Abstract
Aberrant immune responses toward commensal gut bacteria can result in the onset and perpetuation of inflammatory bowel diseases (IBD). Reduced microbiota diversity in conjunction with lower proportion of Gram positive and higher proportion of Gram negative bacteria than in healthy subjects is frequently reported in IBD patients. In a subset of IBD patients, E. coli strains with specific features trigger disease. Important molecular mechanisms underlying this effect have been identified. However, in the majority of patients the exact nature of host-microbe interactions that contribute to IBD development has so far not been defined. The application of metagenomic techniques may help to identify bacterial functions that are involved in the aggravation or alleviation of IBD. Subsequently, the relevance for disease development of bacterial candidate genes may be tested taking advantage of reductionist animal models of chronic gut inflammation. This approach may help to identify bacterial functions that can be targeted in future concepts of IBD therapy.
Keywords: Inflammatory bowel disease (IBD), Crohn disease (CD), ulcerative colitis (UC), intestinal microbiota, Escherichia coli, AIEC, Faecalibacterium prausnitzii
Introduction
Ulcerative colitis (UC) and Crohn Disease (CD) represent the two major entities of inflammatory bowel diseases (IBD). Both disorders are relapsing idiopathic inflammatory conditions that are either restricted to the superficial layers of the colonic wall (UC) or may transmurally affect the entire gastrointestinal tract (CD). Further differences include intestinal obstructions and the presence of fistula, granulomas, fissures and skip lesions which are common features in CD but rarely observed in UC. In contrast, bloody stools and irregular crypt architecture are solely or more often observed in UC patients.1 A systematic review of data published between 1950 and 2010 revealed that prevalence of UC per 100,000 European and North American individuals is approximately 500 and 250, respectively. Numbers for CD are approximately 320 per 100,000 in both Europe and North America. Many of the studies included in this meta-analysis clearly demonstrated that IBD incidence significantly increases with time and the authors concluded that IBD may emerge as a global disease.2
The etiology of CD and UC is not entirely clear but it is generally accepted that aberrant T-cell-mediated immune responses to a subset of commensal gut bacteria results in IBD development in a genetically susceptible host.3 Many of the CD risk genes that have been identified by genome-wide association studies are involved in the recognition by the immune system of intestinal bacteria and in bacterial clearance. Examples are genes coding for the nucleotide-binding oligomerization domain-containing protein 2 (NOD2, also referred to as caspase recruitment domain-containing protein 15, CARD15), the toll-like receptor 4 (TLR4) or the autophagy-related 16-like 1 protein (ATG16L1) and the immunity-related GTPase family, M (IRGM). The identified UC susceptibility genes encoding interleukin-10 (IL-10), the extracellular matrix protein 1 (ECM1) or cadherin-1 (CDH1) suggest that impaired anti-inflammatory pathways and defective mucosal barrier functions contribute to the disease.4-6
The bacterial triggers that induce aberrant host responses and thereby at least contribute to the development of IBD are not exactly known. There is some indication that IBD may be a classical infectious disease. For instance, there is a long-lasting debate whether or not infections with Mycobacterium avium susp. paratuberculosis (MAP) may cause CD. But although a meta-analysis supports a specific association between MAP and CD,7 its exact role in CD remains to be identified.8 In contrast, strong evidence exists for an important role of commensal gut bacteria in IBD since changes in intestinal microbiota composition are frequently reported in CD and UC patients. In addition, many genetically driven mouse models of gut inflammation require the presence of commensal gut bacteria to develop disease. Examples are IL-10−/− mice9 and human leukocyte antigen B27 (HLA-B27)-transgenic rats10 which are protected from colitis in the germ free state. In both models, reconstitution with commensal gut bacteria results in disease development. These observations suggest that commensal bacteria are critically involved in disease etiology. Proposed mechanisms of action are illustrated in Figure 1. However, the exact nature of host-microbe interactions contributing to disease development remains to be exactly defined. Important questions that have to be answered and that are discussed in the following include:
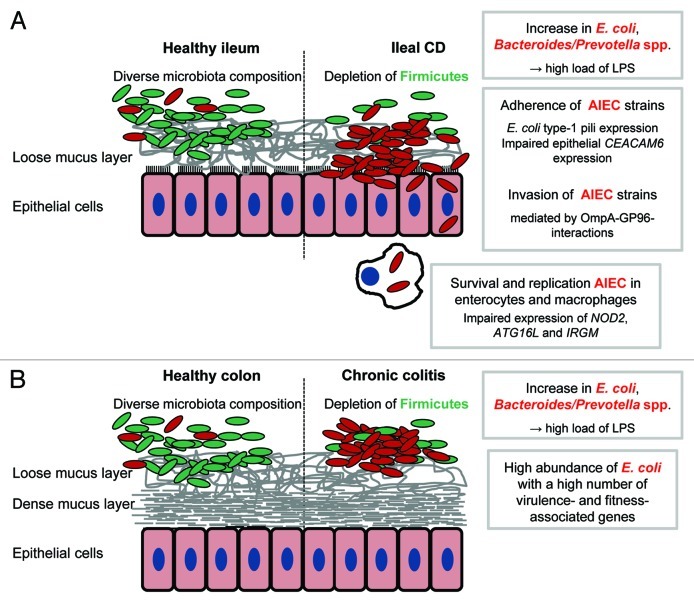
Figure 1. Pro-inflammatory effects of gut bacteria in IBD. Crohn’s disease and ulcerative colitis are characterized by a depletion of Firmicutes in conjunction with an increase of Gram negative bacteria, namely E. coli and Bacteroides/Prevotella spp The shift in microbiota composition is associated with high loads of bacterial antigens (mainly LPS). The recognition by the immune system of these antigens and the subsequent initiation of pro-inflammatory responses may be an important trigger of IBD. In a subset of patients with ileal CD, adherent-invasive E. coli (AIEC) strains are highly abundant and able to invade and to survive and replicate in host cells. An impaired bacterial clearance by the host may be responsible for disease development in patients with mutations in autophagy-associated genes (A). Direct interactions between bacterial and host cells may be less important in chronic colitis because the colonic mucosa is protected by a dense mucus layer. Increased concentrations of bacterial antigens resulting from a high abundance of Gram negative bacteria may drive colitis progression. The exact role in chronic colitis of E. coli strains with a high number of virulence- and fitness-associated genes remains to be identified (B).
(1) Is there a general pattern of IBD-associated microbiota changes that can be observed in CD and UC patients? If this is the case, is it possible to identify the “key players” that are responsible for disease development or prevention?
(2) If specific commensal bacteria contribute to disease aggravation or alleviation, do those bacteria display specific features? Is it possible to identify pro-inflammatory or protective traits that may be targeted in future therapeutic approaches?
(3) Given the fact that alterations in intestinal microbiota composition at least in part result from environmental changes in the inflamed gut: what are the environmental factors that exert a selection pressure on gut microbiota? How do bacteria respond to changes in the ecosystem?
IBD-Associated Changes in Intestinal Microbiota Composition
As mentioned above, UC and CD are distinct entities of IBD with specific anatomical and clinical features and it can be assumed that environmental conditions in the gut differ between the active and remission state of the disease. Such differences may result in disease-specific microbiota characteristics. In fact, is has been demonstrated that active CD and UC can be diagnosed and differentiated based on the patients’ fecal microbiota composition.11 In addition to the disease type, the sampling site may influence the outcome of studies on microbiota in IBD patients. For instance, mucosa-associated bacterial populations in healthy subjects differ significantly from those in feces12,13 and it may well be that these two bacterial populations are differently modulated in the inflamed gut. Therefore, the disease type or state and the sampling site are indicated in the following when studies on microbiota alterations in IBD are discussed and the most important findings are summarized in Table 1.
