and miR-155_SNP with AGTR1 A) have the same binding energies (-12.3 kcal/mol) but demonstrate different extents of repression.
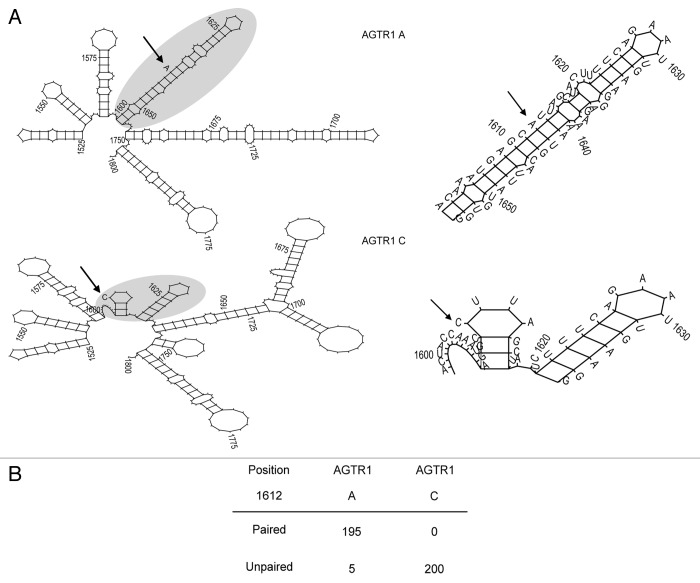
Hence, we had a closer look at the RNA secondary structure of both allelic sequences of the AGTR1 mRNA. Systematic computational RNA secondary structure predictions were performed as described in detail elsewhere.47,48 It is important to note that this methodology includes the calculation and analysis of hundreds of RNA structures and, thus, gives rise to biologically meaningful structure models.49 An example of this kind of analysis is schematically depicted in Figure 3A. It shows the predicted secondary structure of AGTR1 rs5186 variants A and C from position 1525 to 1800 (numbering according to Figure 1A). Both sequences show a similar global structure between positions 1525–1597 and 1754–1800. Conversely, they adopt a different local configuration for both variants within the local region 1598–1753 in which the A variant is located in an intramolecular double-helix whereas the C variant is predicted to form a small hairpin loop. The right panel of Figure 3A shows a magnification of the SNP region and Figure 3B summarizes the results of a systematic analysis based on 200 secondary structure predictions for either variant of position 1612 of the AGTR1 mRNA, respectively. The structure context of the SNP was grouped in two categories according to its involvement in intramolecular interactions. In contrast to variant A in which the SNP stretch was predominantly (97.5%) predicted to be intramolecularly paired, variant C only appeared in unpaired structural elements. For the latter case, 163 out of the 200 predicted structures ( = 81.5%) are represented by Figure 3A. In the remaining 18.5% of the unpaired structures of variant C, the SNP position was located in a bulge. For the A allele, only 2.5% of all structures were found in a loop (i.e., unpaired), and this element only occurred in the predicted structures at a window size of 400 nt but not at a window of 800 nt or 1400 nt. 83% of the paired elements are represented by Figure 3A and in the remaining structures the A was paired but located close to a junction and not within a long stem. Furthermore, comparing structures of the same window size for both variants demonstrated that in all cases the free energy value for the A sequence was lower than for the C sequence suggesting that, from the energetic point of view, the A transcript is more stable.
Figure 3. Theoretical analysis of the local RNA structure of both polymorphic sequences of AGTR1 mRNA. The structural domain containing the SNP is shaded in gray and arrows indicate the rs5186 site. The numbering is according to the position in the AGTR1 mRNA (see Figure 1A). (A) The left panel illustrates the global secondary structure prediction. The RNAs show different structure elements around the polymorphic site. The A is located in a stem whereas the C is predicted to be in a loop. A magnification of each secondary structure around the SNP demonstrating the local structure differences is pictured on the right. (B) Summary of the allele-specific involvement of the SNP in intramolecular folding determined by computational analysis of RNA secondary structure. In total, 200 predicted secondary structures have been analyzed. The number of structures in which the SNP occurs in stem motifs is listed under ‘paired’ vs. the ‘unpaired’ group representing loop and bulge positioning of the SNP. In contrast to the A, which is predominantly located in a paired structure, the C is found only unpaired.
