Abstract
During important cellular processes such as centrosome and spindle positioning, dynein at the cortex interacts with dynamic microtubules in an apparent “end-on” fashion. It is well-established that dynein can generate forces by moving laterally along the microtubule lattice, but much less is known about dynein’s interaction with dynamic microtubule ends. In this paper, we review recent in vitro experiments that show that dynein, attached to an artificial cortex, is able to capture microtubule ends, regulate microtubule dynamics and mediate the generation of pulling forces on shrinking microtubules. We further review existing ideas on the involvement of dynein-mediated cortical pulling forces in the positioning of microtubule organizing centers such as centrosomes. Recent in vitro experiments have demonstrated that cortical pulling forces in combination with pushing forces can lead to reliable centering of microtubule asters in quasi two-dimensional microfabricated chambers. In these experiments, pushing leads to slipping of microtubule ends along the chamber boundaries, resulting in an anisotropic distribution of cortical microtubule contacts that favors centering, once pulling force generators become engaged. This effect is predicted to be strongly geometry-dependent, and we therefore finally discuss ongoing efforts to repeat these experiments in three-dimensional, spherical and deformable geometries.
Keywords: microtubules, centrosome, dynein, positioning, centering, aster, pulling, pushing, slipping, microfabricated chambers, emulsion droplets, GUVs, cytoskeleton, molecular motors
Eukaryotic cells can take various shapes and sizes, ranging from a 10 μm long, rod shape for a fission yeast cell to a millimeter-sized rounded cell for a Xenopus fertilized egg. They however all share the need to position their organelles reliably, both in space and time, for example, to establish the future cell division site. This may be at the center of the cell for nuclear positioning during interphase in Schizosaccharomyces pombe cells,1 or in a de-centered location, such as during nuclear movement in small budding yeast cells2 or spindle positioning in Caenorrhabditis elegans embryos.3
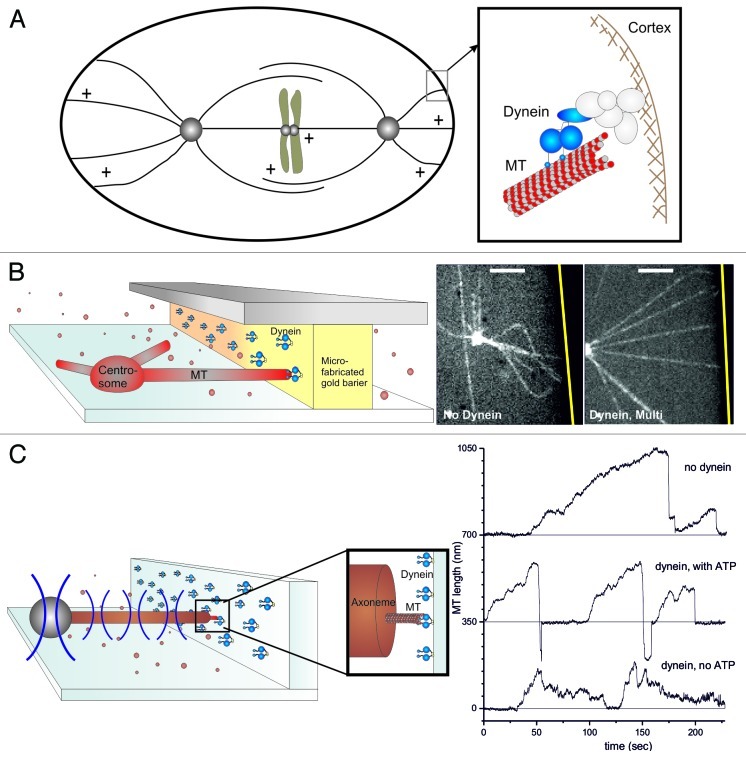
It is well-established that nuclear and spindle positioning processes depend on the presence of a dynamic microtubule (MT) cytoskeleton. MTs are dynamic protein polymers that undergo phases of growth and shrinkage, separated by so-called catastrophes and rescue events, an overall process named dynamic instability.4 To spatially organize the cellular interior, dynamic MTs often (but not always5-8) interact with the cell cortex. These cortical MT contacts contribute to positioning processes, but the exact mechanism appears to differ between cells of different sizes and shapes1,2,5 and remains, in general, not well-understood. Depending on the situation, cortical MTs can exert either pushing or pulling forces. Pushing forces arise from MTs growing against the cortex and from elastic restoring forces,9 while pulling forces are usually assumed to be mediated by dynein motor proteins attached to the cell cortex.10-16 Dynein is a minus-end directed processive motor17,18 that can pull on a MT organizing center (MTOC) through “lateral” cortical contacts with the MT lattice, as seen in dividing Saccharomyces cerevisiae cells.11 In addition, cortical dynein can interact directly with a dynamic MT end, in a so called “end-on” configuration, for example during the asymmetric positioning of the mitotic spindle in C. elegans (Fig. 1A).13 In this situation, the generation of pulling forces may (in part) be due to the shrinkage of MTs themselves.19
Figure 1. Dynein interacting with MT ends can generate pulling forces. (A) Schematic representation of the end-on interaction between cortical dynein and dynamic MTs during spindle positioning in C. elegans embryos. (B–C) In vitro reconstructions of the dynein-MT end-on interaction. (B) Barrier experiment: Dynein molecules are attached to a microfabricated gold barrier; MTs are growing from a purified centrosome, as described in detail previously.20 (Left) Schematic view of the experiment. (Right) Spinning disk confocal fluorescence microscopy images without or with dynein at the barrier. The gold barrier position is marked by a yellow line. Scale bars: 5 μm. (C) Optical tweezers experiment. Dynamic MTs are growing from axonemes attached to a trapped bead, and interact with dynein coated barriers, as described previously.20 Left: Schematic view of the experiment. Right: Growth and shrinkage of MTs interacting with an uncoated barrier (upper trace) or a dynein-coated barrier in presence (middle) or absence (lower) of ATP.20
In this paper, we focus on recent in vitro experiments that shed light on the question of how dynein interacts with a dynamic MT end to regulate its dynamics and to generate pulling forces.20 We discuss recent experimental and theoretical work that shows how these pulling forces may act in concert with MT pushing forces to reliably position MTOCs in (quasi) two-dimensional confined spaces. We further discuss ongoing in vitro efforts, where MTOCs are positioned in three-dimensional systems with flexible boundaries, as is relevant for example, for motile mammalian cells.