Table 1. Studies on changes in intestinal microbiota composition associated with inflammatory bowel disease.
| Study | Disease | Sample material | Methods | Major results | Ref. |
|---|---|---|---|---|---|
| Swidsinski et al., 2008 |
CD, UC |
Feces |
FISH |
Depletion of F. prausnitzii in CD |
11 |
| Ott et al., 2004 |
CD, UC |
Mucosa |
16S rRNA gene clone libraries, SSCP, qPCR |
Reduced microbiota diversity; depletion of Bacteroides spp, Eubacterium spp, Lactobacillus spp |
14 |
| Nishikawa et al., 2009 |
UC |
Mucosa |
T-RFLP |
Reduced microbiota diversity; depletion of members of the clostridial closter XIVa |
15 |
| Frank et al., 2007 |
CD, UC |
Mucosa |
16S rRNA gene clone libraries |
Depletion of Bacteroidetes and Lachnospiraceae; relative increase in proteobacteria |
16, 17 |
| Andoh et al., 2007 |
UC |
Feces |
T-RFLP |
Presence of Ruminoccus spp, Eubacterium spp, Fusobacterium spp, Bacteroides spp, Lactobacillus spp, and proteobacteria which are absent in healthy controls |
18 |
| Andoh et al., 2012 |
CD |
Feces |
T-RFLP |
Depletion of the genera Faecalibacterium and Bifidobacterium; increase of the genus Bacteroides |
19 |
| Manichanh et al., 2006 |
CD |
Feces |
DNA clone library, FISH |
Reduced diversity within the Firmicutes |
20 |
| Willing et al., 2010 |
CD, UC |
Feces |
Pyrosequencing |
Depletion of Faecalibacterium and Roseburia; increased Enterobacteriaceae and Ruminococcus gnavus in CD |
22 |
| Walker et al., 2011 |
CD, UC |
Mucosa |
16S rRNA gene clone libraries |
Reduced microbiota diversity; depletion of Firmicutes.; increased Bacteroidetes (CD and UC) and Enterobacteria (CD) |
24 |
| Joossens et al., 2011 |
CD |
Feces |
DGGE, qPCR |
Depletion of butyrate-producing bacteria |
25 |
| Sokol et al., 2009 |
CD, UC |
Feces |
qPCR |
Depletion of Faecalibacterium prausnitzii |
26 |
| Sokol et al., 2008 |
CD |
Mucosa |
FISH |
Depletion of Faecalibacterium prausnitzii |
27 |
| Baumgart et al., 2007 |
CD |
Mucosa |
16S rRNA clone libraries, qPCR, FISH, characterization of E. coli isolates |
Depletion of Clostridiales; increase of invasive E. coli strains |
30 |
| Martin et al., 2004 |
CD, UC |
Mucosa |
Culture-based analysis; characterization of E. coli isolates |
Important role of adherent E. coli in CD |
31 |
| Martinez-Medina et al., 2009 |
CD |
Mucosa |
qPCR; characterization of E. coli isolates |
High abundance of AIEC |
32 |
| Sasaki et al., 2007 |
CD, UC |
Mucosa |
characterization of E. coli isolates |
Important role of invasive E. coli in CD |
33 |
| Darfeuille-Michaud et al., 1998 |
CD |
Mucosa |
characterization of E. coli isolates |
Important role of adherent E. coli in CD |
34 |
| Darfeuille-Michaud et al., 2004 |
CD, UC |
Mucosa |
characterization of E. coli isolates |
Important role of invasive-adherent E. coli in CD |
36 |
| Swidsinski et al., 2005 |
CD, UC |
Mucosa |
FISH |
Important role of B. fragilis |
43 |
| Seksik et al., 2003 |
CD |
Mucosa |
Dot blot hybridization |
Increased Enterobacteria |
45 |
| Kotlowski et al., 2007 | CD, UC | Mucosa | characterization of E. coli isolates | High abundance of E. coli with potentially pathogenic traits | 47 |
Abbreviations used: CD, Crohn’s Disease; UC, ulcerative colitis; DGGE, denaturing gradient gel electrophoresis; FISH, fluorescence in situ hybridization; qPCR, quantitative PCR; SSCP, single strand conformation polymorphism; T-RFLP, terminal restriction fragment length polymorphism.
A number of studies demonstrated that IBD is associated with reduced intestinal microbiota diversity. A 50% and 30% less diverse mucosa-associated microbiota in the colon of patients with active CD and UC, respectively, has been concluded from bacterial 16S rRNA gene-based single strand conformation polymorphism (SSCP) analysis. The lower microbiota diversity in IBD patients resulted from a reduced abundance of Eubacterium spp, Lactobacillus spp and Bacteroides spp as compared with healthy study participants.14 A significantly less diverse composition of microbiota than in healthy controls was confirmed when 16S rRNA-based terminal restriction fragment length polymorphism (T-RFLP) analysis was applied to study mucosal bacteria in patients with active UC. The bacterial phylotypes that were reduced in UC patients mainly belonged to the clostridia. The authors suggested that a loss of bacteria converting non-digestible dietary fiber into short-chain fatty acids may contribute to UC development.15 The comparison of 16S rRNA clone libraries indicated that bacterial communities at the mucosa of UC and CD patients are characterized by a lower abundance of Bacteroidetes and Lachnospiraceae than in healthy controls. In contrast, more sequences representing Proteobacteria and the class of Bacilli were detected in IBD patients. From quantitative real-time PCR experiments the authors concluded that the relative changes were a consequence of reduced numbers of Bacteroidetes and Lachnospiraceae and stable populations of Proteobacteria.16 In a later study the sequencing data were re-evaluated and the re-classification of sequences confirmed the relative decrease of Lachnospiraceae and the relative increase of Proteobacteria in IBD patients. In addition, differences in microbiota composition were tested for associations with the IBD-susceptibility genes NOD2 and ATG16L1 and the authors proposed that alterations in microbiota may be associated with the selected IBD risk alleles.17
However it may be hypothesized that mucosa-associated bacteria are more critically involved in IBD than the luminal populations, fecal material has also been used to characterize inflammation-associated changes in gut microbiota. Analysis of fecal material with T-RFLP revealed differences between healthy controls and patients with UC in the active and in the remission phase. The presence in UC patients of restriction fragments derived from unclassified bacteria, Ruminococcus spp, Eubacterium spp, Proteobacteria and Bacteroidetes, which were absent in the healthy gut, was responsible for the observed differences. A closer look at the fragments detected in patients with active and inactive UC, respectively, indicated that unclassified bacteria, Ruminococcus spp, Eubacterium spp, Fusobacterium spp, Lactobacillus spp, Proteobacteria and Bacteroidetes are less abundant in inactive UC.18 The use of T-RFLP for the analysis of fecal bacteria in CD patients revealed lower microbiota diversity than in healthy controls. CD was characterized by a decrease of the genus Faecalibacterium, irrespective of the disease phase. In the active phase of CD, the genus Bacteroides was increased and the genus Bifidobacterium decreased.19 A lower fecal microbiota diversity than in healthy controls was also reported in patients with quiescence CD. In this study, the combination of metagenomic clone library sequencing with specific enumeration of selected bacterial groups with fluorescence in situ hybridization (FISH) revealed that CD patients harbored lower numbers of Firmicutes, in particular of members of the Clostridium leptum subgroup. In contrast, one member of the Porphyromonadaceae was exclusively detected in CD patients.20
The analysis of a subset of study subjects in a large-scale metagenomic sequencing analysis confirmed clear-cut differences in intestinal microbiota composition between healthy individuals and patients suffering from IBD. It was possible to clearly separate UC from CD patients based on fecal bacterial composition.21 The specific influence of disease phenotype on intestinal microbiota composition has been addressed in twins. In this study, microbiota composition was analyzed by deep sequencing of 16S rRNA genes extracted from fecal material and biopsy samples. In a first step, the authors defined a fecal core microbiome in healthy subjects. This core microbiome was largely shared between healthy individuals and patients with UC and colonic CD (CCD), respectively. However, fewer members of the core species were detected in UC and CCD patients. Subsequent analyses revealed only few differences between UC patients and healthy individuals at any phylogenetic level tested. In contrast, microbiota in patients with CCD and ileal CD (ICD) differed substantially from that of healthy controls. The differences between ICD patients and healthy individuals were characterized by a lower abundance of Firmicutes and a higher abundance of Proteobacteria. The latter was not observed in CCD patients who, interestingly, had more Firmicutes than healthy controls. At the family level, Bifidobacteriaceae, Coriobacteriaceae, Ruminococcaceae and Anaeroplasmataceae were more frequently detected in CCD than in healthy subjects. In contrast, ICD-associated microbiota was characterized by a reduction of Ruminococcaceae including Faecalibacterium in conjunction with a low abundance of the genera Alistipes, Collinsella and Roseburia. The observed increase of Proteobacteria in ICD patients was mainly attributed to a higher abundance of E. coli. No clear-cut inflammation-associated microbiota pattern was observed when biopsy samples from patients in remission were analyzed. However, Ruminococcus gnavus was found in all samples from ICD patients and represented between approximately 8% (distal colon) and 10% (in the ileum) of the detected pylotypes.22 The statistical re-evaluation of the data set generated in this study confirmed that gut bacterial community structure in healthy subjects and in patients suffering from UC and CCD, respectively, is very similar. In contrast, microbiota composition in ICD patients is more variable. Therefore, the authors proposed that shifts in microbiota composition are less important in CCD and UC than in ICD.23
Differences in mucosal microbiota composition between CD and UC patients have been demonstrated by sequencing more than 10,000 16S rRNA gene fragments obtained by specific PCR amplification. Although there was a large inter-individual variation, specific microbiota patterns have been associated with the two different IBD entities. Both types of disease were characterized by an altered proportion between Firmicutes and Bacteroidetes. Compared with healthy controls, the abundance in CD patients of Firmicutes was reduced at the expense of Bacteroidetes. In addition, a slight increase in Enterobacteria was observed in CD but not in UC patients. In UC patients, slightly more Bacteroidetes than Firmicutes sequences were detected. Significant differences in bacterial colonization of inflamed and non-inflamed mucosal sites were also observed. However, it was not possible to define a specific inflammation-associated signature since the bacterial community patterns strongly differed between individuals.24
In summary, the notion that chronic gut inflammation is associated with significant alterations in intestinal microbiota composition is supported by a number of studies addressing different disease types and states and different bacterial niches in the gut. It is difficult to identify a general disease-associated microbiota pattern. However, reduced microbiota diversity appears to be an important feature of IBD. In addition, mainly CD is characterized by decreased proportions of Gram-positives and increased proportions of Gram-negatives.