In vitro probing experiments show different structural conformations of AGTR1 3'-UTR RNA transcripts
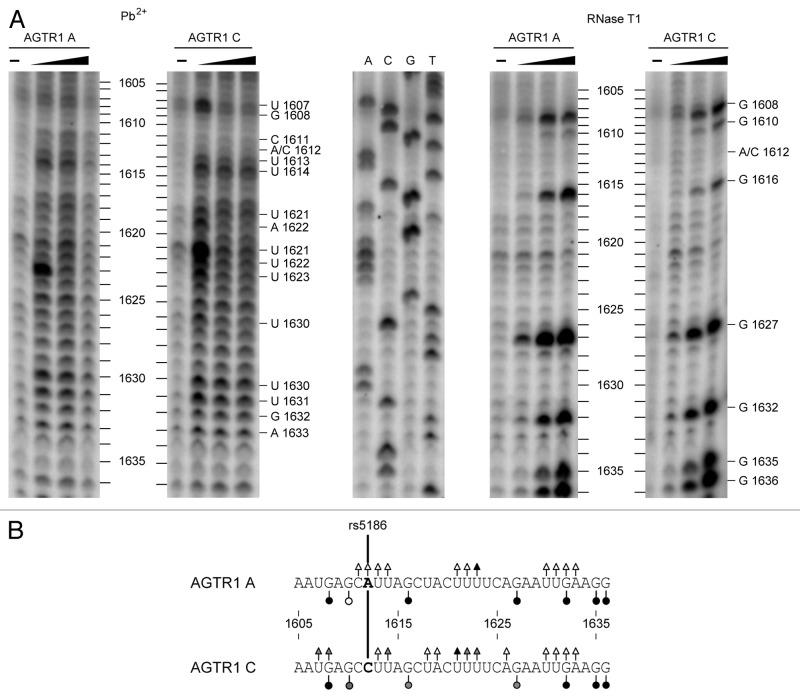
Following the systematic and thorough theoretical analysis of the AGTR1 mRNA secondary structure, which visibly provided distinct results for both alleles, we performed structural probing experiments to further substantiate the theoretical analyses. To this end, long-chain in vitro transcripts of either AGTR1 3′-UTR A or C variants were probed chemically with Pb2+ monitoring unpaired positions of all four nucleotides and enzymatically with RNase T1 which cleaves at unpaired G residues. Subsequent to probing reactions, cleavage sites were visualized by primer extension and electrophoretic analysis. In Figure 4 a typical analysis is shown. This experimental structure probing of the AGTR1 IVT “long” (shown for position 1605–1635) indicates different cleavage patterns in the vicinity of the polymorphic site for probing with Pb2+ as well as in the use of RNase T1 (summarized in Figure 4B). RNA transcript variant C shows stronger and additional hydrolysis sites when compared with transcript variant A (e.g., RNase T1 at position 1610, Pb2+ at positions 1614–1622). Conversely, a stronger cleavage by Pb2+ occurs at position 1623 and by T1 at positions 1616 and 1627 of variant A RNA. A detailed description and discussion of this probing experiment is provided in the legend to Figure 4. In summary, these observations are compatible with structural differences observed in the systematic theoretical structure analysis. Nevertheless, it has to be stressed that our analyses focused on verifying differences seen in an in silico structure model by performing an in vitro assay rather than dissolving the AGTR1 3′-UTR RNA structure in detail. Moreover, we cannot comment on the detailed relationship between accessibility in a probing assay and microRNA-mediated regulation in vivo as too little is known about the pairing mechanisms between a miRNA and its target RNA.
Figure 4. Probing and primer extension analysis of AGTR1 3′-UTR. AGTR1 “long” in vitro transcripts (see Figure 1B) harboring either A or C were hydrolyzed with increasing amounts of lead (II) acetate or RNase T1. The products were reverse transcribed using an AGTR1 sequence specific 32P labeled probe followed by denaturing gel electrophoresis and phosphorimaging. (A) Autoradiograph of a gel resolving a Pb2+ (left) and RNase T1 (right) probing. In the middle a sequencing reaction using AGTR1 3′-UTR A IVT is shown. It has to be noted that AMV reverse transcriptase stops one nucleotide position before the T1 cleaved nucleotide leading to a fragment that is one nucleotide shorter than the fragment in the sequencing reaction. AGTR1 RNA sequences annotated with hydrolysis sites (triangles for Pb2+ probing, circles for T1 probing) and strength of cleavage (open symbol, weak; shaded symbol, moderate; closed symbol, strong) are pictured in (B). The SNP site is indicated by ‘A/C1612’ in (A) and marked in bold in between the two linear sequences (B). The lead cleavage patterns of both sequences (Fig. 4A, left) show major differences at positions 1607/08 and 1621 - 1623. In particular position 1623 of the A variant is cleaved stronger than variant C initially and then signals disappear more rapidly with increasing Pb2+ concentration. This could be interpreted such that the local structure of the A variant is more accessible because, upon initial strong cleavage at these positions, cleavage products are further being degraded more rapidly due to their accessibility. This view could also be true for position 1614. The probing by RNase T1 shows stronger signals for the A transcript at position 1616 and 1627. Conversely, cleavage at position G1610 is stronger for the C variant than for the A variant.
A SNP in proximity but not within a 3'-UTR -located miRNA target site is related to allelic differences of local RNA structure and to miRNA function.
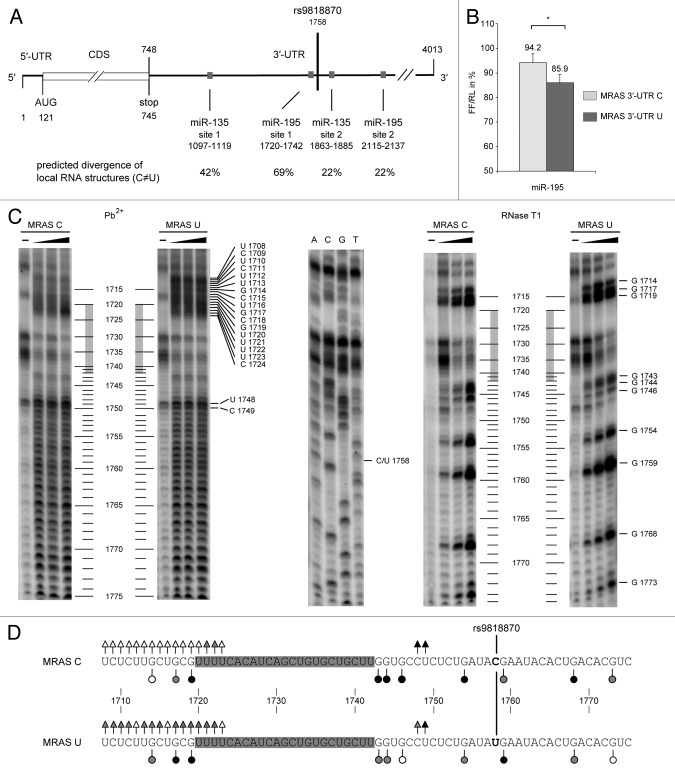
Next we studied the structural and functional role of SNP rs9818870 (C / T polymorphism) that is located within the 3′-UTR -coding sequence of the MRAS gene. This SNP was shown to be associated with coronary artery disease (CAD) in a genome-wide association study (GWAS).50 The M-ras protein is widely expressed but particularly found in the heart (BioGPS). Erdmann et al. further showed that MRAS expression is reduced in injured mouse heart tissue compared with healthy heart tissue.50 Hence we asked whether this SNP could potentially be involved in the pathogenesis of CAD via changes of miRNA regulation. A screen for miRNA binding sites within the 3′-UTR of MRAS mRNA did not indicate that this SNP is located within a known miRNA binding site. However, the 3′-UTR of the MRAS mRNA has two predicted binding sites for miR-195 and miR-135, respectively. One miR-195 binding site is located 16 to 38 nucleotides upstream of rs9818870 and the second one approximately 400 nucleotides downstream of the SNP (Fig. 5A). The involvement of this miRNA in cardiovascular disease has been suggested previously.51 MiR-135 also has a potential impact on the cardiovascular system52 with binding sites approximately 100 nucleotides downstream and 700 nucleotides upstream of the SNP (Fig. 5A).