End-On Interaction Between Dynamic MT Ends and Dynein
Dynein has for a long time been referred to as the molecular motor implicated in the generation of pulling forces at cortical MT contacts. However, little is still known about the details of this interaction in the “end-on” configuration. How are forces generated at dynamic MT ends? Is dynein at the cortex directly involved in the regulation of MT dynamics and length? To address these questions, we recently reproduced in vitro the end-on cortical interaction between dynein and dynamic MTs. In vivo, dynein’s attachment to the cortex is highly regulated and can differ from one organism to another.12,21-23 This cortical attachment can be mimicked in vitro by directly attaching dynein to barriers20 (Fig. 1). We first specifically attached dynein molecules to gold-coated barriers and let them interact with dynamic MTs growing from centrosomes attached to the bottom of a glass surface (Fig. 1B). When dynamic MTs encountered the dynein-coated barriers, their ends were captured; they straightened; and their shrinkage was prevented, resulting in stabilized MTs of a defined length: the length between the organizing center and the dynein-coated barrier (Fig. 1B, right panel). This result was confirmed by another recent in vitro study, where dynamic MTs interacted with dynein-coated beads.24 In these experiments, dynein also proved to be able to tether and stabilize dynamic MTs. Note, however, that here the interaction between dynein and MT ends was not restricted to be end-on, possibly resulting in a mixed observation of end-on and lateral effects.
These first experiments then prompted the question of how dynein molecules prevent MT shrinkage. They could either directly inhibit catastrophes or, alternatively, stall MT shrinkage. To answer this question, we coated in addition to the barrier, the bottom of the coverslip with dynein, which resulted in MTs that were pulled loose from the centrosome and started gliding. When these gliding MTs interacted with the dynein-coated barriers, they rapidly underwent a catastrophe followed by MT shrinkage.20 We found that the catastrophe frequency was not reduced, but rather enhanced by end-on dynein contacts at the barrier. If dynein molecules were solely responsible for this catastrophe frequency increase, one might expect that dynein would enhance the catastrophe frequency also in a lateral gliding configuration, possibly by having a direct depolymerization activity as reported for the depolymerizing motor MCAK.25 We found, however, that dynein is not able to trigger catastrophe in a gliding configuration, nor that it is able to directly depolymerize GMPCPP-stabilized MTs.20 We thus propose that the catastrophe frequency enhancement is due to the presence of both dynein and the barrier. In this scenario, dynein provides a dynamic link that holds or pulls the MT end against the barrier, which hinders growth of the MT, resulting in a catastrophe.26 We also noted that both dynein gliding and MT shrinkage were slowed down by MT-dynein interactions at the barrier. So why do MTs not shrink at their normal velocity when dynein keeps their ends linked to the barrier? It is possible that dynein’s presence at the tip of the MT mechanically prevents MT protofilaments from curling outwards, slowing down or even stalling MT shrinkage, similar to what has been proposed for the Dam 1 complex.27 In an end-on configuration it is possible that two or more dynein molecules interact with a single MT end, enhancing this effect. One can then also understand why dynein velocity is reduced. Without shrinkage, MTs cannot be pulled further toward the barrier, preventing dynein to move toward the MT minus end. The maximum speed of dynein in this configuration is thus limited by the MT shrinkage speed.