Specific Bacteria in IBD
Aiming at identifying bacterial species the higher or lower abundance of which is a typical feature of CD, Joossens and coworkers analyzed the fecal microbiota of 68 CD patients with 16S rRNA-based denaturing gradient gel electrophoresis (DGGE). Bands that contributed to disease-associated microbiota profiles were subjected to 16S rRNA gene sequencing and the respective species were identified based on a 135 bp gene fragment. From these sequence analyses and specific enumeration with quantitative real-time PCR, the authors concluded that an increase of Ruminococcus gnavus and a decreased abundance of Bifidobacterium adolescentis, Dialister invisus, an uncharacterized member of the clostridial cluster XIVa and Faecalibacterium prausnitzii is a typical feature of CD.25 The low abundance of luminal F. prausnitzii is in line with previous observations in patients with active CD and UC, respectively26 and indicative of a special role of this species in the onset or perpetuation of IBD.
In fact, interesting features of F. prausnitzii have been demonstrated.27 Under in vitro conditions, F. prausnitzii strongly induces the expression of anti-inflammatory IL-10 in peripheral blood mononuclear cells. In contrast, the expression of pro-inflammatory IL-12 and interferon-gamma (IFNγ) was only slightly increased. The fact that the supernatant of F. prausnitzii culture medium reduced pro-inflammatory IL-8 production by human cancer cells and abolished the activation of nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) in a reporter gene cell system was indicative of the secretion of an anti-inflammatory factor. The hypothesis that the strain secretes an active compound is supported by the finding that F. prausnitzii culture supernatant is effective against chemically-induced colitis in mice. Surprisingly, not only oral application of F. prausnitzii and its supernatant protected mice from colitis: the intraperitoneal injection resulted in reduced mortality in the mouse colitis model. Unfortunately, the nature of the secreted factor has not been identified in this study. Although these findings are very suggestive for a protective effect of F. prausnitzii, its exact role in gut inflammation remains to be elucidated. For instance, an increased abundance of F. prausnitzii at the mucosal surface of UC patients has also been reported. However, this study confirmed low numbers of this species in the intestine of CD patients but the authors considered this depletion secondary to host immune responses.11
The Role of Gram Negative Bacteria in Ileal IBD
There is strong evidence that Gram negative bacteria and most importantly E. coli strains with specific features play an important role in chronic ileitis. In a mouse model infected with Toxoplasma gondii mimicking at large the active phase of ileal IBD, Gram-negatives including E. coli and Bacteroides/Prevotella spp strongly increase.28 It was demonstrated in a later study taking advantage of gnotobiotic TLR2−/− and TLR4−/− mice that the recognition by the immune system of bacterial lipopolysaccharides (LPS) triggers ileitis in this model.29
Human studies suggest that specific E. coli strains with pro-inflammatory traits are highly prevalent in the inflamed intestine.30-33One of the best examples for the relevance of specific E. coli strains is the abundance of adherent-invasive E. coli (AIEC) in patients with ileal CD. This type of E. coli has extensively been characterized by the group of Darfeuille-Michaud. They isolated from the ileal mucosa of CD patients E. coli strains and recognized that approximately 80% to 85% of the strains were capable of adhering to human intestinal cells under in vitro conditions. Since 22% of the strains displayed α-hemolysin-dependent cytolytic effects, the authors concluded that a combination of adhesive properties and α-hemolysin production is a critical feature of CD-associated E. coli strains.34 Subsequently, the group demonstrated that one of the E. coli strains isolated from a chronic ileal lesion was able to invade cultured intestinal cells and proposed the existence of an E. coli pathovar referred to as adherent-invasive E. coli (AIEC).35 In a later study, a high prevalence of AIEC was demonstrated in CD patients. Many of the strains that have been isolated in this study from ileal specimens from CD patients were able under in vitro conditions to adhere to and to invade intestinal cells. In contrast, E. coli strains isolated from healthy controls and IBD patients with colitis, respectively, did not display adherent-invasive properties. In addition, intracellular survival and replication in cultured macrophages was observed. Based on the ability to adhere to and to invade epithelial cells by actin- and microtubule-dependent mechanisms, the absence of selected invasion-associated genes, and the ability to survive and to replicate in cultured macrophages, these CD-associated strains were considered AIEC. From a high prevalence of AIEC in early ileal lesions in CD patients after ileocolectomy the authors deduced that these strains are involved in the initiation of CD.36
In a series of subsequent experiments, the Darfeuille-Michaud group characterized the nature of AIEC-host interactions in more detail. They worked out that the genome of the AIEC prototype strain LF82 contains many known virulence factors including adhesion- and invasion-associated genes but also genes involved in iron acquisition, motility, capsule and LPS synthesis and serum resistance.37 The attachment of AIEC to host epithelial cells is mediated by interactions between type-1 pili and the carcinoembryogenic antigen-related adhesion molecule 6 (CECAM6) which is expressed at the apical surface of enterocytes. Interestingly, CECAM6 expression by ileal enterocytes is higher in CD than in the healthy situation. In contrast, increased CECAM6 expression is not observed in colonic tissue material from UC patients, indicating that impaired CECAM6 expression is a unique characteristic of ileal IBD. The importance of interactions between CECAM6 and type 1 pili was demonstrated by the use of AIEC mutants in in vitro adhesion assays. In contrast to the wild-type strain, LF82 type-1 pili-negative and FimH adhesin-negative mutants lost their capability to adhere to enterocytes obtained from CD patients. Adherence to enterocytes was not unique for this prototype AIEC strain since it was also demonstrated for other CD-associated isolates. However, the relevance for strong adherence properties of strain-specific type-1 pili was demonstrated when LF81 was compared with a laboratory K12 E. coli strain. K12 showed low adherence when its regular type-1 pili were overexpressed and this was also observed in LF82 mutants expressing the K12-type pili. It was possible to block by the addition of mannose the ability of LF82 to bind to epithelial cells and, therefore, the authors concluded that a high expression of mannosylated CECAM6 in ileal IBD favors the adherence of AIEC. Interestingly, CECAM6 expression by cultured intestinal cells was upregulated after treatment with IFNγ and tumor necrosis factor-α (TNFα). This observation indicates that inflammatory conditions in the gut support AIEC colonization via increased CECAM6 expression.38 Recently, a second mechanism of AIEC adherence to host cells has been identified by the Darfeuille-Michaud group: in both mice and men, AIEC are capable of targeting the M cells of Peyer’s patches.39 Using LF82 mutants deficient for the formation of long polar fimbriae, the authors demonstrated that the ability to adhere to Peyer’s patches and to translocate across M cell monolayers depends on the formation of these structures.
