Abstract
Antibiotic treatment to treat specific infections has the potential to effectively target the offending microbe as well as other microbes that colonize sites within a host. Antibiotic-associated diarrhea (AAD) is a classic example resulting from disruption of host microbial communities; 20% of patients with AAD are likely to become colonized with Clostridium difficile. Restoration of a “normal” microbial community within the host using probiotic bacteria is one approach to circumvent AAD and C. difficile infection. The goals of this study were to assess the interactions between Streptococcus thermophilus, a potential probiotic organism and C. difficile using both in vitro and in vivo systems. Exposure of C. difficile to filtered supernatants from S. thermophilus showed a dose-dependent, bactericidal effect due to lactic acid. Additional studies show that levels of lactic acid (10 mM) that did not inhibit bacterial growth had the potential to decrease tcdA expression and TcdA release into the extracellular milieu. In vivo, treatment with viable S. thermophilus significantly increased luminal levels of lactate in the cecum compared with UV-irradiated S. thermophilus. In the context of infection with C. difficile, mice treated with viable S. thermophilus exhibited 46% less weight loss compared with untreated controls; moreover, less pathology, diarrhea, and lower detectable toxin levels in cecal contents were evident more often in S. thermophillus treated mice. A significant, inverse correlation (Spearman r = -0.942, p = 0.017) between the levels of luminal lactate and abundance of C. difficile were noted suggesting that lactate produced by S. thermophilus is a factor impacting the progression of C. difficile infection in the murine system.
Keywords: Clostridium difficile, Streptococcus thermophilus, probiotic, mouse model
Introduction
Clostridium difficile is a pervasive opportunistic pathogen capable of causing mild to severe colitis especially among aging populations and patients receiving antibiotics.1,2 Increases in C. difficile-associated morbidity is attributed in part to the worldwide emergence of strains (e.g., ribotypes 027, 078) with genomic modifications resulting in high-level fluoroquinolone resistance and potential increases in virulence capacity.2,3 One study suggests that fluoroquinolone resistance of C. difficile contributes to more severe inflammation.4 Nevertheless, the most cost-effective treatment option for C. difficile infection is antibiotics, but these have been associated with an increased rate of recurrence.1
The use of probiotic organisms to alleviate a number of GI-related diseases has increased over the past several years. Along these lines, several randomized control trials designed to evade AAD (and C. difficle) through the use of probiotics have been reported. Several studies support the use of probiotic organisms as an approach for prevention of AAD and CDI.5,6 In at least two of the studies, the use of probiotic preparations resulted in ~20% decrease in the incidence of AAD and C. difficile.6,7 To date, the majority of AAD-associated clinical studies have treated patients with pure Lactobacillus spp, Bifidobacteria spp., or Sacchromyces boulardii. Only one study using a commercial yogurt drink contained Streptococcus thermophilus, a potentially probiotic organism.7 The most common dose used in probiotic studies is 1010 viable organisms. This dose likely comes from consumption of fermented dairy products (i.e., yogurt) where lactobacilli and S. thermophilus can reach 108 cfu/mL. If 100 mL of yogurt is consumed, then approximately, 1010 organisms are ingested (personal communication with Dr. M.E. Sanders).
The use of probiotics for alleviating AAD in clinical research exhibit modest, yet significant decreases in the overall incidence of AAD and possible C. difficile infection; however, the mechanisms responsible for the findings are still unclear.6,7 A probiotic approach may be contraindicated based upon an individual’s health status; however, the criteria for determining what probiotics should be administered to which patients are unclear.8 Therefore, in vivo systems provide an opportunity to understand whether probiotic organisms provide host benefits in certain disease states, and the mechanisms through which benefits are conferred. Furthermore, in vivo systems are an invaluable resource to elucidate safety profiles within a host.
The study presented here examines the use of S. thermophilus as a preventive modality for antibiotic induced C. difficile disease in a murine system; companion experiments were conducted to clarify S. thermophilus factor(s) responsible for observed differences.