Next, we decided to directly measure the pulling force generated by the end-on interaction between dynein and a shrinking MT, by constructing a second in vitro experiment involving an optical trap (Fig. 1C, left). We measured pulling forces up to a few pN (Fig. 1C, right)20 and observed again that dynein was able to trigger catastrophes and slow down subsequent MT shrinkage. Dynein in an end-on interaction with MT ends thus proved to be able to both regulate MT dynamics and mediate the generation of pulling forces. Whether the forces we measured were due to the intrinsic capability of shrinking MTs to generate force,19,28,29 or due to dynein’s power stroke, could not be determined from these experiments. We could, however, show that both the presence of ATP and the shrinkage of MTs were necessary for the generation of pulling forces, leaving open the possibility that both modes of force generation contribute. Another interesting, remaining-open question is how many dynein molecules are necessary for the observed effects. The stalling of MT shrinkage that we observed in our first experiments was most often seen at high densities of dynein at the barrier. This suggests that the ability of dynein to tether MT ends, stabilize them against shrinkage and mediate the generation of pulling forces might be more efficient when multiple dynein molecules are engaged.
The Role of Cortical MT-Dynein Interactions in Positioning Processes
Having established that the end-on interaction between dynein and dynamic MTs can generate pulling forces, we can now ask: what are the relative roles of pushing and pulling forces in positioning strategies for cells of different size and shape? In the literature, different mechanisms by which MTOCs are positioned in cells have been proposed. For example, it is known that simple pushing forces can, under specific conditions, drive centering. Consider a simple situation where a dynamic MT aster is displaced from the center of the cell: due to their dynamic properties, more MTs tend to contact the boundary of the cell that is nearby than the one that is far away, which leads to a net centering force when pushing forces are exerted.30 This effect is expected to be enhanced by the length-dependent critical buckling force of MTs under compression, which limits the force that can be exerted by long MTs.1,9,31 In vitro experiments have shown that MTOCs can indeed center by pushing against the boundaries of a microfabricated square chamber,32 provided that MTs are sufficiently dynamic.33 This mechanism is likely relevant for interphase fission yeast cells, where it is well established that nuclear positioning is the result of pushing forces generated by growing MTs on the cell ends.1 This pushing mechanism however, is not likely to be efficient for larger cells, because of the same length dependence of the buckling force.34 Long MTs in large cells will rather buckle than transmit a significant pushing force to the MTOC.
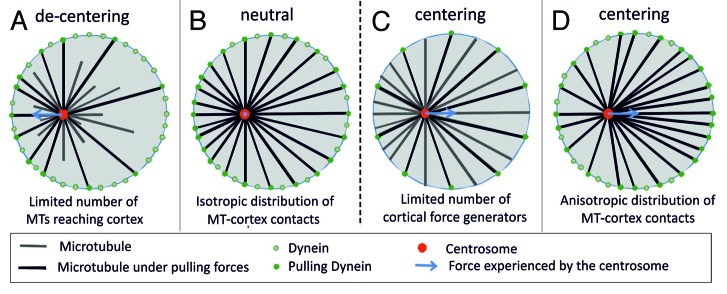
As an alternative, pulling forces have been proposed as a driving mechanism for centering.5 In many systems, these pulling forces arise from dynein at the cortex,3,10,12,35 although there are also cases where dynein seems distributed along the length of the MTs throughout the cytoplasm.5,6,36-38 Focusing here on the situation where pulling forces are only generated at the cortex, one can ask how pulling forces affect positioning processes. It has been argued that the abundant presence of pulling force generators at the cortex might lead to a net de-centering force.5,28,36 The argument here is the same that was used to explain the centering by pushing forces: due to their dynamics more MTs tend to contact a nearby cell boundary than a distant boundary, leading to an excess of force toward the nearby boundary, away from the center (Fig. 2A). At best, all MTs would reach the boundary and the net centering force would be zero (Fig. 2B). To resolve this issue, at least for the specific case of spindle positioning in C. elegans embryo, it was proposed that only a limited number of cortical force generators are available.35,39 In the situation that these interaction sites are always all occupied, which is expected in the presence of an excess number of MTs, one predicts a net centering force due to pulling (Fig. 2C). However, quantitative experiments aimed at testing these or other scenarios are inherently difficult to perform in the complex environment of cells. Therefore we resorted again to in vitro experiments to reveal the inherent capabilities of dynamic MT asters to position themselves in various confined geometries.
Figure 2. Scenario’s for pulling-based centering of MTOCs. (A) Dynamic MTs lead to a length distribution of MTs that favors contacts with nearby boundaries. This leads to a net pulling force away from the center when all cortical contacts generate a pulling force (decentering). (B) When all MTs reach the boundaries, the net pulling force is zero (neutral) independent of the position of the MTOC in the confining space. (C) If only a limited number of cortical contacts generate a pulling force, the net pulling force is directed toward the center (centering). (D) When slipping of MTs along the boundaries of the confining space leads to an anisotropic distribution of MTs, the net pulling force is directed toward the center even when all cortical contacts generate a pulling force (centering).