Figure 5. Schematic depiction of MRAS mRNA including 3′-UTR -located binding sites for miR-195 and miR-135 and effects of miR-195 on MRAS reporter constructs in Hela cells. (A) Schematic representation of the MRAS mRNA. The numbering is according to NCBI Reference Sequence: NM_001085049.1. Shown are the location of rs9818870 and the positions of miR-195 and miR-135 binding sites. The divergence of predicted secondary structures between the two MRAS variants is indicated in percent of all structure predictions. For the binding sites closer to the SNP (miR-195 site 1 and miR-135 site 2) 110 calculated structures were compared and 50 structures were analyzed for miR-135 site 1 and miR-195 site 2. (B) Co-transfection of miR-195 (10–100 nM miRNA) and allele-specific MRAS reporter constructs (10 ng pmirGLO MRAS 3′-UTR constructs) in HeLa cells. Values are means of five independent experiments and represent the lowest relative firefly luciferase activity (FF/RL) in percent normalized to cells that were transfected with plasmids only. P values were determined by analysis of variance (ANOVA) and error bars represent the SEM, *p ≤ 0.05, **p ≤ 0.001, ***p ≤ 0.0001. (C) RNA probing of in vitro transcripts (IVT) representing variant C or U of the MRAS 3′-UTR in the vicinity of the miR-SNP rs9818870 with Pb2+ (left panel) and RNase T1 (right panel), respectively. The middle panel shows a sequencing reaction of MRAS 3′-UTR U IVT. The probing results for both variants are summarized in (D). Cleavage sites of both variants (C variant, upper panel; U variant lower panel) and the extent of cleavage monitoring accessibility for Pb2+ and T1 are indicated by triangles for Pb2+ probing and by circles for T1 probing (open symbol, weak; shaded symbol, moderate; closed symbol, strong). The SNP is indicated by ‘C/U 1758’ in (C) and by a line in bold in between the two sequences in (D). The miR-195 binding site is indicated by a gray bar between positions 1720 and 1742 in (C) and shaded in gray in (D). The Pb2+ cleavage pattern of both sequences show major differences at positions 1708–1720 which are located at the transition of the miR-195 binding site and upstream sequences. Probing by RNase T1 indicates increased accessibility of the C variant downstream of the SNP except for G 1759 and decreased accessibility immediately upstream of the miR-195 target site 1. In summary, both probing experiments reveal detectable structural differences of both allelic variants directly upstream and downstream of the binding site 1 of miR-195.
A systematic computational analysis of secondary structure suggests that the SNP is related to differences of local RNA structure of the two MRAS variants, i.e., structural differences occur at and also in proximity of the SNP which include binding sites for miR-135 and miR-195. In about 69% of all analyzed structures miR-195 binding site number 1 that is located 16 nt upstream of the SNP (Fig. 5A) showed differential local structures between the two variants (Fig. S1). Both miR-135 miRNA binding sites showed structural differences to a decreased extent compared with miR-195. Hence we focused on the role of SNP rs9818870 for local RNA structure of the MRAS mRNA and for gene regulation by miR-195. First, cell culture data in the use of a reporter gene containing the MRAS 3′-UTR showed stronger downregulation of the MRAS U variant by miR-195 compared with the C variant (Fig. 5B). Moreover, RNA structure probing experiments reveal different cleavage patterns of the MRAS variants close to the polymorphic site but also surrounding the miR-195 target site 1 (Fig. 5C). Considering the increased accessibility of the U variant upstream of SNP rs9818870 (Fig. 5D) one may speculate that this site is more critical for binding of miR-195 than sequences 3′ to the SNP. In summary, this analysis is consistent with the view that SNPs even outside of miRNA target sites can influence miRNA-mediated gene regulation via altering local RNA accessibility.