Results
Secreted components by S. thermophilus are bactericidal against C. difficile
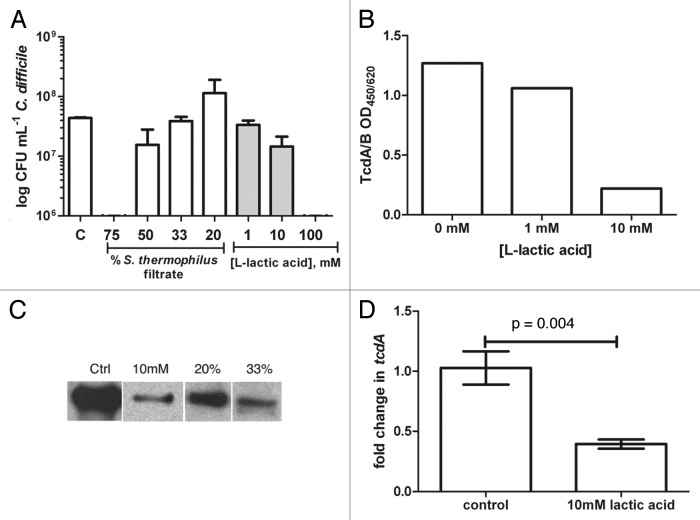
S. thermophilus filtrates were tested for activity against C. difficile growth and showed up to a 5-log reduction after 16h (75% filtrate). The activity was dose dependant since less filtrate was permissive of C. difficile growth after 16h comparable to control growth (Fig. 1A). The major constituents secreted by several lactic acid bacteria include lactic acid, hydrogen peroxide, and bacteriocins. To assess whether LMD-9 produced bacteriocin affected C. difficile, filtrates (25% v/v) from a bacteriocin mutant (ΔblpD-blpF;9) were tested in parallel in vitro. LMD-9 derived bacteriocin(s) had no significant impact on the growth of C. difficile (TL24) after 16h (1.9 × 108 (LMD-9) vs. 9.0 × 107 (ΔblpD-blpF); p = 0.18). Lack of bacteriocin activity from LMD-9 against C. difficile is in line with previous data showing lack of activity against another clostridial species, C. acetobutylicum.9
Figure 1.Clostridium difficile growth is reduced by secreted components from Streptococcus thermophilus, namely lactic acid. (A) BHI media was supplemented with filtered supernatants from S. thermophilus (% v/v) or L-lactic acid (mM), inoculated (105 TL24) incubated for 16h and enumerated using plate counts; increased concentrations (75%) of filtrates or lactic acid (100mM) were bactericidal to C. difficile. Bacterial counts were comparable between control and 33% filtrate (p = 0.62), 20% filtrate (p = 0.52), 1mM lactic acid (p = 0.25) or 10mM (p = 0.052) using a student’s t-test. (B) Supernatants from C. difficile cultures supplemented with lactic acid exhibited dose-dependent decreases in TcdA/B. (C) Similarly, immunoblot analysis showed that TcdA levels decreased in the presence of S. thermophilus filtrates. (D) Decreased expression of tcdA compared with control 8h post-inoculation occurred in the presence of 10mM lactic acid. Results are displayed with standard errors of the mean.
The effect of hydrogen peroxide was assessed by comparing S. thermophilus filtrates (25% v/v) treated with catalase prior to inoculation with C. difficile. C. difficile growth was similar in catalase treated and untreated filtrates after 16h (1.9 × 108 (LMD-9) vs. 2.4 × 108 (LMD-9 + cat); p = 0.29). Since neither bacteriocin nor H2O2 affected C. difficile in vitro, additional experiments were designed to focus on how lactic acid affects C. difficile. In vitro, S. thermophilus LMD-9 produced an average of 10 g/L of lactic acid (90 mM) using the method described below. Experiments using L-lactic acid alone show high doses (100 mM) are bactericidal to C. difficile while 10-fold lower levels permit growth (Fig. 1A). Similar effects of L-lactic acid (1 and 10mM) were observed with VPI10463 while 100mM completely ablated growth after 16h (data not shown).
The media pH before and after growth of C. difficile was not dramatically different (pH range 5.5–6.5) with the exception of 100mM lactic acid (pH 4.5). Media containing 1–10mM lactic acid ranged from pH 6–6.5, similar to media without any additions. In the presence of 20–50% S. thermophilus filtrates, the pH ranged from 5.3–6.5 prior to inoculation with C. difficile. Addition of filtrates to 75% of the total volume lowered the pH to ≤ 5. When pH values were ≤ 5, significant decrements on C. difficile growth and viability were evident compared with control (Fig. 1A, control vs. 75% filtrate: p < 0.0001; control vs. 100 mM lactic acid: p = 0.0005 using an unpaired student’s t-test). No significant differences in C. difficile numbers were evident compared with control when the media pH was within the range of 5.5–6.5.
