Abstract
PCTAIRE kinases (PCTK) are a highly conserved, but poorly characterized, subgroup of cyclin-dependent kinases (CDK). They are characterized by a conserved catalytic domain flanked by N- and C-terminal extensions that are involved in cyclin binding. Vertebrate genomes contain three highly similar PCTAIRE kinases (PCTK1,2,3, a.k.a., CDK16,17,18), which are most abundant in post-mitotic cells in brain and testis. Consistent with this restricted expression pattern, PCTK1 (CDK16) has recently been shown to be essential for spermatogenesis. PCTAIREs are activated by cyclin Y (CCNY), a highly conserved single cyclin fold protein. By binding to N-myristoylated CCNY, CDK16 is targeted to the plasma membrane. Unlike conventional cyclin-CDK interactions, binding of CCNY to CDK16 not only requires the catalytic domain, but also domains within the N-terminal extension. Interestingly, phosphorylation within this domain blocks CCNY binding, providing a novel means of cyclin-CDK regulation. By using these functional characteristics, we analyzed “PCTAIRE” sequence containing protein kinase genes in genomes of various organisms and found that CCNY and CCNY-dependent kinases are restricted to eumetazoa and possibly evolved along with development of a central nervous system. Here, we focus on the structure and regulation of PCTAIREs and discuss their established functions.
Keywords: CDK, CDK16, PCTAIRE, PCTK, cyclin, development, evolution, spermatogenesis
Introduction
Cyclin-dependent kinases (CDKs) comprise a large family of proline-directed serine/threonine protein kinases that, together with MAP kinases, GSKs and CDK-like kinases, belong to the CMGC branch of the kinome tree.1 CDKs are regulated by binding to an activating subunit, a cyclin, as well as by phosphorylation and binding to inhibitory proteins.2 Some of the CDKs (CDK1, 2, 3, 4, 6) are involved in cell cycle regulation and have been studied intensively for more than 30 y. The majority of the CDK family members, however, do not control proliferation but are involved in other cellular processes, including transcription (CDK7, 8, 9, 10), translation (CDK1, 11), mRNA processing (CDK10, 11, 12, 13) or control differentiation and function of neurons (CDK5).3-5 The CDK family counts 26 members, classified as CDK1–20 (there are two CDK11 isoforms, CDK11A and B) and five additional more distant members called CDK-like (CDKL) kinases.6 The CDKs range in size from ~300 amino acid residues, which just encompass the catalytic domain, to proteins of more than 1,500 residues with N- and/or C-terminal extensions of variable lengths, whose functions are not well characterized.4 The function and regulation of CDKs 14–20 as well as CDKL-1–5 are only poorly understood. This is due to the fact that activating cyclins for these kinases are unknown or poorly described, and no essential functions for most of these kinases have been described. During the last couple of years, substantial progress has been made in our understanding of the function and regulation of CDKs 14–18, which are better known as PFTAIRE (PFTK) and PCTAIRE (PCTK) kinases. In Drosophila, CDK14 is essential for development and participates in WNT signaling,7,8 while the CDK16 homolog PCT-1 plays an important role in motor neurons in nematodes. In mammals, CDK16 is essential for spermatogenesis,9 suggesting that PFTAIRE/PCTAIRE kinases are involved in many important, yet different, cellular functions. Here, we review PCTAIRE and PFTAIRE kinases and argue that true PCTAIRE kinases, i.e., CDK16 homologs, are restricted to animals, suggesting an essential role in the nervous system.