Discussion
Former studies on the miRNA-mediated regulation of AGTR1 gene expression indicated a differential regulation of the A vs. the C allele by miR-155 in cell culture models.33,35 In both studies, reporter genes harboring the A variant were downregulated to a greater extent than the C variant after co-transfection of pre-miR-155 (Ambion). Using the long-chain 3′-UTR reporter constructs and pre-miR-155 we observed comparable effects although the magnitude of repression was not as pronounced as published before. It should be noted that these previous studies were performed in hamster cells (CHO)33 whereas we chose human cells (HeLa) which does not seem to allow a valid comparison. When we applied miR-1, which targets the 3′-UTR of the AGTR1 mRNA upstream of the SNP (Fig. 1A), we observed a repression of reporter gene expression of approximately 50% (data not shown), which is within the usual range of regulation of gene expression by miRNAs. Therefore, the less pronounced but significant downregulation observed in our study could indicate an inherent feature of the miR-155 AGTR1 interaction in human cells. Then again, we cannot explain why we did not observe a concentration dependent decrease in luciferase activity when we applied 25–150 nM miRNA. Nevertheless, we decided not to use pre-miR-155 in our further analyses because the sequence and chemical modification information is proprietary. We can only assume that the company inserted modifications or exchanged nucleotides to stabilize the miRNA target binding, probably even within the seed region. Therefore it is not a suitable tool to study miR-SNPs occurring in the seed region and we designed alternative miRNA tools with known sequence and chemical composition (Fig. 1D) for the remaining experiments.
An association of the C variant of AGTR1 rs5186 with hypertension has been indicated by several studies.32 In addition to reporter gene silencing assays, Sethupathy et al.35 analyzed AGTR1 protein levels and miR-155 levels in a pair of monozygotic twins discordant for trisomy 21 and performed bioinformatics-based analyses of disease-related aspects in several populations. Occurrence of a seed-mismatch between the AGTR1 C target RNA and miR-155, leading to a thermodynamically destabilized interaction, was suggested as the cause of reduced miR-155 regulation and elevated AGTR1 levels.33,35 We performed more detailed biochemical and cell biology studies using AGTR1 reporter constructs that only harbor the miR-155 target site and miRNA strands that are not chemically modified. In addition to existing data33,35 this allowed us to expand the earlier proposed hypothesis by the new observation that RNA secondary structure alterations caused by the miR-SNP play an important role by affecting the accessibility of the AGTR1 target site for miR-155. In the following, we did not continue to study the physiological effects caused by the SNP on AGTR1 function but rather used this system to elucidate the molecular pathomechanism underlying the role of miR-SNPs for dysregulated miRNA control, i.e., to draw more refined conclusions on miRNA-target interactions.
After having switched from the commercially available pre-miR-155 to the unmodified miRNA tools with known sequence, we saw a differential regulation of the two polymorphic variants with miR-155 for both reporter constructs. This difference was more pronounced when using the long target site constructs, leading us to the assumption of RNA secondary structure being involved in the extent of the miRNA-mediated regulation. However, in other studies miRNA target interactions were analyzed by using minimal reporter constructs harboring only short sequences corresponding to the length of the miRNA binding site.53,54 This suggests that in those studies either target site structure did not measurably contribute to miRNA-target interactions or possible combined effects on the miRNA regulation through secondary structure have not been taken into consideration. We hypothesize that for analyzing a specific miRNA-mediated regulation of gene expression the use of small miRNA target segments within reporter gene-based assays has to be considered an artificial system which is not fully compatible with the in vivo situation because structural influences are not considered. In order to minimize this problem we propose to carefully select segments of a given long-chain 3′-UTR that most likely represent the authentic local RNA secondary structure adopted by the relevant wild type sequence stretch.
We performed computational structure prediction analyses on a number of candidate miR-SNPs and observed that some do not show significant differences in RNA structure (data not shown) while others show only minor differences (e.g., ESR1, Fig. S2). However, some SNPs (e.g., AGTR1, MRAS) are related to clear changes in secondary structure. In our opinion the latter should be subject of further experimental validation and this additional feature of miR-SNPs should be included in bioinformatical analyses.
The influence of rs5186 on the regulation of AGTR1 expression by miR-155 is an example of a functional miR-SNP that is located within a miRNA binding site. If RNA structure alterations induced by the SNP are jointly responsible for dysregulation of the C variant it is conceivable that SNPs adjacent to miRNA target sites can affect miRNA function as well. To test this hypothesis we performed analyses with rs9818870 in the MRAS 3′-UTR (Fig. 5; Fig. S1). This SNP is not located directly within a miRNA target site, but in proximity to a miR-195 binding site. Indeed, RNA folding analyses of the polymorphic RNAs suggest SNP-induced changes in local structure supported by in vitro probing analyses showing differential cleavage patterns around this site as well as functional differences in miRNA regulation. In a recent study, Mishra et al.55 performed analyses on a SNP (rs34764978) in the dihydrofolate reductase (DHFR) mRNA. This C > U conversion at position 223 of the 3′-UTR affects DHFR expression by interfering with miR-24 function, resulting in DHFR overexpression and methotrexate resistance. Surprisingly, this SNP occurs outside of the miR-24 binding site by a distance of 14 nucleotides but influences miRNA mediated regulation. This misregulation cannot be explained by SNP-induced mismatches between miRNA target site and the miRNA itself. As a possible explanation, Mishra et al. suggest that the SNP may affect binding of the miR-24-Argonaute protein complex to the DHFR mRNA. Regarding our MRAS results one might also consider that the observed dysregulation for the 3′-UTR U variant could be due to alterations of target site accessibility for miR-24 through structural changes induced by the SNP. To test this assumption we performed mfold analyses and compared the secondary structure of the miR-24 binding site in both variants (200 structures each, Fig. S3). In appr. 50% of the predicted polymorphic RNA structures for both variants the miR-24 target site was located in a stem structure element whereas the remaining structures show some diversity between the two RNA variants. For example in case of the C mRNA, the miR-24 target site was found preferentially in a junction whereas in the mRNA harboring the U residue at position 223 it was located in a loop. Therefore it is conceivable that the SNP causes structural alterations which are involved in the observed differential miRNA-mediated regulation of DHFR expression. Together with our MRAS data, this implies in a more general sense that studies of disease-related SNPs should not only search for miRNA target sites directly at the position of the SNP but also a few hundred base pairs up- or downstream.