Lactic acid reduces levels of C. difficile toxins and expression of tcdA in vitro
The major virulence factors produced by C. difficile include two large toxins, TcdA and TcdB. Supernatants from C. difficile grown in the presence of lactic acid were tested for combined TcdA/B levels and revealed a dose-dependent decrease in toxin production that was independent of bacterial growth (Fig. 1A and B). Similarly, growth of TL24 in the presence of S. thermophilus filtrates (20% and 33% v/v) decreased the levels of TcdA produced despite comparable levels of bacteria (Fig. 1A and C). The same experimental approach assessed toxin production at the transcriptional level in BHI containing 10mM lactic acid compared with media alone. Similar to decreased levels of TcdA, a modest, but significant decrease in tcdA expression by C. difficile TL24 occurred after 8h in media containing lactic acid (Fig. 1D).
S. thermophilus significantly alleviates Clostridium difficile infection in a murine model
In order to understand the impact of S. thermophilus on antibiotic associated C. difficile infection, viable S. thermophilus was administered to mice daily at a dose similar to probiotics used in humans either in purified capsule form (e.g., ~1010 CFU per 70 kg human,6) or within a food matrix (e.g., fermented yogurt drink containing ~1010 CFU/100mL,7), within the first 72h of starting the antibiotic cocktail and was continued daily throughout the course of infection unless animals experienced diarrhea, upon which treatment with S. thermophilus was discontinued. Initial studies used VPI10463 at 104 CFU (LD50), although only 30% died with S. thermophilus treatment (vs. 50% of control; data not shown), this trend was not significant and the subsequent focus of our studies were performed using a clinical isolate.
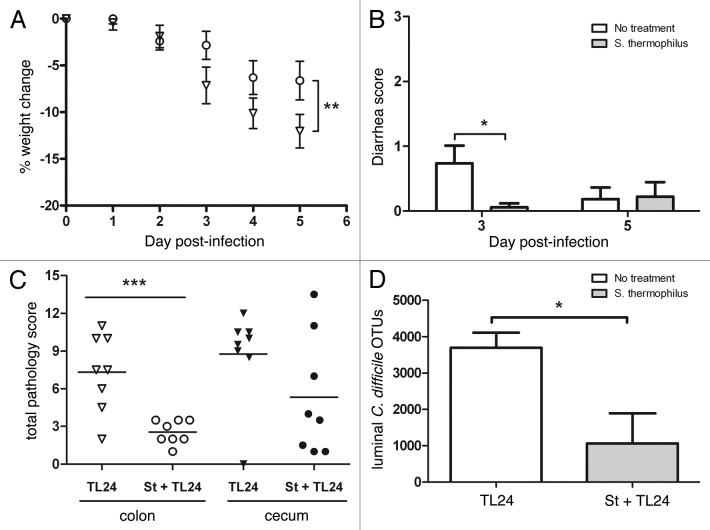
An intestinal isolate from a patient with C. difficile colitis was used to infect mice to provide sufficient morbidity with little mortality. Using this strain, mice treated with S. thermophilus lost significantly less weight (46%) due to C. difficile infection compared with controls (Fig. 2A). Similarly, mice treated with S. thermophilus had significantly less diarrhea compared with untreated mice three days post-infection (Fig. 2B). In a separate experiment, UV-irradiated, nonviable S. thermophilus was given to mice and showed no differences in weight or diarrhea compared with the infected controls (data not shown). Cecal contents and tissue samples were taken from treated and untreated mice and analyzed for levels of histopathology, TcdA/B, and numbers of organisms. Histopathology scoring showed that mice treated with S. thermophilus had significantly less C. difficile-associated pathology in the colon and less cecal pathology (Fig. 2C). Toxin levels in the cecal contents provide a measure of TcdA/B released by C. difficile as an indicator of disease progression in mice where lower levels are observed after the acute phase of infection. Although TcdA/B levels using ELISA were not significantly different, mice (n = 4/group/day) treated with S. thermophilus tended to have lower detectable levels of toxins on days three (OD450 1.8 ± 0.17(TL24) vs. 1.2 ± 0.32(St+TL24); p = 0.12) and five (OD450 1.2 ± 0.25 (TL24) vs. 0.53 ± 0.17 (St+TL24); p = 0.059). In addition, mice treated with S. thermophilus had significantly less C. difficile compared with control mice in the cecal lumen based on operational taxanomic units (OTUs) generated during sequencing (Fig. 2D).