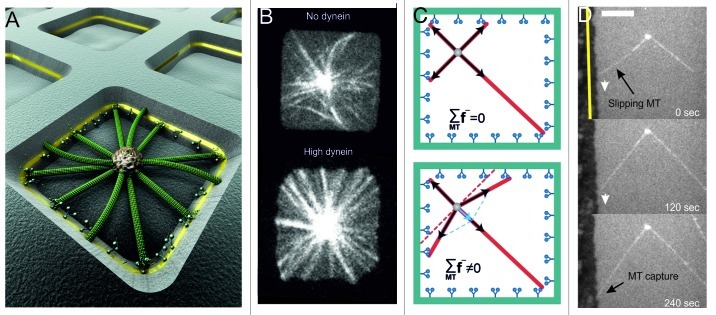
As discussed above, cortical dynein in an end-on configuration is able to both (1) regulate MT dynamics, thereby tuning the length of MTs between the MTOC and the barrier, and (2) mediate pulling on shrinking MTs, thereby pulling on MTOCs. With this in mind we confined dynamic MT asters grown from centrosomes to square microfabricated chambers with dynein-coated sidewalls (Fig. 3A).20 Dynein numbers could be modified by changing the thickness of a gold layer incorporated in the chamber walls. In this configuration, both pushing and pulling forces could act during positioning: pushing forces coming from growing MTs that were not yet captured by dynein molecules, pulling forces coming from captured MTs. For short MTs, i.e., with lengths shorter than the half-width of the chamber d, we found that centrosomes always performed a diffusion-like movement independent of whether dynein was present at the sidewalls. This result is understandable, since at this length, MTs were rarely interacting with the sidewalls of the chambers. For intermediate MT lengths, i.e., with lengths approximately equal to d, dynein caused a destabilizing effect: about 50% of the centrosomes were centered when no dynein was present vs. about 20% at high dynein numbers. In the last case, MTOCs were constantly moving, thereby exploring the whole chamber. Surprisingly, we found that for long MTs, i.e with lengths longer than d, the presence of dynein led to the reliable centering of MT asters, reaching 95% of success for high dynein numbers (Fig. 3B). In these cases, the centrosomes did not move, and all MTs appeared straight and in contact with the chamber walls. In the absence of dynein at the walls, MTs were often observed to buckle, and centering was overall less successful, with only about 40% of the centrosomes being centered. So how did cortical dynein lead to such reliable centering, assuming that we were not saturating a limited number of dynein interaction sites?35 Let’s imagine a situation in which all MTs, grown isotropically from the centrosome, have reached the barrier and are subject to an identical pulling force. As argued above (Fig. 2B), the net pulling force on the centrosome should then be zero (Fig. 3C, top). How then can the centrosome be pulled toward the center? The answer comes from the observation of time-lapse images of MTs growing toward dynein-coated barriers. Pushing MTs can be seen slipping along the barrier before being captured by dynein (Fig. 3D). If we account for this slipping of pushing MTs against the sidewalls of chambers, the distribution of MTs becomes anisotropic, leading to a net centering pulling force (Figs. 2D and 3C, bottom).
Figure 3. Dynein-mediated pulling and MT slipping lead to centrosome centering in a square chamber. (A) Artistic view of the experiment. Microtubules grow from a centrosome in a microfabricated chamber as described previously.20 Dynein molecules are attached to a gold layer in the walls of the chambers. (B) Spinning disk confocal fluorescence images of MTs grown from centrosome in square chambers (side lengths: 15 μm), in absence (upper), or presence (lower) of dynein at the walls.20 (C) Cartoon showing the net pulling force without (upper) and with (lower) MT slipping in a square geometry. (D) Evidence for MT slipping in vitro. A MT grows against a dynein coated barrier, slips and is then captured by dynein at the barrier. The gold barrier is indicated by the yellow line. Scale bar: 10 μm.
Based on this observation, we developed a mechanical model20 taking into account MT dynamics, with growing MTs exerting pushing forces and shrinking MTs exerting pulling forces. This model confirmed the importance of MT slipping for a centering effect on MT asters confined in semi, two-dimensional square chambers. As MT slipping leads to different MT reorganizations depending on the shape of the confining geometry, this dynein-mediated positioning capability is dependent on the shape of the confinement.40 The model predicts that in a circular shape, MTOCs are in fact more efficiently centered than in a square shape, mainly because of more extensive slipping of MTs in the direction that will center the aster.40 Interestingly, the model also predicts that combined pushing/pulling does not work in a quasi, one-dimensional geometry, corresponding to, for example, the strongly elongated shape of fission yeast.40 In this case, MT slipping does not cause any reorientation of the MTs, and the original arguments for why pulling should lead to de-centering remain valid. Possibly, this provides an explanation for why in fission yeast cells, nuclear positioning is achieved by MT pushing forces and not by pulling forces.1 In the case of a stadium shape, which is relevant for mitotic spindle positioning in cells such as C. elegans embryos, it can be shown that positioning by pulling forces is more precise along the short axis than along the long axis of the cell.40 This may mean that any additional mechanism employed to achieve slight asymmetric positioning in this system does not need to overcome a strong centering force along the longitudinal direction of the embryo.