Predicting secondary structures of polymorphic targets can be used as an in silico tool to pre-screen the possible influence of a SNP on local RNA structure and, hence, on RNA-RNA recognition and miRNA regulation. However, it should be noted that the defined sort of recorded local structural elements, such as for example stems, loops, bulges, or perfect duplexes, cannot be further correlated with the extent of miRNA-mediated regulation of gene expression because of a lack of detailed knowledge of the structure-function relationship underlying miRNA-target recognition. This is somewhat different from the partially known structure-function relationship thought to be relevant for perfectly matching strands of naturally occurring and artificial antisense nucleic acids or even siRNA and their targets. Mechanistic rules governing interactions between partially matching strands, however, seem to be very complex and are not yet revealed on a general level.
Besides RNA-RNA interactions, proteins might also play a role in miRNA target site recognition, either via blocking or facilitating access to the target region. Such a correlation has been predicted via bioinformatic analyses for several RNA binding proteins.56 In the context of this study it is interesting to note that Pumilio was recently discovered to influence miRNA mediated regulation.57,58 Kedde et al.58 showed that binding of Pumilio to the p27 3′-UTR alters local RNA structure which is related to augmented miRNA binding resulting in increased downregulation of the target. Leibovich et al.57 performed computational analyses which suggest an assistance of Pumilio in miRNA regulation of inaccessible miRNA targets by opening their secondary structure. Interestingly, we found that the AGTR1 3′-UTR also contains the Pumilio recognition sequence (located in the 3′-UTR at position 716–723, data not shown) approximately 600 nucleotides downstream of rs5186 indicating that miRNA regulation of AGTR1 mRNA may also involve changes of local RNA structure upon binding of Pumilio or related RNA binding proteins. This further demonstrates that our in silico and in vitro analyses contribute to elucidate the impact of miR-SNPs and the underlying functional mechanisms of disease development.
Our results as well as many other studies show that regulation of gene expression by miRNAs is a very complex process. Besides other features, accessibility and thermodynamics are two important factors for miRNA-target interaction. The presence of miR-SNPs in miRNA targets adds another layer of complexity to miRNA binding and regulation and, hence, to the understanding of molecular disease mechanisms. We suggest to not only consider the influence of a SNP on the miRNA target interaction through introduced mismatches but also on miRNA target accessibility through secondary structure alterations caused by SNPs inside or in the vicinity of miRNA target sites when searching for a possible disease association.
Material and Methods
Cloning of reporter gene constructs
To analyze AGTR1 and miR-155 interactions the pMIR-REPORTTM Luciferase vector (Ambion, #AM5795) was used and target sites were inserted within the firefly luciferase 3′-UTR. DNA oligonucleotides corresponding to AGTR1 mRNA sense and antisense were designed containing the miR-155 target site, four flanking nucleotides, an internal BlpI restriction site for the screening of positive clones and terminal SpeI and HindIII restriction sites (sense 5′ CTAGTAGCACTTCACTACCAAATGAGC[A/C]TTAGCTACGCTCAGCA 3′, antisense 5′ AGCTTGCTGAGCGTAGCTAA[T/G]GCTCATTTGGTAGTGAAGTGCTA 3′). The oligonucleotides (biomers.net GmbH) were annealed and cloned into the multiple cloning site to generate the short target site constructs. For the 3′-UTR constructs, the AGTR1 3′-UTR A insert was amplified out of a human prostate cDNA library (CLONTECH, # HL4037AH) by using gene specific forward (5′ GAGACTAGTCATGTTCGAAACCTGTCCATAAAG 3′) and reverse primer (5′ TCAGACGCGTGAACACAGCTGTCTACAGCAGCTGTG 3′) harboring SpeI and SacI restriction sites and the PCR products were inserted into the vector. For cloning of the polymorphic 3'-UTR construct, site-directed mutagenesis (adapted from ref. 59) was performed with 5' CCAAATGAGCCTTAGCTACTTTTC 3' as mutagenic primer.
MRAS 3'-UTR sequences harboring SacI and SalI restriction sites were amplified from human prostate cDNA library and Hec1A cDNA and cloned into pmirGLO Dual-Luciferase vector (Promega, # E1330) using 5' AGAGGAGCTCCAGGCCTGAGGCCCTGG 3' as forward, 5' TCAGGTCGACCGTGTTTCTCTTCTTTATTAATCAGTC 3' as reverse and 5' GGTGCCTCTCTGATATGAATACACTG 3' as mutagenic primer.
Probing and primer extension
PCR products harboring the RNA polymerase T7 recognition site were amplified for AGTR1 with 5′ GAAATTAATACGACTCACTATAGGGCATGTTCGAAACCTGTCCATAAAG 3′ (MRAS: 5′ GAAATTAATACGACTCACTATAGGGCAGGCCTGAGGCCCTGG 3′) as forward, 5′ TCAGACGCGTGAACACAGCTGTCTACAGCAGCTGTG 3′ (MRAS: 5′ TCAGGTCGACTTGGTGGAACAAATCTTTTATTTTC 3′) as reverse primer and the pMIR AGTR1 3′-UTR (MRAS: pmirGLO MRAS 3′-UTR) constructs as template and used for in vitro transcription (T7 RiboMAX™ Express Large Scale RNA Production System, Promega, #P1320). In vitro transcripts (IVTs) were purified by phenol-chloroform-extraction, gel filtration (G-50 Nick Column, GE Healthcare, #17–0855–02) and ethanol precipitation. Subsequently, IVTs were denatured (10 min 70°C) and refolded in 1 × folding buffer (20 mM Tris/HCl pH 7.4, 0.1 M NaCl and 1 mM EDTA) at room temperature for 120 min. For the subsequent probing reactions 1 pmol IVT was utilized.