The second important trait of AIEC is the invasion of host epithelial cells. This process is mediated by interactions between the outer membrane protein A (OmpA) of AIEC and the chaperone GP96 which is considered a stress response protein of the endoplasmatic reticulum. This protein is highly upregulated in the chronically inflamed ileal mucosa and mainly located in the apical membrane of ileal epithelial cells. As concluded from in vitro experiments with LF82 and intestinal epithelial cells, AIEC target this protein to invade host cells: the invasion of LF82 can be reduced in GP96 expressing cells by the use of GP96-specific antibodies and gp96 siRNA. Subsequently, an ompA deletion mutant was tested for its ability to invade intestinal cells and this mutant displayed an approximately 60% lower invasion ability than the wild-type control strain.40
Taking advantage of a mouse embryonic fibroblast (MEF) in vitro system, host factors controlling the replication of AIEC, namely LF82, in host cells were identified.41 The authors hypothesized that impaired clearance of intracellular bacteria in CD patients promotes replication of AIEC in epithelial cells since the autophagy-associated genes ATG16L1 and IRGM are considered disease risk genes. They tested the ability of LF82 and of other commensal and pathogenic E. coli strains to replicate in wild-type MEF and in autophagy-deficient atg5−/− MEF and observed that exclusively the intracellular replication of LF82 was impaired by functional autophagy. The relevance of functional autophagy for intracellular LF82 replication in human epithelial cell lines was demonstrated when autophagy in these cells was induced or blocked. Rapamycin-induced autophagy resulted in reduced intracellular LF82 numbers in a dose-dependent manner. Similar results were obtained when autophagy was induced by amino acid starvation, LPS- and IFN-γ-treatment, respectively, of the epithelial cells. When autophagy-deficient cells were used in the experiments, LF82 was capable of increased intracellular replication. Strong evidence for the critical role of ATG16L1 and IRGM in AIEC-induced gut inflammation was provided by knock-down experiments. When either ATG16L1 or IRGM expression by intestinal cells was reduced by the use of siRNA, intracellular LF82 replication was increased in comparison to control cells. Very recently, similar effects of impaired autophagy on bacterial replication within macrophages have been reported. In these cells, increased AIEC replication associated with impaired NOD2, ATG16L1 and IRGM expression resulted in an elevated production of pro-inflammatory cytokines. Since induction of autophagy resulted in decreased AIEC replication in conjunction with a reduced pro-inflammatory response, the authors proposed that autophagy stimulation may be a promising strategy to treat CD.42
These studies on AIEC demonstrate that in a sub-population of patients CD shows characteristics of an infectious disease and that CD etiology in the respective patients can be characterized at the molecular level. However, it has to be taken into account that CD-associated AIEC have been detected in only approximately 20% to 50% of CD patients.32,36 Therefore, it is likely that AIEC are not implicated in disease development in a large proportion of CD patients. Possible candidate bacteria that may be involved in AIEC-independent CD include Bacteroides spp. Their special role can be deduced from the FISH-based investigation of mucosal biopsy specimens. In this study, adherent enterobacteria were only detected in three out of 20 CD patients and no clear indication for bacterial invasion was observed. The most striking observation in this study was that mucosa-attached biofilms dominated by B. fragilis were detected in 90% of the CD patients. Similar observations have been made in 60% of the UC patients and the authors considered a dense colonization by B. fragilis of the mucosal surface a prominent feature of IBD.43
Gram Negative Bacteria in Chronic Colitis
As mentioned above, a number of studies support the notion that E. coli with specific pro-inflammatory traits are critically involved in the onset and aggravation of ileal IBD. Less clear is the role of enterobacteria in chronic colitis. However, high numbers in mouse colitis models and in human subjects suffering from colitis indicate that E. coli is implicated in this type of disease. For instance, a shift toward a so-called pro-inflammatory intestinal microbiota composition was reported in mice with dextran-sodium sulfate (DSS)-induced colitis. The inflammation-associated changes were mainly characterized by an increase by four orders of magnitude of E. coli.44 A high abundance of fecal enterobacteria was also reported in patients with CD of the colon, irrespective of the disease state45 and numbers of cultured E. coli in colonic biopsy samples were 3–4 logs higher in both CD and UC patients than in healthy controls.46 In the latter study, E. coli strains isolated from IBD patients have been characterized in more detail. E. coli strains belonging to the virulence-associated phylogenetic groups B2 and D were more frequently isolated from IBD patients than from healthy controls. A typical attribute of the B2 and D isolates was a high prevalence of genes coding for serine protein autotransporters and adhesins.
The hypothesis that virulent E. coli strains are implicated in chronic colitis is supported by findings in patients with active left-sided colitis. In this study, B2 E. coli strains were isolated from 60% of the IBD patients but from only 11% of the healthy controls. In 86% of the strains isolated from the subjects with active colitis, at least one gene encoding extraintestinal pathogenic E. coli (ExPEc)-characteristic traits including adhesins, iron acquisition systems, protectins or autotransporters has been detected. The proportion of strains tested positive in patients with quiescence colitis and in healthy controls was only 13% and 11%, respectively. Based on multilocus sequence typing, the authors proposed a common ancestry and clonal nature of the strains with virulent properties. Therefore, the authors concluded that the increase of B2 E. coli in the inflamed colon is not a random event and that these E. coli strains actively participate in IBD.47
A strong proliferation of E. coli in conjunction with reduced microbiota diversity was also observed in IL-10−/− mice with chronic colitis. The most intriguing finding in this study was that the E. coli populations in the inflamed gut were exclusively represented by one single E. coli strain. This O7:H7:K1 strain was assigned to the virulence-associated phylogenetic group B2. Various virulence- or fitness-associated genes were detected in the strains’ genome including adhesins and invasins, protectins, iron acquisition systems and autotransporters. These traits enabled the strain to outnumber other E. coli competitors in the intestine of gnotobiotic mice. However, no evidence was found that the O7:H7:K1 caused or triggered gut inflammation in the genetically susceptible host. Therefore, it was concluded that the high number of fitness-associated genes provided a high adaptation capacity to the hostile environment in the inflamed gut and that the strain’s bloom was rather a consequence than the cause of disease.48
The notion that a bloom of E. coli is a secondary phenomenon in chronic colitis is supported by findings in the dnKO mouse model with deficiencies in IL-10- and transforming growth factor β-dependent signaling.49 Colitis in these mice was highly responsive toward antibiotics that decreased numbers of Bacteroidetes including members of the Bacteroidaceae, Porphyromonadaceae, and Prevotellaceae and of Firmicutes including Lachnospiraceae and Ruminococcaceae. Numbers of Enterococcaceae and Lactobacillaceae increased under these conditions. When antibiotic-treated dnKO mice were gavaged with fecal material from untreated dnKO mice, colitis evolved within 3 weeks after inoculation. In order to identify the bacteria responsible for this effect, different microbiota fractions were generated by culture-dependent methods. The application of a fraction enriched in Gram-positive anaerobes did not result in colitis. In contrast, a fraction that included obligate anaerobic Gram-negatives caused severe inflammation when applied to dnKO mice. Since it turned out that Bacteroides spp were important representatives of the Gram-negative fraction, five isolates from colitic mice were tested for their ability to induce colitis in antibiotic-treated dnKO mice. It turned out that B. vulgatus and B. thetaiotaomicron induced severe colitis whereas the other isolates caused milder forms of gut disease. Interestingly, a bloom of Bacteroides spp was not observed in the colitic animals. In contrast, Enterobacteriaceae contributed approximately 50% to the total cultivable bacteria in feces. Treatment with antibiotics alleviated colitis and eliminated the Enterobacteriaceae from the intestine of the experimental animals. However, when antibiotic-treated mice were gavaged with a prominent E. coli isolate from colitic mice, no gut inflammation evolved. Therefore, it was concluded that Bacteroides spp but not Enterobacteriaceae including E. coli are critically involved in the onset of colitis in the dnKO mouse model.