Discussion
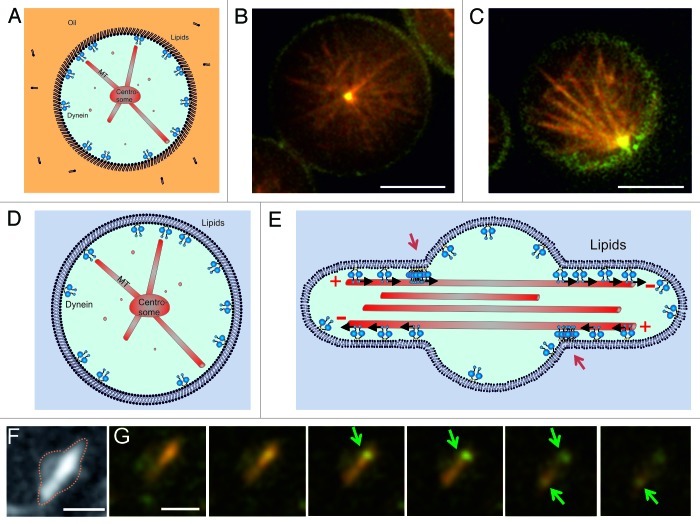
In vitro experiments in microfabricated chambers have provided us a with a useful testing ground for studying the intrinsic capabilities of dynamic force-generating MT asters to position themselves in confined spaces of different sizes and shapes. They have revealed a new geometry-dependent mechanism by which cortical pulling forces may contribute to efficient centering, which is likely to be relevant for life cell situations. Aside from trying to find ways to test the relevance of this mechanism inside living cells, there are also further steps that can be taken to bring the in vitro experiments closer to the cellular reality. For example, we have started experiments in which we confine MT asters to spherical emulsion droplets with dynein at the boundaries (Fig. 4A). This system better represents the three-dimensional nature of living cells and at the same time allows for potentially relevant mobility of dynein at the “cortex.” In a next step, we can go from droplets to giant unilamellar vesicles (GUVs), allowing for deformation of the confining boundary, and thus, vesicle shape, in response to forces generated at the cortex (Fig. 4D). This may be relevant for example for motile cells, which constantly change their shape in response to cytoskeletal reorganizations. The interplay between MT organization, cell shape and cortical force generation may lead to unexpected effects on both the nature of the interaction between MTs and cortical dynein (lateral vs. end-on) and the resulting positioning of MT asters. In droplets, preliminary results suggest that dynein is again able to both stabilize and destabilize the aster’s central position. Further analysis will have to reveal whether this again depends on the length of the MTs compared with the size of the droplet (Fig. 4B and C). Note that dynein is now linked to a phospholipid at the surface of the droplet, which in principle is able to diffuse freely along this surface. Preliminary results with free MTs confined to deformable vesicles show that GUVs with membrane-bound dynein and freely growing MTs exhibit a Ф-like shape (similar to what has been reported before for MTs confined in GUVs,41-44 with dynein concentrated at the entrance of the protrusions (Fig. 4F and G). This indicates that in these experiments dynein is both active, membrane-bound and interacting with the MTs, as explained in the cartoon in Figure 4E. Note that in this situation, the Ф shape deformation of the liposome forces dynein to interact mainly laterally with MTs. This effect is likely to change when MTs are grown from centrosomes.
Figure 4. Dynein-mediated centrosome positioning in emulsion droplets and liposomes. (A–C) Centrosome positioning in emulsion droplets. (A) Cartoon of the experiment. Dynein molecules are linked to phospholipids at the surface of the droplet. (B–C) Preliminary experiments show that dynein molecules attached to phospholipids can either center (B) or decenter (C) asters. MTs (red) growing from a purified centrosome interact with dynein (green) at the edge of the droplets. Shown is a single Z-plane of a spinning disk confocal fluorescence stack. Scale bars: 10 µm. (D) Centrosome positioning in GUVs. Cartoon of the desired experiment. (E) Cartoon explaining the accumulation of dynein at the entrance of the protrusions created by free MTs. Red arrows point to the accumulations. (F–G) Free taxol-stabilized MTs grown in GUVs, with membrane-bound dynein molecules. Scale bar: 3 µm (F) Z-projection of fluorescent MTs. (G) Individual Z-planes of the GUV seen in (F). Shown is a superposition of the MT (red) and dynein (green) signals. Arrows indicate the positions of dynein accumulation at the entrance of the protrusions. Z spacing = 0.3 μm. Scale bar: 3 µm.
Conclusion
In summary, we have shown that cortical dynein can tether MT plus ends, control MT length, regulate their dynamics and mediate the generation of pulling forces. These findings are relevant for understanding the role of cortical dynein in the positioning of MTOCs, but may be equally relevant for other cellular processes, where MT ends interact with dynein, such as the dynein-mediated poleward motion of chromosomes.45,46 Interestingly, it was recently shown that in budding yeast, dynein accumulation at contact points between shrinking MT ends and the cortex appears indeed to be crucial for shrinkage-coupled spindle pole body movement.47,48 It is important to keep in mind, however, that in vivo other factors are likely providing additional control of MT dynamics and forces. The in vitro reconstitution of MTOC positioning in microfabricated chambers has allowed us to reveal a simple reliable mechanism for pulling-based centering that is different from mechanisms that have been proposed before.36-38 In cells, it remains to be confirmed how dominant this mechanism is in different cases. However, the slipping of MT ends along the cortex has been observed in living cells37,49,50 (see, Fig. 3B in 51), and MT slipping has in fact been suggested to be responsible for the different MT distributions observed in different cell shapes.52 In the future, it will be interesting to further test this idea, e.g., by correlating MT distributions with the positioning of MTOCs in living cells. In addition, one can foresee further improvements of in vitro approaches, in combination with theoretical modeling, to capture additional relevant properties of real cellular systems.