Hydrolysis by RNase T1 was performed for 4 min at room temperature in a 10 µl reaction containing 1 µg tRNA (from yeast, Sigma, #83853–25MG) and varying units of RNase T1 (0, 0.25, 1 and 2 units, Fermentas, # EN0541). Hydrolysis products were purified by phenol-chloroform-extraction and ethanol precipitation.
For lead probing refolded IVTs were incubated for 15 min at room temperature in a volume of 10 µl containing 20 mM HEPES pH 7.1, 10 mM Mg(CH3COO)2 and 0.1 M KCH3COO. Afterwards, 5 µg tRNA and varying amounts of lead(II) acetate (Pb2+, Sigma,# 32307) were added (0, 10, 20 and 40 mM final concentration). The probing reaction was stopped after 10 min at room temperature by adding 5 µl 0.1 M EDTA and 60 µl ethanol, followed by ethanol precipitation (adapted from ref. 60).
Sequence-specific RT-primers bind downstream of the AGTR1 SNP (5′ CGGTTCAGTCCACATAATGC 3′) and, in case of MRAS, downstream of the MRAS SNP and the miR-195 binding site 1 (5′ TGGATGGTGTCCACATTAGGT 3′). The primers were 32P-labeled at their 5′-end and purified by gel filtration (G-50 Nick-column, GE Healthcare). RT reactions with either RNase T1- or lead hydrolysis products were performed for 45 min at 42°C using 1 mM dNTPs, 2.5 mM RT-primer and 1.5 units AMV Reverse Transcriptase (Promega, # M5108) in a 10 µl reaction volume. In parallel, sequencing reactions with denatured IVT (2 min at 95°C) were performed for each of the 4 bases using 31.5 µM dNTP (Fermentas, # R0181) and 2.5 µM ddNTPs (Fermentas, # R0072) and 125 µM of the remaining three dNTPs, respectively (adapted from ref. 61). After cDNA-synthesis samples were denatured in formamide-containing loading buffer for 3 min at 95°C and resolved on a 10% polyacrylamide sequencing gel (21 × 40 cm) under denaturing conditions (8M urea) for 70 min at 80W and 52°C. Subsequently gels were vacuum-dried for 1 h. Signals were analyzed with a PhosphorImager (Typhoon™ 8600 Variable Mode Imager, GE Healthcare).
miRNA annealing
Single-stranded miRNA guide and passenger strands (miR-155 and miR-155_SNP, Figure 1D; miR-195 guide: 5′ UAGCAGCACAGAAAUAUUGGC 3′, miR-195 passenger: 5′ CCAAUAUUGGCUGUGCUGCUCC 3′) were synthesized by biomers.net GmbH. Double-stranded miRNA was generated by incubating the two strands at a final concentration of 20 µM in 1 × RNA annealing buffer (6 mM TRIS-HCl pH 7.4, 20 mM KCl, 0.4 mM MgCl2). The annealing reaction was performed by denaturing the oligonucleotides (3 min at 95°C) and subsequent slow cooling in a heat block. The hybridization product was analyzed by native PAGE.
Transfections
Transfection experiments were performed with HeLa cells in 96 well plates using LipofectamineTM 2000 (Invitrogen, # 11668–019) transfection reagent. Plasmid DNA (25 ng per well) harboring either the miR-155 target site or the 3′-UTR of AGTR1 (A and C allele) was co-transfected with varying amounts (25–150 nM) of Pre-miRTM-155 (Ambion, # AM17100, diluted in RNase-free water as specified by the manufacturer), miR-155 or miR-155_SNP. For normalization a control-plasmid coding for Renilla luciferase (pRL-CMV, Promega, # E2261, 0.25 ng per well) was added. For MRAS reporter gene assays pmirGLO MRAS 3′-UTR constructs (either C or U variant) were co-transfected with 0–100 nM miR-195. The transfection mixture was prepared in Opti-MEM (Invitrogen, #31985–047) with a final concentration of 6–12 µg/ml LipofectamineTM 2000, depending on the concentration of RNA.
Four hours after transfection, the transfection mixture was replaced by full medium (DMEM, PAA, # E15–843 supplemented with 10% FCS, PAA Laboratories, # A15–151). After further 20 h of incubation cells were lysed with 1x Passive Lysis Buffer (Promega, # E1941) and subsequently, the dual-luciferase assay was performed according to the manufacturer’s protocol (Promega, # E1960) using a microplate reader (anthos Lucy3 Luminometer).
Computational analysis of RNA secondary structure
In silico folding of RNA sequences was performed using an adaptation of the mfold package (version 2.3)62,63 that has been modified to work with the Accelrys Genetics Computer Group (GCG). The calculations were performed with the polymorphic sequence segments containing the SNP at varying internal positions and by defining stepwise (20–50 nt) moving segments of a size of 400, 800 and 1400 nt. The resulting structures were compared globally and locally at the SNP position and/or the respective miRNA binding site and grouped according to the involvement of the SNP-containing sequence segment in intramolecular folding.
Supplementary Material
Acknowledgments
We wish to thank Winfried Wünsche for excellent technical assistance, Simon Dornseifer for initial help with the mfold analyses, and Alessandra Mescalchin and Jeanette Erdmann for fruitful discussions.