Figure 2.C. difficile infection using TL24 in mice is reduced by treatment with S. thermophilus. (A) Weight change was calculated relative to day 0 weight for each mouse. S. thermophilus treated mice (circles; n = 18) lost significantly less weight (p = 0.004 using Wilks’ Lambda multivariate test) than untreated control mice (triangles; n = 19). Calculated data include final weights from mice that succumbed to infection or were euthanized for sample collection. (B) S. thermophilus treated mice had significantly less diarrhea than untreated mice (*p = 0.02 using an unpaired t-test) on day three, but not day five. (C) The colon of mice treated with S. thermophilus had significantly less pathology (***p = 0.0009 using a unpaired t-test) while cecal tissues from S. thermophilus treated mice showed less (p = 0.13) than untreated mice three days post-infection. (D) Cecal contents from mice treated with S. thermophilus had significantly less C. difficile operational taxonomic units (OTUs) (*p = 0.05; n = 3/group) compared with untreated mice from metagenomic based data.
S. thermophilus produces lactic acid in vivo
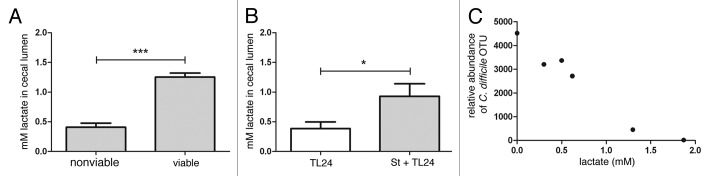
In order to delineate whether S. thermophilus retained the capacity to produce measurable amounts of lactic acid in vivo, mice were treated with antibiotics as described above. After antibiotic treatment, mice were treated with viable or UV-irradiated S. thermophilus daily for three days. Following the final day of treatment, intestinal tissue and cecal contents were harvested and luminal lactic acid (lactate) measured. Mice that received viable S. thermophilus had significantly higher luminal lactate compared with mice that received UV-irradiated S. thermophilus suggesting that viable S. thermophilus is necessary for production of lactic acid in vivo (Fig. 3A).
Figure 3.S. thermophilus treatment increases levels of cecal lactate and is correlated with lower levels of C. difficile. (A) Antibiotic treated, uninfected mice receiving subsequent treatment with viable S. thermophilus had significantly higher levels of cecal lactate vs. mice receiving UV-irradiated (nonviable) S. thermophilus (***p = 0.0009; n = 3/group). (B) Mice treated with S. thermophilus before and after C. difficile infection maintained significantly higher cecal lactate levels (*p = 0.04) vs. untreated, infected mice. (C) An inverse correlation between cecal lactate and abundance of C. difficile OTUs from cecal luminal contents were calculated (two-tailed, Spearman r = -0.942, p = 0.017).
Luminal lactate is inversely correlated with C. difficile levels
Luminal cecal lactate levels were significantly higher in infected mice treated with S. thermophilus compared with untreated controls. Interestingly, luminal lactate had a significant, inverse correlation with the abundance of C. difficile in the cecal lumen reflecting in vitro findings (Fig. 3B and Fig. 3C).
Discussion
C. difficile is a ubiquitous bacterial species comprised of toxigenic and nontoxigenic strains associated with humans and animals. Of the toxigenic strains, epidemic BI/NAP1/027 isolates are linked to both hospital- and community-acquired C. difficile.2 The prophylactic use of probiotics during antibiotic treatment has been shown to significantly reduce the incidence of AAD as the primary outcome and therefore, C. difficile infection as a secondary outcome measure.6,7 Both probiotic study designs excluded patients that presented with diarrhea or had received antibiotics previously, thus segregating a population with a stable intestinal community. In both cases, probiotic treatment significantly decreased the incidence of AAD and C. difficile. Conversely, at least one probiotic organism, S. boulardii, has been assessed in combination with conventional antibiotics to treat C. difficile infection.10,11 Interestingly, treatment with S. boulardii benefited patients with prior recurrent C. difficile infection compared with those with an initial C. difficile infection. The opposing patient populations (e.g., stable intestinal community vs. antibiotic treatment of recurrent C. difficile infection) highlight the necessity to test individual probiotics in the context of specific diseases.