Materials and Methods
Materials
Lipids and surfactant
1,2-dioleoyl-sn-glycero-3-phospho-l-serine (sodium salt) (DOPS), 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(biotinyl) (biotin PE), 1,2-di-(9Z-octadecenoyl)-sn-glycero-3-phosphocholine (DOPC), were purchased from Avanti Polar Lipids. Span 80 was purchased from Sigma.
Buffers
Chemical reagents were obtained from Sigma, unless stated otherwise. Tubulins were purchased from Cytoskeleton Inc. The dynein construct used was a biotinylated GST-dynein331 construct that contains a GFP, as described previously.17 Centrosomes were purified with the generous help of Claude Celati from human lymphoblastic KE37 cell lines.
Emulsion droplets
Oil/Lipid mixture
Lipids (DOPS, biotin PE) in chloroform were dried under nitrogen flow and dissolved in mineral oil in a molar ratio DOPS/biotin PE of 99:1 at a total concentration of 0.5 mg/mL. Span 80 was added at 2 wt%. The mixture was then sonicated for 30 min.
Buffer I
Tubulin (34 µM), Tubulin488 (3.4 µM), GTP (5 mM), Streptavidin (40 nM), ATP (1 mM), glucose (50 mM), dynein (60 nM) and purified centrosomes were mixed in MRB80 buffer (80 mM K-Pipes, 4 mM MgCl2, 1 mM EGTA) with an oxygen scavenging system (glucose oxidase 0.1 mg/mL, DTT 4 mM, catalase 0.1 mg/mL). Centrosome concentration was adjusted so that most of the droplets contain one centrosome.
Emulsion droplet formation
Emulsion droplets of Buffer I in Oil/Lipid mixture were prepared using a flow focusing microfluidic chip as described elsewhere.53 Liquids were driven with a pressure controller MFCS-FLEX-4C-1,000 mbar (Fluigent). Droplets were imaged with a spinning disk confocal head from Yokogawa and a cooled EM-CCD camera (Hamamatsu Photonics; C9100).
Giant, unilamellar liposomes
Inside buffer
Sucrose (280 mM), tubulin (30 µM), RhodamineTubulin (3 µM), GTP (5 mM), Streptavidin (40 nM), ATP (1 mM), glucose (50 mM), dynein (60 nM) and taxol (12.5 µM) were mixed in MRB80 buffer (80 mM K-Pipes, 4 mM MgCl2, 1 mM EGTA) with an oxygen scavenging system (glucose oxidase 0.1 mg/mL, DTT 4 mM, catalase 0.1 mg/mL).
Outside buffer
Glucose (300 mM) was dissolved in MRB80 buffer.
Liposome formation
Giant unilamellar vesicles were prepared with the so called “Agarose method,” as described elsewhere.54
Acknowledgments
The theoretical work referred to in this Extra View was developed in collaboration with Nenad Pavin and Frank Jülicher. The experimental work referred to was performed in collaboration with Ron Vale and Samara Reck-Peterson. We thank S. Reck-Peterson’s group and in particular Sirui Zou for help with dynein purification. We thank M. Bornens’ group and in particular Claude Celati for help with the centrosome purification. We thank P. Tabeling’s group and in particular Bingqing Shen for help with microfluidics. We thank G. Koenderink's group and in particular Feng-Ching Tsai for help with liposome formation. We thank G. Koenderink for a critical reading of the manuscript. M.D. and L.L. gratefully acknowledge support from Human Frontier Science Program. This work is part of the research program of the Foundation for Fundamental Research on Matter (FOM), which is part of the Netherlands Organisation for Scientific Research (NWO).