Glossary
Abbreviations:
- AGTR1
angiotensin II receptor, type 1
- CAD
coronary artery disease
- CDS
coding sequence
- DHFR
dihydrofolate reductase
- ESR1
estrogen receptor 1
- FF
firefly luciferase
- GWAS
genome-wide association study
- IRGM
immunity-related GTPase family, M
- IVT
in vitro transcript
- miRNA
microRNA
- MRAS
muscle RAS oncogene homolog
- RL
Renilla luciferase
- RISC
RNA induced silencing complex
- SLITRK1
LIT and NTRK-like family, member 1
- SNP
single nucleotide polymorphism
- UTR
untranslated region
Funding
This work was supported by the medical faculty of the University of Lübeck [E22–2009].
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/20497
References
- 1.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–6. doi: 10.1016/S0092-8674(01)00616-X. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 4.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Stricker HM, Gou D, Liu L. MicroRNA: past and present. Front Biosci. 2007;12:2316–29. doi: 10.2741/2234. [DOI] [PubMed] [Google Scholar]
- 9.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 10.Couzin J. MicroRNAs make big impression in disease after disease. Science. 2008;319:1782–4. doi: 10.1126/science.319.5871.1782. [DOI] [PubMed] [Google Scholar]
- 11.Jeyaseelan K, Herath WB, Armugam A. MicroRNAs as therapeutic targets in human diseases. Expert Opin Ther Targets. 2007;11:1119–29. doi: 10.1517/14728222.11.8.1119. [DOI] [PubMed] [Google Scholar]
- 12.Li M, Li J, Ding X, He M, Cheng SY. microRNA and cancer. AAPS J. 2010;12:309–17. doi: 10.1208/s12248-010-9194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson KM, Weiss GJ. MicroRNAs and cancer: past, present, and potential future. Mol Cancer Ther. 2008;7:3655–60. doi: 10.1158/1535-7163.MCT-08-0586. [DOI] [PubMed] [Google Scholar]
- 14.Yu Z, Li Z, Jolicoeur N, Zhang L, Fortin Y, Wang E, et al. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res. 2007;35:4535–41. doi: 10.1093/nar/gkm480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu M, Jolicoeur N, Li Z, Zhang L, Fortin Y, L’Abbe D, et al. Genetic variations of microRNAs in human cancer and their effects on the expression of miRNAs. Carcinogenesis. 2008;29:1710–6. doi: 10.1093/carcin/bgn073. [DOI] [PubMed] [Google Scholar]
- 16.Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A. 2007;104:3300–5. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet. 2006;38:1452–6. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 18.Chen K, Song F, Calin GA, Wei Q, Hao X, Zhang W. Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis. 2008;29:1306–11. doi: 10.1093/carcin/bgn116. [DOI] [PubMed] [Google Scholar]
- 19.Mishra PJ, Bertino JR. MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics. 2009;10:399–416. doi: 10.2217/14622416.10.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bandiera S, Hatem E, Lyonnet S, Henrion-Caude A. microRNAs in diseases: from candidate to modifier genes. Clin Genet. 2010;77:306–13. doi: 10.1111/j.1399-0004.2010.01370.x. [DOI] [PubMed] [Google Scholar]
- 21.Glinsky GV. An SNP-guided microRNA map of fifteen common human disorders identifies a consensus disease phenocode aiming at principal components of the nuclear import pathway. Cell Cycle. 2008;7:2570–83. doi: 10.4161/cc.7.16.6524. [DOI] [PubMed] [Google Scholar]
- 22.Landi D, Gemignani F, Naccarati A, Pardini B, Vodicka P, Vodickova L, et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis. 2008;29:579–84. doi: 10.1093/carcin/bgm304. [DOI] [PubMed] [Google Scholar]
- 23.Horikawa Y, Wood CG, Yang H, Zhao H, Ye Y, Gu J, et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14:7956–62. doi: 10.1158/1078-0432.CCR-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kontorovich T, Cohen Y, Nir U, Friedman E. Promoter methylation patterns of ATM, ATR, BRCA1, BRCA2 and p53 as putative cancer risk modifiers in Jewish BRCA1/BRCA2 mutation carriers. Breast Cancer Res Treat. 2009;116:195–200. doi: 10.1007/s10549-008-0121-3. [DOI] [PubMed] [Google Scholar]
- 25.Brendle A, Lei H, Brandt A, Johansson R, Enquist K, Henriksson R, et al. Polymorphisms in predicted microRNA-binding sites in integrin genes and breast cancer: ITGB4 as prognostic marker. Carcinogenesis. 2008;29:1394–9. doi: 10.1093/carcin/bgn126. [DOI] [PubMed] [Google Scholar]
- 26.Abelson JF, Kwan KY, O’Roak BJ, Baek DY, Stillman AA, Morgan TM, et al. Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science. 2005;310:317–20. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 27.Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibé B, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–8. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 28.Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet. 2011;43:242–5. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 29.Baudin B. Polymorphism in angiotensin II receptor genes and hypertension. Exp Physiol. 2005;90:277–82. doi: 10.1113/expphysiol.2004.028456. [DOI] [PubMed] [Google Scholar]
- 30.Oro C, Qian H, Thomas WG. Type 1 angiotensin receptor pharmacology: signaling beyond G proteins. Pharmacol Ther. 2007;113:210–26. doi: 10.1016/j.pharmthera.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosskopf D, Michel MC. Pharmacogenomics of G protein-coupled receptor ligands in cardiovascular medicine. Pharmacol Rev. 2008;60:513–35. doi: 10.1124/pr.108.000612. [DOI] [PubMed] [Google Scholar]
- 32.Bonnardeaux A, Davies E, Jeunemaitre X, Féry I, Charru A, Clauser E, et al. Angiotensin II type 1 receptor gene polymorphisms in human essential hypertension. Hypertension. 1994;24:63–9. doi: 10.1161/01.hyp.24.1.63. [DOI] [PubMed] [Google Scholar]