From Orphans to Eccentric Family Members
The PCTAIRE and PFTAIRE subgroups of CDKs contain three (PCTK1–3) and two members (PFTK1,2), respectively. Until recently, these kinases were considered “orphan CDKs,” because their putative cyclin partners were unknown and, hence, named according to a characteristic alphα-helical sequence that corresponds to the cyclin-interaction “PSTAIRE” helix in CDK1 or 2. Recently, however, all members of the CDK family were re-classified; PFTKs are now known as CDK14 and 15, and PCTKs as CDK16–18.6 Mammalian CDK16 and 18 show highest expression in postmitotic tissues like brain and testis, while CDK17 is expressed predominantly in the brain.10,11 CDK16 is abundant in Purkinje and pyramidal cells of the hippocampus,11,12 and CDK16 transcripts and protein are present in postmeiotic spermatids. CDK16 protein is absent in mature spermatozoa, because it is sequestered into the residual body during the terminal stages of spermatogenesis.11,13 Lower levels of CDK16 can be detected in various tissues and in tumor-derived as well as immortalized cell lines.14
The subcellular localization of CDK16 is still not completely resolved. When bound to its activator cyclin Y (CCNY), CDK16 is targeted to the plasma membrane but this localization has still to be confirmed in vivo.9 The endogenous protein was, so far, found to be localized in the cytoplasm14 and the nucleolus of neurons.15 As discussed further below, CDK16 is phosphorylated at several residues, which regulates its ability to interact with, and become activated by, CCNY. Thus, it is possible that only a small fraction of PCTK1/CDK16 is at the cell membrane at any time and may therefore be difficult to detect there.
Transcripts of rat PCTK2/CDK17 are predominantly found in the hippocampal and olfactory bulb regions of the brain.10 CDK17 kinase activity purified from the brain was found to be sensitive to high salt concentration, suggesting that it might require an activating subunit. Subsequently, CDK17 was shown to interact with TRAP (Tudor repeat associated with PCTK2)16 as well as IK3–1/Cables1,17 but none of these interactors were able to stimulate the kinase activity of CDK17 in vitro.10,16-18 PCTK3/CDK18, the least well-studied member of this CDK subfamily, is also mainly expressed in brain and testis.10 In a genome-wide, protein-protein interaction screen, CDK18 was found to interact with cyclin K,19 but the functional significance of this interaction remained unclear.
PFTK1/CDK14 shares around 50% identity with the PCTAIRE family members. This homology is largely restricted to the CDK domain and only a small portion of the N-terminal extension is related to the PCTAIRE sequences.20 CDK14 is expressed ubiquitously with high expression in the brain, testis and in many organs during development.11,12,20,21 In the adult mouse testis, CDK14 expression is restricted to late pachytene/diplotene spermatocytes, which are just about to undergo the first meiotic division.12 Human CDK14 is also expressed in several organs, including brain and testis, but high levels of human CDK14 mRNA levels could also be detected in the heart, kidney, pancreas and ovary.22 CDK14 protein is expressed in the cytoplasm22 but can also be found in the nucleus of motor neurons.20 Like CDK16, ectopically expressed CDK14 is cytoplasmic unless co-expressed with CCNY, which targets it to the plasma membrane.23
PFTK2/CDK15 is very poorly characterized, and little is known about its expression and regulation. Evolutionarily, CDK15 seems to be of a newer origin, since most animal species have only one PFTAIRE, which is more similar to CDK14 (Table 1). Like the other CDKs of this subfamily, CDK15 appears to be primarily expressed in the human brain as judged by publicly available gene expression data (BioGPS).
Table 1. Comparison of human CDK1 (as reference), CDK14-18 and CCNY primary sequences with other species.