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21753
References
- 1.Tran PT, Marsh L, Doye V, Inoué S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149:863–74. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kimura A, Onami S. Local cortical pulling-force repression switches centrosomal centration and posterior displacement in C. elegans. J Cell Biol. 2007;179:1347–54. doi: 10.1083/jcb.200706005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 5.Wühr M, Dumont S, Groen AC, Needleman DJ, Mitchison TJ. How does a millimeter-sized cell find its center? Cell Cycle. 2009;8:1115–21. doi: 10.4161/cc.8.8.8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamaguchi MS, Hiramoto Y. Analysis of the Role of Astral Rays in Pronuclear Migration in Sand Dollar Eggs by the Colcemid-UV Method. Dev Growth Differ. 1986;28:143–56. doi: 10.1111/j.1440-169X.1986.00143.x. [DOI] [PubMed] [Google Scholar]
- 7.Rodionov VI, Borisy GG. Microtubule treadmilling in vivo. Science. 1997;275:215–8. doi: 10.1126/science.275.5297.215. [DOI] [PubMed] [Google Scholar]
- 8.Tolić-Nørrelykke IM. Push-me-pull-you: how microtubules organize the cell interior. Eur Biophys J. 2008;37:1271–8. doi: 10.1007/s00249-008-0321-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howard J. Elastic and damping forces generated by confined arrays of dynamic microtubules. Phys Biol. 2006;3:54–66. doi: 10.1088/1478-3975/3/1/006. [DOI] [PubMed] [Google Scholar]
- 10.Burakov A, Nadezhdina E, Slepchenko B, Rodionov V. Centrosome positioning in interphase cells. J Cell Biol. 2003;162:963–9. doi: 10.1083/jcb.200305082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–41. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dujardin DL, Vallee RB. Dynein at the cortex. Curr Opin Cell Biol. 2002;14:44–9. doi: 10.1016/S0955-0674(01)00292-7. [DOI] [PubMed] [Google Scholar]
- 13.Grill SW, Gönczy P, Stelzer EH, Hyman AA. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 2001;409:630–3. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- 14.Koonce MP, Köhler J, Neujahr R, Schwartz JM, Tikhonenko I, Gerisch G. Dynein motor regulation stabilizes interphase microtubule arrays and determines centrosome position. EMBO J. 1999;18:6786–92. doi: 10.1093/emboj/18.23.6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vallee RB, Varma D, Dujardin DL. ZW10 function in mitotic checkpoint control, dynein targeting and membrane trafficking: is dynein the unifying theme? Cell Cycle. 2006;5:2447–51. doi: 10.4161/cc.5.21.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto A, Tsutsumi C, Kojima H, Oiwa K, Hiraoka Y. Dynamic behavior of microtubules during dynein-dependent nuclear migrations of meiotic prophase in fission yeast. Mol Biol Cell. 2001;12:3933–46. doi: 10.1091/mbc.12.12.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–48. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gennerich A, Carter AP, Reck-Peterson SL, Vale RD. Force-induced bidirectional stepping of cytoplasmic dynein. Cell. 2007;131:952–65. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozlowski C, Srayko M, Nedelec F. Cortical microtubule contacts position the spindle in C. elegans embryos. Cell. 2007;129:499–510. doi: 10.1016/j.cell.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 20.Laan L, Pavin N, Husson J, Romet-Lemonne G, van Duijn M, López MP, et al. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 2012;148:502–14. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen-Ngoc T, Afshar K, Gönczy P. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat Cell Biol. 2007;9:1294–302. doi: 10.1038/ncb1649. [DOI] [PubMed] [Google Scholar]
- 22.Collins ES, Balchand SK, Faraci JL, Wadsworth P, Lee WL. Cell Cycle-Regulated Cortical Dynein/Dynactin Promotes Symmetric Cell Division by Differential Pole Motion in Anaphase. Mol Biol Cell. 2012 doi: 10.1091/mbc.E12-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niclas J, Allan VJ, Vale RD. Cell cycle regulation of dynein association with membranes modulates microtubule-based organelle transport. J Cell Biol. 1996;133:585–93. doi: 10.1083/jcb.133.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendricks AG, Lazarus JE, Perlson E, Gardner MK, Odde DJ, Goldman YE, et al. Dynein tethers and stabilizes dynamic microtubule plus ends. Curr Biol. 2012;22:632–7. doi: 10.1016/j.cub.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–9. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- 26.Janson ME, de Dood ME, Dogterom M. Dynamic instability of microtubules is regulated by force. J Cell Biol. 2003;161:1029–34. doi: 10.1083/jcb.200301147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franck AD, Powers AF, Gestaut DR, Gonen T, Davis TN, Asbury CL. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat Cell Biol. 2007;9:832–7. doi: 10.1038/ncb1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dogterom M, Kerssemakers JWJ, Romet-Lemonne G, Janson ME. Force generation by dynamic microtubules. Curr Opin Cell Biol. 2005;17:67–74. doi: 10.1016/j.ceb.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Lombillo VA, Stewart RJ, McIntosh JR. Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature. 1995;373:161–4. doi: 10.1038/373161a0. [DOI] [PubMed] [Google Scholar]