- 33.Martin MM, Buckenberger JA, Jiang J, Malana GE, Nuovo GJ, Chotani M, et al. The human angiotensin II type 1 receptor +1166 A/C polymorphism attenuates microrna-155 binding. J Biol Chem. 2007;282:24262–9. doi: 10.1074/jbc.M701050200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Martin MM, Lee EJ, Buckenberger JA, Schmittgen TD, Elton TS. MicroRNA-155 regulates human angiotensin II type 1 receptor expression in fibroblasts. J Biol Chem. 2006;281:18277–84. doi: 10.1074/jbc.M601496200. [DOI] [PubMed] [Google Scholar]
- 35.Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, et al. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3′ untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet. 2007;81:405–13. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ceolotto G, Papparella I, Bortoluzzi A, Strapazzon G, Ragazzo F, Bratti P, et al. Interplay between miR-155, AT1R A1166C polymorphism, and AT1R expression in young untreated hypertensives. Am J Hypertens. 2011;24:241–6. doi: 10.1038/ajh.2010.211. [DOI] [PubMed] [Google Scholar]
- 37.Mishra PK, Tyagi N, Kumar M, Tyagi SC. MicroRNAs as a therapeutic target for cardiovascular diseases. J Cell Mol Med. 2009;13:778–89. doi: 10.1111/j.1582-4934.2009.00744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kretschmer-Kazemi Far R, Sczakiel G. The activity of siRNA in mammalian cells is related to structural target accessibility: a comparison with antisense oligonucleotides. Nucleic Acids Res. 2003;31:4417–24. doi: 10.1093/nar/gkg649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sczakiel G, Far RK. The role of target accessibility for antisense inhibition. Curr Opin Mol Ther. 2002;4:149–53. [PubMed] [Google Scholar]
- 41.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–20. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 42.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–84. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 43.Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y. Potent effect of target structure on microRNA function. Nat Struct Mol Biol. 2007;14:287–94. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 44.Robins H, Li Y, Padgett RW. Incorporating structure to predict microRNA targets. Proc Natl Acad Sci U S A. 2005;102:4006–9. doi: 10.1073/pnas.0500775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–17. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36(Database issue):D149–53. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patzel V, Steidl U, Kronenwett R, Haas R, Sczakiel G. A theoretical approach to select effective antisense oligodeoxyribonucleotides at high statistical probability. Nucleic Acids Res. 1999;27:4328–34. doi: 10.1093/nar/27.22.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Far RK, Nedbal W, Sczakiel G. Concepts to automate the theoretical design of effective antisense oligonucleotides. Bioinformatics. 2001;17:1058–61. doi: 10.1093/bioinformatics/17.11.1058. [DOI] [PubMed] [Google Scholar]
- 49.Overhoff M, Alken M, Far RK, Lemaitre M, Lebleu B, Sczakiel G, et al. Local RNA target structure influences siRNA efficacy: a systematic global analysis. J Mol Biol. 2005;348:871–81. doi: 10.1016/j.jmb.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 50.Erdmann J, Grosshennig A, Braund PS, König IR, Hengstenberg C, Hall AS, et al. Italian Atherosclerosis, Thrombosis, and Vascular Biology Working Group. Myocardial Infarction Genetics Consortium. Wellcome Trust Case Control Consortium. Cardiogenics Consortium New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009;41:280–2. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sõber S, Laan M, Annilo T. MicroRNAs miR-124 and miR-135a are potential regulators of the mineralocorticoid receptor gene (NR3C2) expression. Biochem Biophys Res Commun. 2010;391:727–32. doi: 10.1016/j.bbrc.2009.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–7. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du Q, Thonberg H, Zhang HY, Wahlestedt C, Liang Z. Validating siRNA using a reporter made from synthetic DNA oligonucleotides. Biochem Biophys Res Commun. 2004;325:243–9. doi: 10.1016/j.bbrc.2004.09.222. [DOI] [PubMed] [Google Scholar]
- 55.Mishra PJ, Humeniuk R, Mishra PJ, Longo-Sorbello GS, Banerjee D, Bertino JR., Jr. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc Natl Acad Sci U S A. 2007;104:13513–8. doi: 10.1073/pnas.0706217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jacobsen A, Wen J, Marks DS, Krogh A. Signatures of RNA binding proteins globally coupled to effective microRNA target sites. Genome Res. 2010;20:1010–9. doi: 10.1101/gr.103259.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leibovich L, Mandel-Gutfreund Y, Yakhini Z. A structural-based statistical approach suggests a cooperative activity of PUM1 and miR-410 in human 3′-untranslated regions. Silence. 2010;1:17. doi: 10.1186/1758-907X-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12:1014–20. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 59.Picard V, Ersdal-Badju E, Lu A, Bock SC. A rapid and efficient one-tube PCR-based mutagenesis technique using Pfu DNA polymerase. Nucleic Acids Res. 1994;22:2587–91. doi: 10.1093/nar/22.13.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benecke H. RNA/Protein & Protein/Protein-Interaktionen innerhalb des U11/U12 di-snRNP Partikels des U12-abhängigen Spleißosoms aus HeLa-Zellen. Max-Planck-Institut für biophysikalische Chemie, Abteilung Zelluläre Biochemie. Göttingen, 2004. [Google Scholar]
- 61.Eckardt S, Romby P, Sczakiel G. Implications of RNA structure on the annealing of a potent antisense RNA directed against the human immunodeficiency virus type 1. Biochemistry. 1997;36:12711–21. doi: 10.1021/bi9707234. [DOI] [PubMed] [Google Scholar]
- 62.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–40. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 63.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.