The study presented here sought to examine the impact of S. thermophilus on experimental C. difficile infection with a BI strain in a murine system. S. thermophilus was initially chosen because its genome has been sequenced, molecular tools exist to manipulate the organism, and is widely consumed in foodstuffs (e.g., yogurt, cheese) attesting to its safety. Similar to previous studies,12 infections with a BI strain resulted in lower mortality rates in vivo compared with VPI10463. Animals treated with viable S. thermophilus prior to infection with C. difficile (TL24) lost significantly less weight indicative of lower diarrheal severity, had lower pathological findings and lower burdens of C. difficile compared with untreated controls. A main metabolite of S. thermophilus is lactic acid, which is shown to impact C. difficile TcdA production and tcdA expression in vitro; moreover, treatment with viable S. thermophilus is essential for increased lactate in vivo and alleviation of C. difficile infection.
The in vivo relationship between C. difficile and lactate is supported by in vitro data where higher lactate levels are associated with growth suppression or ablation. In vivo cecal lactate levels are approximately 10–100 times lower than the in vitro levels that exhibit an impact on growth. Cecal lactate measurements account for the total lactate in the volume sampled and not local lactate levels resulting from S. thermophilus treatment. Therefore, it is plausible that localized, high lactate concentrations in the lumen would negatively impact the survival of C. difficile and account for significantly lower detectable levels (Fig. 2D). In addition, lactate generated by S. thermophilus may be utilized by other bacteria and/or host cells. Indeed, gnotobiotic rats treated with S. thermophilus exhibited increased luminal lactate and expression of two monocarboxylic acid transporters (i.e., SLC16A1 and SLC5A8).13
The experimental results presented here reflect studies that examined the impact of colonization and cytotoxicity in gnotobiotic mice with human stool isolates prior to C. difficile challenge.14 The authors found that C. difficile-infected mice mono-associated with Bifidobacterium bifidum or Escherichia coli had lower cecal levels of cytotoxin (i.e., TcdB) and C. difficile. Although the data presented in this work did not specifically look at TcdB, both studies suggest a role for specific bacteria and/or bacterial metabolites that suppress C. difficile toxin production and growth. Along these lines, a number of other studies have defined an array of inhibitory effects exhibited by probiotic, lactic acid bacteria on C. difficile growth and pathogenesis.15-18 Notably, previous reports show normal C. difficile growth but decreased toxin production in co-culture systems with B. bifidum and Lactobacilli spp.; however, the involvement of lactic acid was not addressed in either report.15,18
Materials and Methods
Bacterial strains and growth
Streptococcus thermophilus (LMD-9), which is fully sequenced, was grown in Elliker’s broth supplemented with 1% beef extract (42°C, 18h) resulting in 109 CFU/mL. Cultures for use as an in vivo treatment were pelleted, washed, and resuspended in broth to a final concentration of 108/mL. The clinical isolate of Clostridium difficile (TL24) in these studies was isolated from patient stool and was characterized to have the following features: REA type BI-1 (kindly performed by Dr. D. Gerding’s laboratory), ribotype 027 (kindly assayed by Dr. R.J. Carman at TechLab), moxifloxacin sensitive, positive for tcdA, tcdB, cdtA/B, and sequence analysis of tcdC shows a base pair deletion at position 117 and a base change at 120 (C120T). C. difficile VPI10463, which produces elevated levels of TcdA and TcdB, was used in experiments as a comparator strain. Initial in vivo studies utilized VPI10463 to replicate C. difficile infection in a murine system as described elsewhere.12 Similarly, initial in vitro studies used VPI10463 and then included isolate TL24. C. difficile were grown in pre-reduced chopped meat broth (18h, 37°C) for use in vivo or overnight in BHI broth for in vitro studies. C. difficile was enumerated on BHI agar containing 0.5% neutral red (1% w/v in ethanol) using the broth microdilution and the drop plate method.19
In vivo infection model
A mouse model of C. difficile-associated disease12 was used for in vivo studies with the following modifications: omission of kanamycin from the antibiotic cocktail and an increase in clindamycin (32 mg/kg). Kanamycin was omitted since another aminoglycoside, gentamicin, was included in the cocktail. An increased dose of clindamycin was chosen to reflect daily dosages that would be used to clinically treat bacterial infections. All experiments were conducted according to the protocol approved by the UVA ACUC. Male mice (8 w.o., Jackson Laboratories) were used in all studies. S. thermophilus treatment (107 CFU/100μL/mouse) was administered within 72h of the antibiotic cocktail and every 24h thereafter by oral gavage and discontinued if diarrhea was evident. Animals were infected with the the clinical isolate, TL24 (105 CFU/100uL/mouse). Animals were observed and weighed at the same time, daily. The severity of diarrhea was scored using the following scale: 0, no symptoms; 1, clumped cobb; 2, slight wet tail; 3, wet tail. On days three and five, mice with similar clinical signs were euthanized for sample collection (cecal contents and tissue). Samples were either frozen and stored at -80°C or fixed in Bouin’s solution; paraffin embedded, and stained using hematoxylin/eosin (services provided by UVA Research Histology Core). Slides were blinded and scored by an independent reader using published parameters.20 The results presented are from two experimental replicates.