A more complex role of enterobacteria in the onset of colitis was concluded from studies in the TRUC mouse model. TRUC mice are simultaneously deficient for the transcription factor T-bet and the recombination activating gene 2 (Rag2) and spontaneously develop UC-like colitis. Since colitis severity in the TRUC mice was alleviated by the application of gentamycin and metronidazole but not by vancomycin, a special role of Gram-negatives in this model was deduced. Culture-dependent methods revealed that Klebsiella pneumoniae and Proteus mirabilis were present in TRUC mice but absent in healthy controls. However, there was no correlation between inflammation severity and intestinal numbers of these two species. The hypothesis that the presence of K. pneumoniae and P. mirabilis is a general feature of gut inflammation was rejected because they were not detected in mice with DSS-induced colitis. To explore whether the presence of these two species causes colitis in a genetically susceptible host, germ-free TRUC mice were inoculated with K. pneumoniae and P. mirabilis. However, although both strains successfully colonized the intestine of the di-associated mice, the animals did not develop colitis. The opposite was observed when K. pneumoniae and P. mirabilis were applied to Rag2−/− and wild-type mice with a complex microbiota. In these animals that do not develop gut inflammation under specific pathogen-free housing conditions, K. pneumoniae and P. mirabilis and a combination of both induced colitis. The nature of bacteria-bacteria interactions that are required for the pro-inflammatory properties of K. pneumoniae and P. mirabilis have not been addressed in this study. However, the authors proposed that the immune system may be shaped by intestinal microbiota in a way that facilitates host responses to K. pneumoniae and P. mirabilis. Alternatively, horizontal gene transfer and the acquisition by the enterobacterial species of pro-inflammatory traits from the commensal microbiota or vice versa may be involved in the pro-inflammatory effects of the two strains.50
A lower accessibility of intestinal cells in the colon than in the small intestine may be one explanation for the less prominent role of E. coli in the onset of colonic than ileal IBD. The epithelial surface in the intestine is covered by mucus which is produced by goblet cells. The mucus in the colon consists of two layers composed of highly glycosylated gel-forming MUC2 mucins. The stratified inner layer is firmly attached to the epithelium and forms a dense network. With increasing distance to the epithelium, the inner layer is converted by host and probably by bacterial proteases into the loose outer layer which in mice is twice as thick than the inner layer. One critical feature of the inner layer is that it is devoid of bacteria and, therefore, protects the epithelial cells from contact with gut microbes.51,52 A mucus double layer has also been described in the small intestine. However, the majority of small intestinal mucus is removable and the remaining inner layer is very thin and of discontinuous nature.53 It is therefore possible that gut bacteria can more easily interact with host cells in the small intestine than in the colon.
The protective function in gut inflammation of the colonic inner mucus layer has been demonstrated in DSS-treated MUC1- and MUC2-deficient mice, respectively. In wild-type mice, thickness of the layer decreased in the course of chemically-induced colitis and this decrease correlated with inflammation severity. When colitis was induced by DSS in MUC2−/− mice which lack a typical mucus layer, the animals develop more severe colitis in conjunction with bacterial translocation and sepsis. Interestingly, when DSS was applied to MUC1−/− mice which have a reduced thickness of the inner mucus layer, a less severe inflammation than in wild-type mice was observed. The latter finding was explained by a possible role of MUC1 in immune functions and a reduced ability of Muc1−/− mice to recruit T-cells to the inflamed mucosa.54
It is possible that the mucus layer protects the epithelium from contact with intestinal bacteria but allows the penetration of bacterial antigens. When these antigens are sensed by so-called pattern recognition receptors, immune responses are initiated. At the mucosal interface in the intestine, TLRs belong to the important sensors of highly conserved bacterial motifs. Examples are TLR2, TLR4 and TLR5 that recognize lipoteichoic acid, LPS and flagellin, respectively. The signal cascades that are initiated upon TLR activation result in the expression of pro-inflammatory cytokines and chemokines.55 The hypothesis that TLR ligands play an important role in IBD is supported by findings in the T. gondii mouse model of ileitis and in mice with DSS-induced colitis. In mice with acute ileitis, intestinal concentrations of TLR2, TLR4 and TLR5 ligands were approximately 370-fold, 3300-fold and 38-fold, respectively, higher than in healthy control animals. The increase in colitic mice was 4-fold for TLR2 ligands and 550-fold for TLR4 ligands. Flagellin concentrations were not significantly changed in the inflamed colon.56 These findings suggest that mainly the TLR4 ligand LPS is an important trigger of gut inflammation. Further evidence for the important role of TLR signaling in IBD comes from studies in TLR2 and TLR4 knockout mice. DSS-induced colitis in these mice is less severe than in wild-type mice and numbers of neutrophils and regulatory T-cells in the inflamed tissue are reduced. Based on this observation and since alleviated colitis was accompanied by a less pronounced shift toward a so-called pro-inflammatory microbiota, the authors concluded that bacteria-derived ligands exacerbate colitis through TLR2 and TLR4 signaling.44
In addition to TLR2- and TLR4-dependent signaling, the activation of TLR5 by E. coli flagellin has been implicated in the aggravation of colitis. DSS-treated mice orally challenged with 108 cells of the AIEC prototype strain LF82 displayed a substantially increased inflammation severity and weight loss in conjunction with reduced survival rates when compared with control animals that were treated with an avirulent K12 E. coli strain. When either ompA or fliC was deleted in LF82, the mutants lost their ability to aggravate inflammation. Since the mutants were not capable of flagella expression, the authors concluded a critical role of flagellin detection by the innate immune system in the pro-inflammatory effects of LF82. This notion was supported by the fact that the two flagellin receptors TLR5 and IPAF were strongly upregulated at the mRNA level in colonic specimens from mice colonized with the virulent LF82 but not with the avirulent mutants. This finding was accompanied by higher pro-inflammatory IL-1β and IL-6 mRNA levels in response to the LF82 wild-type.57 However, the exact role of TLR5-dependent signaling in colitis remains to be elucidated since it has been demonstrated that DSS-induced colitis is more severe in TLR5-deficient mice rectally treated with purified flagellin than in identically treated wild-type mice.58 A more protective role of TLR5-dependent host responses toward gut bacteria may also be concluded from the fact that TLR5−/− mice are prone to spontaneously developing transmural colitis. Interestingly, this phenotype was not observed in mice simultaneously deficient for TLR4 and TLR5 suggesting that TLR4 ligands trigger colitis in TLR5−/− mice.59
Bacterial Responses Toward the Environmental Conditions in the Inflamed Intestine
It has been mentioned above that the frequently observed increase of E. coli in the inflamed colon may result from the disease but that it does not necessarily contribute to its onset. The same may be true for other changes in intestinal microbiota composition including alterations in F. prausnitzii populations. Since gut bacteria most likely adapt to inflammatory conditions, the identification of inflammation-associated environmental factors that influence intestinal microbiota may help to develop new strategies for IBD therapy. However, knowledge of the environmental differences between the healthy and the inflamed gut with possible implication in microbiota changes is scarce.