- 30.Dogterom M, Yurke B. Microtubule Dynamics and the Positioning of Microtubule Organizing Centers. Phys Rev Lett. 1998;81:485–8. doi: 10.1103/PhysRevLett.81.485. [DOI] [Google Scholar]
- 31.Inoué S. Mitotic organization and force generation by assembly/disassembly of microtubules. Cell Struct Funct. 1996;21:375–9. doi: 10.1247/csf.21.375. [DOI] [PubMed] [Google Scholar]
- 32.Holy TE, Dogterom M, Yurke B, Leibler S. Assembly and positioning of microtubule asters in microfabricated chambers. Proc Natl Acad Sci USA. 1997;94:6228–31. doi: 10.1073/pnas.94.12.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faivre-Moskalenko C, Dogterom M. Dynamics of microtubule asters in microfabricated chambers: the role of catastrophes. Proc Natl Acad Sci USA. 2002;99:16788–93. doi: 10.1073/pnas.252407099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janson ME, Dogterom M. Scaling of microtubule force-velocity curves obtained at different tubulin concentrations. Phys Rev Lett. 2004;92:248101. doi: 10.1103/PhysRevLett.92.248101. [DOI] [PubMed] [Google Scholar]
- 35.Grill SW, Hyman AA. Spindle positioning by cortical pulling forces. Dev Cell. 2005;8:461–5. doi: 10.1016/j.devcel.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 36.Zhu J, Burakov A, Rodionov V, Mogilner A. Finding the cell center by a balance of dynein and myosin pulling and microtubule pushing: a computational study. Mol Biol Cell. 2010;21:4418–27. doi: 10.1091/mbc.E10-07-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu J, Misra G, Russell RJ, Ladd AJ, Lele TP, Dickinson RB. Effects of dynein on microtubule mechanics and centrosome positioning. Mol Biol Cell. 2011;22:4834–41. doi: 10.1091/mbc.E11-07-0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 39.Grill SW, Howard J, Schäffer E, Stelzer EH, Hyman AA. The distribution of active force generators controls mitotic spindle position. Science. 2003;301:518–21. doi: 10.1126/science.1086560. [DOI] [PubMed] [Google Scholar]
- 40.Pavin N, Laan L, Ma R, Dogterom M, Julicher F. Positioning of microtubules organizing centers by cortical pushing and pulling forces. New J Phys. submitted. [Google Scholar]
- 41.Hotani H, Inaba T, Nomura F, Takeda S, Takiguchi K, Itoh TJ, et al. Mechanical analyses of morphological and topological transformation of liposomes. Biosystems. 2003;71:93–100. doi: 10.1016/S0303-2647(03)00113-8. [DOI] [PubMed] [Google Scholar]
- 42.Emsellem V, Cardoso O, Tabeling P. Vesicle deformation by microtubules: A phase diagram. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1998;58:4807–10. doi: 10.1103/PhysRevE.58.4807. [DOI] [Google Scholar]
- 43.Fygenson DK, Marko JF, Libchaber A. Mechanics of microtubule-based membrane extension. Phys Rev Lett. 1997;79:4497–500. doi: 10.1103/PhysRevLett.79.4497. [DOI] [Google Scholar]
- 44.Pinot M, Chesnel F, Kubiak JZ, Arnal I, Nedelec FJ, Gueroui Z. Effects of confinement on the self-organization of microtubules and motors. Curr Biol. 2009;19:954–60. doi: 10.1016/j.cub.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 45.Savoian MS, Goldberg ML, Rieder CL. The rate of poleward chromosome motion is attenuated in Drosophila zw10 and rod mutants. Nat Cell Biol. 2000;2:948–52. doi: 10.1038/35046605. [DOI] [PubMed] [Google Scholar]
- 46.Sharp DJ, Rogers GC, Scholey JM. Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat Cell Biol. 2000;2:922–30. doi: 10.1038/35046574. [DOI] [PubMed] [Google Scholar]
- 47.Ten Hoopen R, Cepeda-García C, Fernández-Arruti R, Juanes MA, Delgehyr N, Segal M. Mechanism for Astral Microtubule Capture by Cortical Bud6p Priming Spindle Polarity in S. cerevisiae. Curr Biol. 2012;22:1075–83. doi: 10.1016/j.cub.2012.04.059. [DOI] [PubMed] [Google Scholar]
- 48.Xiang X. Nuclear positioning: Dynein needed for microtubule shrinkage-coupled movement. Curr Biol. 2012;22:R496–9. doi: 10.1016/j.cub.2012.04.044. [DOI] [PubMed] [Google Scholar]
- 49.Komarova Y, De Groot CO, Grigoriev I, Gouveia SM, Munteanu EL, Schober JM, et al. Mammalian end binding proteins control persistent microtubule growth. J Cell Biol. 2009;184:691–706. doi: 10.1083/jcb.200807179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Minc N, Bratman SV, Basu R, Chang F. Establishing new sites of polarization by microtubules. Curr Biol. 2009;19:83–94. doi: 10.1016/j.cub.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Rourke SM, Christensen SN, Bowerman B. Caenorhabditis elegans EFA-6 limits microtubule growth at the cell cortex. Nat Cell Biol. 2010;12:1235–41. doi: 10.1038/ncb2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Picone R, Ren X, Ivanovitch KD, Clarke JD, McKendry RA, Baum B. A polarised population of dynamic microtubules mediates homeostatic length control in animal cells. PLoS Biol. 2010;8:e1000542. doi: 10.1371/journal.pbio.1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ménétrier-Deremble L, Tabeling P. Droplet breakup in microfluidic junctions of arbitrary angles. Phys Rev E Stat Nonlin Soft Matter Phys. 2006;74:035303. doi: 10.1103/PhysRevE.74.035303. [DOI] [PubMed] [Google Scholar]
- 54.Tsai F-C, Stuhrmann B, Koenderink GH. Encapsulation of active cytoskeletal protein networks in cell-sized liposomes. Langmuir. 2011;27:10061–71. doi: 10.1021/la201604z. [DOI] [PubMed] [Google Scholar]