In vitro culture assays
All assays were performed in a Bactron I anaerobic chamber (Shel Labs, Cornelius, OR). C. difficile (105 CFU) was inoculated into broth containing filtrates, B-Elliker broth, or L-lactic acid. Supernatants from S. thermophilus were filtered (0.2μm) and diluted (v/v) with BHI to support growth of C. difficile; control wells contained B-Elliker broth and BHI in the same ratios as used for S. thermophilus filtrates. Stocks of L-lactic acid were diluted in BHI to a final concentration of 10 – 100 mM. After inoculation and growth (16–24h, 37°C), bacteria were enumerated on BHI agar as described above. S. thermophilus filtrates were treated with catalase (0.1mg/mL; 37°C for 1h) prior to dilution with BHI and inoculation with C. difficile.
RNA isolation and expression analysis
In parallel with in vitro studies, a separate set of wells were set up for RNA analysis of C. difficile TL24 after inoculation with 105 CFU into BHI containing 10mM lactic acid. Bacteria were harvested 6h after inoculation, pelleted (10,000 × g, 2 min), and resuspended in RNAlater. Total RNA was isolated using Protocol 5 in the RNAprotect Bacteria Reagent Handbook and the Qiagen RNeasy mini-kit. RNA was quantified and converted to cDNA using Superscript II and random primers (Invitrogen). Real-time PCR was run on cDNA to detect tcdA21 and 16S rRNA (966F-5′gcaacgcgaagaaccttacc and 1069R-5′gctgacgacagccatgca) using SYBR PreMix ExTaqII (Takara Bio, Inc.). Data was analyzed by the 2-ΔΔCt method using 16S rRNA as the housekeeping gene. RNA from untreated TL24 (0h) was used as the control.Triose phosphate isomerase (tpi) was also tested as a housekeeping gene and as previously reported, was found to be variable.22
Detection of C. difficile toxins
A commercially available kit (C. difficile Toxin A/B II, Techlab) was used for detection of toxin(s) in the cecal contents of mice or from in vitro supernatants. Cecal contents were weighed and normalized to the sample weighing the least using the sample diluent provided in the kit. Normalized samples were diluted and tested according to manufacturer’s directions. In vitro supernatants were tested at 1:10 dilution using the TcdA/B kit or using equal volumes, were separated using the NuPAGE Novex Tris-Acetate Gel/SDS buffer system (Invitrogen), transferred to nitrocellulose, and probed for TcdA (anti-TcdA, clone PCG4, Novus Biologicals, LLC.; secondary antibody, anti-mouse-HRP, Cell Signaling Technology, Inc.).
DNA isolation and detection of C. difficile
DNA was isolated from normalized cecal contents using the Qiagen Stool extraction kit according to manufacturer’s instructions; after adding the lysis buffer, the temperature was increased from 70°C to 95°C to lyse recalcitrant organisms. DNA from bacteria associated with intestinal mucosa was isolated according to a previously published method using bead beating and subsequent DNA isolation.23 Metagenomic data was obtained from 16S rRNA amplicons that included the V1→V3 regions. Amplicons were generated using the provisional 16S 454 protocol described by the Human Microbiome Project (http://www.hmpdacc.org/tools_protocols/tools_protocols.php) and then sequenced using 454 GS FLX technology. QIIME analysis of 454 sequence reads annotated C. difficile as Peptostreptococcus incertae sedis; the most recent version of the Ribosomal Database Project (November 2011) classifies C. difficile sequences at the genus level into Clostridium cluster XI under the family Peptostreptococcaceae. Therefore, one detectable sequence with > 95% identity to Peptostreptococcus incertae sedis is equal to one C. difficile operational taxonomic unit (OTU).
Detection of cecal lactate
Aliquots (100µL) of normalized cecal contents used in the toxin assay were concentrated using a 10 kDa molecular weight cut-off spin concentrator (Millipore). S. thermophilus supernatants were also assayed to determine the amount of lactic acid produced in vitro using this method. L-lactic acid (lactate) in the eluate fractions was quantified using the colorimetric Lactate assay kit (Biovision) according to manufacturer’s instructions.