An effect on intestinal microbiota of oral iron supplementation in IBD patients suffering from iron deficiency anemia can be deduced from findings in TNFΔARE/WT mice. Feeding these animals a diet containing 180 mg/kg iron sulfate which is in the normal range of standard rodent chow, resulted in the spontaneous development of severe CD-like ileitis. Interestingly, the mice remained healthy when oral iron was depleted and when iron was applied systemically. In this model, iron depletion was associated with reduced endoplasmatic reticulum stress responses and apoptotic processes in the ileal epithelium. In addition, the iron-free diet profoundly influenced cecal microbiota composition as determined by deep sequencing of bacterial 16S rRNA genes. The changes were characterized by an increased abundance of sequences assigned to the genera Bifidobacterium, Succinivibrio, Turicibacter and Clostridium. In contrast sequences representative for Desulfovibrio and Bacteroides were less frequently detected. The authors concluded that the presence of luminal iron influences intestinal microbiota composition and that iron-dependent microbiota alterations may be a factor that promotes stress-associated apoptosis of epithelial cells in the inflamed gut.60
One endogenous factor that may affect microbiota composition in the chronically inflamed gut is an impaired bile acid metabolism. Bile acid malabsorption61,62 and differences in bile acid excretion in conjunction with altered bacterial bile acid conversion63,64 have been reported in IBD patients. Since bile acids selectively inhibit the growth of intestinal anaerobes but not of aerobic gut bacteria,65 alterations in intestinal bile acid metabolism may be involved in inflammation-associated changes in gut microbiota. This assumption is supported by findings in the IL-10−/− mouse model of chronic colitis.66 In this model, differences in intestinal bile acid concentrations were indicative of a lower bacterial capacity than in healthy wild-type control mice to transform primary into secondary bile acids. Alterations in the intestinal bile acid profile were associated with reduced intestinal microbiota diversity in conjunction with an increased abundance of highly bile acid-resistant bacterial species including Robinsiella peroriensis, Clostridium inocuum, Enterococcus gallinarum and, most drastically, E. coli. In addition, the bacterial conversion of cholesterol to coprostanol was impaired in the colitic mice but the relevance of this observation for changes in intestinal microbiota composition was difficult to interpret.
It can be assumed that gut bacteria in the inflamed intestine have to deal with stress factors that derive from host immune responses. In fact, it has been demonstrated that active colitis in IBD patients is associated with elevated mucosal levels of nitric oxide, which correlate with disease severity.67 It can therefore be hypothesized that bacterial species that are well equipped to cope with increased oxidative stress have a selection advantage under chronic inflammation and that more sensitive bacteria are repressed. The notion that defense mechanisms against oxidative stress are important bacterial traits improving survival in the inflamed gut is supported by findings in E. coli. In IL-10−/− mice monoassociated with the commensal E. coli strain NC101, moderate cecal inflammation was observed 3 and 7 weeks after association, respectively. In contrast, wild-type control mice did not display any signs of disease. Microarray-based transcriptional profiling of E. coli cells harvest from the inflamed and healthy ceca of IL-10−/− and wild-type control mice, respectively, revealed that 64 genes were upregulated and 150 genes downregulated in E. coli from the inflamed gut. Many upregulated genes (27%) were identified to play a role in E. coli stress responses and genes with the highest expression differences including ibpA, ibpB and oxyS have been implicated in E. coli protection from oxidative damage. The authors hypothesized that these genes are critical for the survival of E. coli in the inflamed gut and constructed ibpAB deletion mutants. The NC101 wild-type and mutant strains were subsequently tested for their ability to colonize the intestine of IL-10−/− and healthy control mice. In contradiction to the original hypothesis, the mutant was able to reach higher intestinal cell numbers than the wild-type strain. In IL-10−/− mice, the higher cell numbers of the mutant strain correlated with increased E. coli antibodies in plasma and increased colon histopatology in conjunction with higher IL-12 and IL-23 production by colonic explants. In addition, NC101-stimulated mesenteric lymph node (MLN) cells from IL-10−/− mice monoassociated with the mutant strain produced more interferon IFNγ than cells obtained from mice monoassociated with the E. coli wild-type strain. When a complemented mutant strain capable of ibpAB expression was used in monoassociated IL-10−/− mice, bacterial cell numbers in the intestine, inflammation severity and IFNγ production by MLN cells were comparable to that in mice monoassociated with wild-type NC101. The authors concluded from their findings that ibpAB expression by E. coli NC101 in the intestine reduces bacterial survival and triggers host responses toward the strain. From co-colonization experiments using the ibpAB mutant and wild-type strains in IL-10−/− mice, the authors concluded that the mutant strain has a selection advantage and that gut inflammation in the IL-10−/− mouse better correlates with intestinal E. coli cell numbers than with the proportion of ibpAB expressing bacteria. However, in experiments addressing the earlier time points of inflammation development the role of elevated intestinal cell numbers was not clearly demonstrated. It was not possible in this study to clarify why ibpAB expression by E. coli NC101 is upregulated in mice with chronic gut inflammation. The authors proposed that a lack of bacteria-bacteria interactions in the gnotobiotic mouse model, differences between luminal and mucosal E. coli populations in gene expression, different ibpAB expression levels with different implications for NC101 fitness and possible effects of the ibpAB deletion on the expression of other E. coli genes may contribute to the unexpected findings. Taken together, this study clearly demonstrated that gut inflammation causes stress in E. coli NC101 and that the strain responds to this stress with the upregulation of defense genes.68
The notion that inflammation induces stress in gut bacteria has been supported by proteome analyses in two different E. coli strains harvested from colitic and healthy mice.69 In this study, germ-free mice were either associated with the colitogenic E. coli UNC or with E. coli Nissle 1917 which is marketed as a probiotic product and which has been effective in keeping IBD patients in remission. Bacterial cells were harvested from cecal contents of mice with DSS-induced gut inflammation and from untreated mice with a healthy gut. The DSS-treated mice did not differ in inflammation severity. Cell numbers of Nissle and UNC under the different conditions were equal in terms of biological relevance. The application of 2-dimensional difference gel electrophoresis for the detection of differentially expressed E. coli proteins revealed a higher expression by Nissle of 11 proteins in the inflamed than in the healthy gut. Twenty-four proteins were downregulated under inflamed conditions. UNC responded to gut inflammation with the upregualtion of 16 and the downregulation of 35 proteins. In both strains, induction in response to gut inflammation of stress-associated proteins and a repression of proteins involved in carbohydrate metabolism was observed. The lower expression of proteins of the central carbohydrate metabolism resulted in reduced production of E. coli fermentation products including formate, lactate, and succinate which may be indicative of energy deprivation in the inflamed gut. Since inflammation-driven induction of NfuA was observed in both E. coli strains, this Fe-S biogenesis protein involved in E. coli adaptation to oxidative stress and reduced iron availability was investigated in more detail. Destruction of FE-S clusters under in vitro conditions by the addition of an iron chelator resulted in the activation of the nfuA promoter indicating that Fe-S cluster repair is required in the inflamed gut. The relevance of NfuA for growth under oxidative stress was demonstrated in nfuA deletion mutants incubated in the presence of a superoxide generator: the mutants were not able to cope with these conditions and reached lower cell densities than the wild-type control strain. The so far uncharacterized protein YggE, which is implicated in the reduction of intracellular concentrations of reactive oxygen species, was 4-fold (healthy gut) to 8-fold (inflamed gut) higher expressed in Nissle than in UNC. Based on this observation, it was hypothesized that Nissle responds more efficiently to oxidative stress than UNC. In fact, a substantial induction of the yggE promoter in response to oxidative stress was observed for Nissle but not for UNC when reporter gene assays were performed. In line with this finding, the in vitro growth of UNC was more strongly repressed in the presence of oxidative stress than the growth of Nissle. Taken together, this study supports the assumption that intestinal inflammation is associated with increased oxidative stress and that bacteria that are able to cope with these conditions have a selection advantage.