| species | CDK1 | CDK16 | CDK17 | CDK18 | CCNY | CDK14 | CDK15 | comment |
|---|---|---|---|---|---|---|---|---|
| Human (Homo sapiens) |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
| Chicken (Gallus gallus) |
90% |
- |
99% |
70% |
96% |
54% (92%) |
58% |
CDK14: 322 additional N-terminal residues |
| Zebra fish (Danio rerio) |
82% |
75% |
91% |
64% |
86% |
64% |
61% |
|
| Frog (Xenopus laevis, tropicalis) |
86% |
75% |
91% |
71% Xt |
77% |
79% |
60% Xt |
CDK15: two incomplete sequences for X.t. |
| Lancelet (Branchiostoma floridae) |
77% |
44% (52%) |
- |
- |
61% |
- |
- |
Incomplete genome |
| Sea squirt (Ciona intestinalis) |
73% |
- |
53% (63%) |
- |
60% |
45% |
- |
|
| Sea urchin (Strongylocentrotus purpuratus) |
70% |
- |
49% (54%) |
- |
61% |
- |
51% |
|
| Fly (Drosophila melanogaster) |
72% |
- |
- |
- |
54% |
42% (58%) |
- |
CDK14: 82 additional N-terminal residues |
| Shoulder tick (Ixodes scapularis) |
71% |
59% (67%) |
- |
- |
60% |
- |
- |
|
| Water flea (Daphnia pulex) |
69% |
- |
57% (69%) |
- |
60% |
40% (49%) |
- |
CDK14: 75 additional N-terminal residues |
| Round worm (Caenorhabditis elegans) |
58% (64%) |
- |
40% (47%) |
- |
51% |
33% (40%) |
- |
|
| Flatworm (Schistosoma mansoni) |
45% (61%) |
41% (47%) |
- |
- |
36% |
15% (55%) |
- |
CDK14: 424 N- and 606 C- additional residues |
| Leech (Helobdella robusta) |
64% (67%) |
30% (50%) |
- |
- |
53% |
(65%) |
- |
CDK14:only partial sequence |
| Snail (Lottia gigantea) |
76% |
- |
62% (69%) |
- |
58% |
47% (59%) |
- |
|
| Hydra magnipapillata |
61% (64%) |
- |
41% (65%) |
- |
54% |
37% (55%) |
- |
CDK17: 102 additional C-terminal residues; CDK14: only partial sequence |
| Sea anemone (Nematostella vectensis) |
66% |
- |
52% (59%) |
- |
61% |
46% (53%) |
- |
|
| Placozoa (Trichoplax adhaerens) |
70% |
- (48%) |
- |
- |
43% |
49% (57%) |
- |
‘PCTAIRE’ kinase missing domain I |
| Sponge (Amphimedon queenslandica) |
67% |
- |
- (50%) |
- |
38% |
43% |
- |
‘PCTAIRE’ kinase missing domain I |
| Collared flagellate (Monosiga brevicollis) |
64% |
- (40%) |
- |
- |
- (no myr (35%)) |
- |
- |
‘PCTGIRE’ protein missing domain I |
| Slime mold (Dictyostelium discoideum) |
61% |
- |
- |
- |
24% (33%) |
- |
- |
CDK5 with ‘PCTAIRE’ sequence (66% with human CDK5) |
| Yeast (Saccharomyces cerevisiae) | 57% (60%) | - | - | - | - | - | - |
Sequence alignment was done using VNTI software (BLOSUM62 matrix) and shows identity of amino acid sequences between human and other organisms. Values in the brackets indicate calculations when sequences were clipped to the length of the human protein.
The cell type-specific expression, membrane localization as well as the mode of regulation, which is described in more detail below, distinguishes CDK16 and its related kinases from most other CDKs, suggesting that these “eccentric” CDKs carry out important membrane proximal signaling events in differentiated cell types.
Conventional Cyclin-CDK Interactions
In the absence of cyclin, CDKs are typically “loners:” monomeric and inactive. Cyclin binding, however, induces structural changes that allow CDKs to interact with and phosphorylate substrates.24 In addition, cyclins are believed to contribute to substrate specificity and for guiding their partner to specific subcellular sites of action.25,26 Finally, regulated cyclin synthesis and proteolysis are important means for regulating the activities of some CDKs, especially those involved in cell cycle regulation.27,28
Cyclins comprise a large family of more than 29 proteins,4 varying in size from 35–90 kDa. While the first cyclins, i.e., those involved in cell cycle regulation,29,30 were identified and defined by their cell cycle-dependent oscillations, the expression levels of many other cyclins do not change during the cell division cycle. Family membership is structurally defined by the presence of the so-called cyclin-box, a domain of ~100 amino acid residues that forms a “cyclin fold,” a stack of five α-helices. Conventional cyclins usually have two such folds; the N-terminal one corresponds to the cyclin-box,31 which is necessary for CDK binding and activation, and the C-terminal fold, which is structurally related to the first one and is required for the proper folding of the cyclin molecule. This stack of folds makes cyclins rather rigid molecules that hardly change their conformation after binding to CDKs.24,32 The N- and/or C-terminal extensions outside the CDK binding domain serve various purposes, including control of subcellular localization or protein stability. The cyclin fold is, however, also present in other proteins, such as pRb, TFIIB and Cables,33,34 which are unlikely to function as CDK activators.