Statistics
Data was analyzed using SPSS v.19 software or GraphPad Prismv.5. A repeated measure ANOVA was used to compare weight change across six occasions from the point of infection. Additional tests and levels of significance are reported in figure legends.
Conclusion
The results presented here build on a working in vivo model of C. difficile infection for testing conventional and novel probiotics for use in patients with AAD or C. difficile. Similar to several lactic acid bacteria, S. thermophilus is generally recognized as safe by the FDA and is used in commercial yogurt production. Because S. thermophilus is easily attainable through dietary choices, deciphering the complexities involved in the prevention or alleviation of C. difficile infection are critically important clinically, and within the field of probiotic research.
At a more basic level, this system permits examination of the intestinal microbial community after antibiotic exposure and/or use of probiotic organisms to alter susceptibility to infection with C. difficile. It is likely that both inter- and intra-bacterial metabolic processes are crucial to suppressing C. difficile toxin metabolism and therefore disease development; this notion is supported by the work presented here and elsewhere.14 Lactic acid is only one factor that was considered in this study; it is possible that S. thermophilus treatment also alters local immune responses or the intestinal microbiota after antibiotic treatment. Studies to define the intestinal microbial community and local intestinal immune response from in vivo studies presented here are forthcoming and will provide information on the impact of S. thermophilus in experimental C. difficile infection.
Disclosure of Potential Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
This research was supported by a Young Investigator Grant in Probiotics Research (to G.L.K.) from the Global Probiotics Council and by the National Institutes of Health Grant U01AI075526 (to R.L.G.) and the North Carolina Agricultural Foundation (to E.D. and T.R.K.). The authors thank Pascal Hols for kindly providing the bacteriocin negative mutant of S. thermophilus for use as a control in this study. The authors wish to thank Dr Relana Pinkerton for assistance with a portion of the statistical analysis.
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/21757
References
- 1.Baxter R, Ray GT, Fireman BH. Case-control study of antibiotic use and subsequent Clostridium difficile-associated diarrhea in hospitalized patients. Infect Control Hosp Epidemiol. 2008;29:44–50. doi: 10.1086/524320. [DOI] [PubMed] [Google Scholar]
- 2.O’Connor JR, Johnson S, Gerding DN. Clostridium difficile infection caused by the epidemic BI/NAP1/027 strain. Gastroenterology. 2009;136:1913–24. doi: 10.1053/j.gastro.2009.02.073. [DOI] [PubMed] [Google Scholar]
- 3.Cartman ST, Heap JT, Kuehne SA, Cockayne A, Minton NP. The emergence of ‘hypervirulence’ in Clostridium difficile. Int J Med Microbiol. 2010;300:387–95. doi: 10.1016/j.ijmm.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Pawlowski SW, Archbald-Pannone L, Carman RJ, Alcantara-Warren C, Lyerly D, Genheimer CW, et al. Elevated levels of intestinal inflammation in Clostridium difficile infection associated with fluoroquinolone-resistant C. difficile. J Hosp Infect. 2009;73:185–7. doi: 10.1016/j.jhin.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McFarland LV. Evidence-based review of probiotics for antibiotic-associated diarrhea and Clostridium difficile infections. Anaerobe. 2009;15:274–80. doi: 10.1016/j.anaerobe.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Gao XW, Mubasher M, Fang CY, Reifer C, Miller LE. Dose-response efficacy of a proprietary probiotic formula of Lactobacillus acidophilus CL1285 and Lactobacillus casei LBC80R for antibiotic-associated diarrhea and Clostridium difficile-associated diarrhea prophylaxis in adult patients. Am J Gastroenterol. 2010;105:1636–41. doi: 10.1038/ajg.2010.11. [DOI] [PubMed] [Google Scholar]