The two studies in gnotobiotic mouse models provided insight into the response at the molecular level of gut bacteria to the hostile environmental conditions in the inflamed gut. However, reductionist animal models do not necessarily reflect the human situation. In addition, influences of other gut bacteria that are normally present in the gut on bacteria with pro-inflammatory traits and on the host cannot be addressed with such approaches. Extended gnotobiotic models mimicking the human intestinal microbiota may help to overcome these limitations. Such a model of a simplified human microbiota consisting of Anaerostipes caccae, Bacteroides thetaiotaomicron, Bifidobacterium longum, Blautia producta, Clostridium butyricum, Clostridium ramosum, Escherichia coli and Lactobacillus plantarum has been established in rats. The members of this bacterial community were selected according to their prevalence in humans, the metabolites formed during the fermentation of undigested feed material or host-derived substrates and the availability of the complete genome sequence. The bacteria of the simplified human microbiota fulfilled a number of microbiota-associated characteristics including degradation of mucins, conversion of bilirubin to urobilirubin, degradation of β-aspartylglycin and short-chain fatty acid formation. It has been demonstrated during the development of this model that the simplified human gut microbiota responses toward changes in dietary composition. However, the applicability of this model to study the role of bacteria in gut inflammation remains to be demonstrated.70
The application of high-throughput metagenomic methods for the exploration of the human intestinal microbiota in the inflamed gut is an additional approach to explore bacterial response at the molecular level toward gut inflammation. For instance, 25% fewer bacterial genes than in healthy subjects were detected in the intestine of IBD patients which may be indicative of a reduced diversity of intestinal microbiota functions.21 In a pilot study that demonstrated the feasibility of a so-called metagenomic systems biology approach, shifts in the gut microbiome of obese subjects and IBD patients were demonstrated. Many genes and functions were enriched in both study groups and the authors proposed well-conserved responses of gut microbiota toward diseases of different nature. The genes that were enriched in response to disease encode enzymes of the phosphotransferase system involved in carbohydrate uptake, the production of NO2 and in choline and p-cresol metabolism. Since the study focused on metabolic pathway analysis, possible responses of gut bacteria toward e.g., host immune reactions have not been considered in this study.71
Summary and Conclusion
It has been demonstrated that chronic gut inflammation is associated with alterations in intestinal microbiota composition. Although reduced microbiota diversity in conjunction with decreased abundance of Gram-positives and increased abundance of Gram-negatives is frequently reported in IBD patients, it is difficult to define a general disease-associated pattern of intestinal microbiota composition. The observation that specific E. coli strains are critically involved in the onset and perpetuation of ileitis in a subset of CD patients offers the opportunity to develop new therapeutic strategies to cure or alleviate inflammation in these patients. Additional future concepts of IBD treatment may include the propagation of bacteria with protective properties in the inflamed gut. One interesting example is F. prausnitzii which is reported to decrease under chronic gut inflammation. More importantly, there is strong evidence that this species produces a soluble factor with anti-inflammatory effects. However, further studies are needed to explore the exact nature of this factor and to test whether it has the potential to serve as a therapeutic agent. The application of high-throughput metagenomic techniques may help to identify more candidate factors or functions that possibly influence onset and perpetuation of IBD. In hypothesis-driven approaches taking advantage of simplified animal models, the role of such factors in disease development can subsequently be tested.
In conclusion, the extensive analysis of gut microbiota composition has put forward our understanding of the role of commensal gut bacteria in IBD. However, knowledge about the mechanisms that are involved in the bacterial contribution to chronic gut inflammation is scarce. The combination of holistic deep sequencing-based approaches for the identification of bacterial functions that are enriched or depleted in the inflamed gut with reductionist animal models of gut inflammation may be a promising way to identify new therapeutic targets for IBD treatment.
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/22156
References
- 1.Baumgart DC, Sandborn WJ. Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet. 2007;369:1641–57. doi: 10.1016/S0140-6736(07)60751-X. [DOI] [PubMed] [Google Scholar]
- 2.Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54, e42, quiz e30. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Sartor RB. Mechanisms of disease: pathogenesis of Crohn’s disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- 4.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogler G. Interaction between susceptibility and environment: examples from the digestive tract. Dig Dis. 2011;29:136–43. doi: 10.1159/000323876. [DOI] [PubMed] [Google Scholar]
- 6.Thompson AI, Lees CW. Genetics of ulcerative colitis. Inflamm Bowel Dis. 2011;17:831–48. doi: 10.1002/ibd.21375. [DOI] [PubMed] [Google Scholar]
- 7.Feller M, Huwiler K, Stephan R, Altpeter E, Shang A, Furrer H, et al. Mycobacterium avium subspecies paratuberculosis and Crohn’s disease: a systematic review and meta-analysis. Lancet Infect Dis. 2007;7:607–13. doi: 10.1016/S1473-3099(07)70211-6. [DOI] [PubMed] [Google Scholar]
- 8.Packey CD, Sartor RB. Commensal bacteria, traditional and opportunistic pathogens, dysbiosis and bacterial killing in inflammatory bowel diseases. Curr Opin Infect Dis. 2009;22:292–301. doi: 10.1097/QCO.0b013e32832a8a5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–31. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taurog JD, Richardson JA, Croft JT, Simmons WA, Zhou M, Fernández-Sueiro JL, et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J Exp Med. 1994;180:2359–64. doi: 10.1084/jem.180.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swidsinski A, Loening-Baucke V, Vaneechoutte M, Doerffel Y. Active Crohn’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm Bowel Dis. 2008;14:147–61. doi: 10.1002/ibd.20330. [DOI] [PubMed] [Google Scholar]
- 12.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zoetendal EG, von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans AD, de Vos WM. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68:3401–7. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Fölsch UR, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–93. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishikawa J, Kudo T, Sakata S, Benno Y, Sugiyama T. Diversity of mucosa-associated microbiota in active and inactive ulcerative colitis. Scand J Gastroenterol. 2009;44:180–6. doi: 10.1080/00365520802433231. [DOI] [PubMed] [Google Scholar]
- 16.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–5. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank DN, Robertson CE, Hamm CM, Kpadeh Z, Zhang T, Chen H, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:179–84. doi: 10.1002/ibd.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andoh A, Sakata S, Koizumi Y, Mitsuyama K, Fujiyama Y, Benno Y. Terminal restriction fragment length polymorphism analysis of the diversity of fecal microbiota in patients with ulcerative colitis. Inflamm Bowel Dis. 2007;13:955–62. doi: 10.1002/ibd.20151. [DOI] [PubMed] [Google Scholar]
- 19.Andoh A, Kuzuoka H, Tsujikawa T, Nakamura S, Hirai F, Suzuki Y, et al. Multicenter analysis of fecal microbiota profiles in Japanese patients with Crohn’s disease. J Gastroenterol. 2012 doi: 10.1007/s00535-012-0605-0. [DOI] [PubMed] [Google Scholar]
- 20.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–11. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–54, e1. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 23.Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One. 2012;7:e30126. doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker AW, Sanderson JD, Churcher C, Parkes GC, Hudspith BN, Rayment N, et al. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011;11:7. doi: 10.1186/1471-2180-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, et al. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011;60:631–7. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 26.Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–9. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 27.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–6. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heimesaat MM, Bereswill S, Fischer A, Fuchs D, Struck D, Niebergall J, et al. Gram-negative bacteria aggravate murine small intestinal Th1-type immunopathology following oral infection with Toxoplasma gondii. J Immunol. 2006;177:8785–95. doi: 10.4049/jimmunol.177.12.8785. [DOI] [PubMed] [Google Scholar]