- 7.Hickson M, D’Souza AL, Muthu N, Rogers TR, Want S, Rajkumar C, et al. Use of probiotic Lactobacillus preparation to prevent diarrhoea associated with antibiotics: randomised double blind placebo controlled trial. BMJ. 2007;335:80. doi: 10.1136/bmj.39231.599815.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petrof EO. Probiotics and gastrointestinal disease: Clinical evidence and basic science. Antiinflamm Antiallergy Agents Med Chem. 2009;8:260–9. doi: 10.2174/187152309789151977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fontaine L, Hols P. The inhibitory spectrum of thermophilin 9 from Streptococcus thermophilus LMD-9 depends on the production of multiple peptides and the activity of BlpG(St), a thiol-disulfide oxidase. Appl Environ Microbiol. 2008;74:1102–10. doi: 10.1128/AEM.02030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Surawicz CM, McFarland LV, Greenberg RN, Rubin M, Fekety R, Mulligan ME, et al. The search for a better treatment for recurrent Clostridium difficile disease: use of high-dose vancomycin combined with Saccharomyces boulardii. Clin Infect Dis. 2000;31:1012–7. doi: 10.1086/318130. [DOI] [PubMed] [Google Scholar]
- 11.McFarland LV, Surawicz CM, Greenberg RN, Fekety R, Elmer GW, Moyer KA, et al. A randomized placebo-controlled trial of Saccharomyces boulardii in combination with standard antibiotics for Clostridium difficile disease. JAMA. 1994;271:1913–8. doi: 10.1001/jama.1994.03510480037031. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Katchar K, Goldsmith JD, Nanthakumar N, Cheknis A, Gerding DN, et al. A mouse model of Clostridium difficile-associated disease. Gastroenterology. 2008;135:1984–92. doi: 10.1053/j.gastro.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Rul F, Ben-Yahia L, Chegdani F, Wrzosek L, Thomas S, Noordine ML, et al. Impact of the metabolic activity of Streptococcus thermophilus on the colon epithelium of gnotobiotic rats. J Biol Chem. 2011;286:10288–96. doi: 10.1074/jbc.M110.168666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corthier G, Dubos F, Raibaud P. Modulation of cytotoxin production by Clostridium difficile in the intestinal tracts of gnotobiotic mice inoculated with various human intestinal bacteria. Appl Environ Microbiol. 1985;49:250–2. doi: 10.1128/aem.49.1.250-252.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banerjee P, Merkel GJ, Bhunia AK. Lactobacillus delbrueckii ssp. bulgaricus B-30892 can inhibit cytotoxic effects and adhesion of pathogenic Clostridium difficile to Caco-2 cells. Gut Pathog. 2009;1:8. doi: 10.1186/1757-4749-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naaber P, Smidt I, Stsepetova J, Brilene T, Annuk H, Mikelsaar M. Inhibition of Clostridium difficile strains by intestinal Lactobacillus species. J Med Microbiol. 2004;53:551–4. doi: 10.1099/jmm.0.45595-0. [DOI] [PubMed] [Google Scholar]
- 17.Trejo FM, Minnaard J, Perez PF, De Antoni GL. Inhibition of Clostridium difficile growth and adhesion to enterocytes by Bifidobacterium supernatants. Anaerobe. 2006;12:186–93. doi: 10.1016/j.anaerobe.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Trejo FM, Pérez PF, De Antoni GL. Co-culture with potentially probiotic microorganisms antagonises virulence factors of Clostridium difficile in vitro. Antonie Van Leeuwenhoek. 2010;98:19–29. doi: 10.1007/s10482-010-9424-6. [DOI] [PubMed] [Google Scholar]
- 19.Chen CY, Nace GW, Irwin PL. A 6 x 6 drop plate method for simultaneous colony counting and MPN enumeration of Campylobacter jejuni, Listeria monocytogenes, and Escherichia coli. J Microbiol Methods. 2003;55:475–9. doi: 10.1016/S0167-7012(03)00194-5. [DOI] [PubMed] [Google Scholar]
- 20.Pawlowski SW, Calabrese G, Kolling GL, Platts-Mills J, Freire R, AlcantaraWarren C, et al. Murine model of Clostridium difficile infection with aged gnotobiotic C57BL/6 mice and a BI/NAP1 strain. J Infect Dis. 2010;202:1708–12. doi: 10.1086/657086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houser BA, Hattel AL, Jayarao BM. Real-time multiplex polymerase chain reaction assay for rapid detection of Clostridium difficile toxin-encoding strains. Foodborne Pathog Dis. 2010;7:719–26. doi: 10.1089/fpd.2009.0483. [DOI] [PubMed] [Google Scholar]
- 22.Metcalf D, Sharif S, Weese JS. Evaluation of candidate reference genes in Clostridium difficile for gene expression normalization. Anaerobe. 2010;16:439–43. doi: 10.1016/j.anaerobe.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 23.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–75. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]