- 29.Heimesaat MM, Fischer A, Jahn HK, Niebergall J, Freudenberg M, Blaut M, et al. Exacerbation of murine ileitis by Toll-like receptor 4 mediated sensing of lipopolysaccharide from commensal Escherichia coli. Gut. 2007;56:941–8. doi: 10.1136/gut.2006.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 2007;1:403–18. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 31.Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, et al. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology. 2004;127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 32.Martinez-Medina M, Aldeguer X, Lopez-Siles M, González-Huix F, López-Oliu C, Dahbi G, et al. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn’s disease. Inflamm Bowel Dis. 2009;15:872–82. doi: 10.1002/ibd.20860. [DOI] [PubMed] [Google Scholar]
- 33.Sasaki M, Sitaraman SV, Babbin BA, Gerner-Smidt P, Ribot EM, Garrett N, et al. Invasive Escherichia coli are a feature of Crohn’s disease. Lab Invest. 2007;87:1042–54. doi: 10.1038/labinvest.3700661. [DOI] [PubMed] [Google Scholar]
- 34.Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology. 1998;115:1405–13. doi: 10.1016/S0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- 35.Boudeau J, Glasser AL, Masseret E, Joly B, Darfeuille-Michaud A. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn’s disease. Infect Immun. 1999;67:4499–509. doi: 10.1128/iai.67.9.4499-4509.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412–21. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 37.Miquel S, Peyretaillade E, Claret L, de Vallée A, Dossat C, Vacherie B, et al. Complete genome sequence of Crohn’s disease-associated adherent-invasive E. coli strain LF82. PLoS One. 2010;5:5. doi: 10.1371/journal.pone.0012714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barnich N, Carvalho FA, Glasser AL, Darcha C, Jantscheff P, Allez M, et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J Clin Invest. 2007;117:1566–74. doi: 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chassaing B, Rolhion N, de Vallée A, Salim SY, Prorok-Hamon M, Neut C, et al. Crohn disease--associated adherent-invasive E. coli bacteria target mouse and human Peyer’s patches via long polar fimbriae. J Clin Invest. 2011;121:966–75. doi: 10.1172/JCI44632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rolhion N, Barnich N, Bringer MA, Glasser AL, Ranc J, Hébuterne X, et al. Abnormally expressed ER stress response chaperone Gp96 in CD favours adherent-invasive Escherichia coli invasion. Gut. 2010;59:1355–62. doi: 10.1136/gut.2010.207456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lapaquette P, Glasser AL, Huett A, Xavier RJ, Darfeuille-Michaud A. Crohn’s disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell Microbiol. 2010;12:99–113. doi: 10.1111/j.1462-5822.2009.01381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lapaquette P, Bringer MA, Darfeuille-Michaud A. Defects in autophagy favour adherent-invasive Escherichia coli persistence within macrophages leading to increased pro-inflammatory response. Cell Microbiol. 2012;14:791–807. doi: 10.1111/j.1462-5822.2012.01768.x. [DOI] [PubMed] [Google Scholar]
- 43.Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43:3380–9. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heimesaat MM, Fischer A, Siegmund B, Kupz A, Niebergall J, Fuchs D, et al. Shift towards pro-inflammatory intestinal bacteria aggravates acute murine colitis via Toll-like receptors 2 and 4. PLoS One. 2007;2:e662. doi: 10.1371/journal.pone.0000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, et al. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut. 2003;52:237–42. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kotlowski R, Bernstein CN, Sepehri S, Krause DO. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut. 2007;56:669–75. doi: 10.1136/gut.2006.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petersen AM, Nielsen EM, Litrup E, Brynskov J, Mirsepasi H, Krogfelt KA. A phylogenetic group of Escherichia coli associated with active left-sided inflammatory bowel disease. BMC Microbiol. 2009;9:171. doi: 10.1186/1471-2180-9-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wohlgemuth S, Haller D, Blaut M, Loh G. Reduced microbial diversity and high numbers of one single Escherichia coli strain in the intestine of colitic mice. Environ Microbiol. 2009;11:1562–71. doi: 10.1111/j.1462-2920.2009.01883.x. [DOI] [PubMed] [Google Scholar]
- 49.Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, et al. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4659–65. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–9. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Atuma C, Strugala V, Allen A, Holm L. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am J Physiol Gastrointest Liver Physiol. 2001;280:G922–9. doi: 10.1152/ajpgi.2001.280.5.G922. [DOI] [PubMed] [Google Scholar]
- 54.Petersson J, Schreiber O, Hansson GC, Gendler SJ, Velcich A, Lundberg JO, et al. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am J Physiol Gastrointest Liver Physiol. 2011;300:G327–33. doi: 10.1152/ajpgi.00422.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wells JM, Rossi O, Meijerink M, van Baarlen P. Epithelial crosstalk at the microbiota-mucosal interface. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4607–14. doi: 10.1073/pnas.1000092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Erridge C, Duncan SH, Bereswill S, Heimesaat MM. The induction of colitis and ileitis in mice is associated with marked increases in intestinal concentrations of stimulants of TLRs 2, 4, and 5. PLoS One. 2010;5:e9125. doi: 10.1371/journal.pone.0009125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carvalho FA, Barnich N, Sauvanet P, Darcha C, Gelot A, Darfeuille-Michaud A. Crohn’s disease-associated Escherichia coli LF82 aggravates colitis in injured mouse colon via signaling by flagellin. Inflamm Bowel Dis. 2008;14:1051–60. doi: 10.1002/ibd.20423. [DOI] [PubMed] [Google Scholar]
- 58.Ivison SM, Himmel ME, Hardenberg G, Wark PA, Kifayet A, Levings MK, et al. TLR5 is not required for flagellin-mediated exacerbation of DSS colitis. Inflamm Bowel Dis. 2010;16:401–9. doi: 10.1002/ibd.21097. [DOI] [PubMed] [Google Scholar]
- 59.Vijay-Kumar M, Sanders CJ, Taylor RT, Kumar A, Aitken JD, Sitaraman SV, et al. Deletion of TLR5 results in spontaneous colitis in mice. J Clin Invest. 2007;117:3909–21. doi: 10.1172/JCI33084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Werner T, Wagner SJ, Martínez I, Walter J, Chang JS, Clavel T, et al. Depletion of luminal iron alters the gut microbiota and prevents Crohn’s disease-like ileitis. Gut. 2011;60:325–33. doi: 10.1136/gut.2010.216929. [DOI] [PubMed] [Google Scholar]
- 61.Heuman R, Sjödahl R, Tobiasson P, Tagesson C. Decreased absorption of ingested unconjugated chenodeoxycholic acid in patients with Crohn’s disease. Scand J Gastroenterol. 1983;18:23–6. doi: 10.3109/00365528309181553. [DOI] [PubMed] [Google Scholar]
- 62.Nyhlin H, Merrick MV, Eastwood MA. Bile acid malabsorption in Crohn’s disease and indications for its assessment using SeHCAT. Gut. 1994;35:90–3. doi: 10.1136/gut.35.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ejderhamn J, Rafter JJ, Strandvik B. Faecal bile acid excretion in children with inflammatory bowel disease. Gut. 1991;32:1346–51. doi: 10.1136/gut.32.11.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kruis W, Kalek HD, Stellaard F, Paumgartner G. Altered fecal bile acid pattern in patients with inflammatory bowel disease. Digestion. 1986;35:189–98. doi: 10.1159/000199367. [DOI] [PubMed] [Google Scholar]
- 65.Binder HJ, Filburn B, Floch M. Bile acid inhibition of intestinal anaerobic organisms. Am J Clin Nutr. 1975;28:119–25. doi: 10.1093/ajcn/28.2.119. [DOI] [PubMed] [Google Scholar]
- 66.Wohlgemuth S, Keller S, Kertscher R, Stadion M, Haller D, Kisling S, et al. Intestinal steroid profiles and microbiota composition in colitic mice. Gut Microbes. 2011;2:159–66. doi: 10.4161/gmic.2.3.16104. [DOI] [PubMed] [Google Scholar]
- 67.Keshavarzian A, Banan A, Farhadi A, Komanduri S, Mutlu E, Zhang Y, et al. Increases in free radicals and cytoskeletal protein oxidation and nitration in the colon of patients with inflammatory bowel disease. Gut. 2003;52:720–8. doi: 10.1136/gut.52.5.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patwa LG, Fan TJ, Tchaptchet S, Liu Y, Lussier YA, Sartor RB, et al. Chronic intestinal inflammation induces stress-response genes in commensal Escherichia coli. Gastroenterology. 2011;141:1842–51, e1-10. doi: 10.1053/j.gastro.2011.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schumann S, Alpert C, Engst W, Loh G, Blaut M. Dextran sodium sulfate-induced inflammation alters the expression of proteins by intestinal Escherichia coli strains in a gnotobiotic mouse model. Appl Environ Microbiol. 2012;78:1513–22. doi: 10.1128/AEM.07340-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Becker N, Kunath J, Loh G, Blaut M. Human intestinal microbiota: characterization of a simplified and stable gnotobiotic rat model. Gut Microbes. 2011;2:25–33. doi: 10.4161/gmic.2.1.14651. [DOI] [PubMed] [Google Scholar]
- 71.Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci U S A. 2012;109:594–9. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]