By primary sequence analysis, the kinase domains of CDKs are very similar, showing 59% consensus homology over the whole CDK family [calculated by comparing CDK domain sequences of all known human CDKs using VNTI software (Invitrogen, Life Technologies Corp.) and BLOSUM6235]. Three-dimensional structures are available for many CDK monomers (CDKs 2, 4, 5, 8 and 9) as well as cyclin/CDK dimers (cyclins A, B, C, H, T and p25/Pho80 in complex with their CDKs),36-42 which provided a molecular understanding of how cyclins activate CDKs.24,43,44
Like all kinases, cyclin-dependent kinases have a two-lobed structure. The smaller N-terminal lobe consists of mainly β-sheets, while the larger C-terminal lobe is rich in α-helices. The N-lobe also contains the glycine-rich inhibitory segment (G-loop) and the PSTAIRE helix. The cleft between the two lobes forms the active site with the ATP-binding pocket and the substrate-binding interface. In the monomeric form of CDK, proper ATP and substrate binding are precluded by the structure of the N-terminal lobe and the position of the activation loop, also known as the T-loop. Cyclin makes contacts to both lobes, causing the two lobes to move apart from each other and the T-loop away from the catalytic site. These sterical changes readjust critical residues for the proper positioning of ATP for the phosphotransfer reaction32 and make a conserved threonine (Thr160 in CDK2) within the T-loop accessible for phosphorylation by CDK-activating kinase (CAK).45 This phosphorylation is required to lock the T-loop, and thus, the cyclin-CDK pair in an open conformation.46 In addition to T-loop phosphorylation, the activity of CDKs can also be negatively regulated by phosphorylation of G-loop residues that affects the formation of the catalytic center and substrate binding.47
Cyclin/CDKs can exist as soluble dimers but are also part of larger protein complexes and are involved in the regulation of many different cellular processes over very different time scales. Cell cycle-regulatory cyclins interact stably with their CDK partner, and regulated cyclin proteolysis is a widely employed mechanism to terminate their activity. Proteolysis thus underlies the oscillations of cyclin protein levels and is an essential component of cell cycle regulation.48 In addition, these CDK complexes are controlled by upstream inhibitory kinases (Wee1, Myt1), activating kinases (CAK) and interaction with CDK inhibitory proteins.2,49
Cyclin-CDK pairs involved in transcriptional control and mRNA splicing, e.g., cyclin C/CDK8 and cyclin L/CDK11, appear to be mainly regulated by their assembly into large multi-protein complexes.50-52 Via their C-terminal cyclin fold, “transcriptional” cyclins make additional interactions with the catalytic subunit, resulting in a more open conformation suitable to accommodate additional proteins.41,42 The formation of these complexes usually stabilizes one of the partners, like the CDK subunit in the cyclin T1/CDK953 or the cyclin subunit in the cyclin C/CDK8 complex.54 Thus, this class of cyclin/CDK pairs appears to be regulated by complex formation-induced stabilization of subunits. In contrast to most of the CDKs, which require phosphorylation of a conserved threonine in the T-loop,42,50,55 CDK8 is proposed to be solely activated by interaction with its cyclin.39 Additional mechanisms of regulation of this subclass of CDKs include autophosphorylation (CDK9, 11)41 and activation-dependent dimerization (CDK11).56
Another class of CDKs appears to function close to the cell membrane. Membrane proximal signaling is often very short-lived, suggesting that “membrane”-CDKs might be controlled by yet another mechanism. The first membrane-CDK to be defined was CDK5, which is expressed in many cell types and tissues, but due to the more restricted expression patterns of its activators, p35 and p39, CDK5 activity is restricted to neurons and few other cell types.57 Structurally, CDK5 is similar to CDK2, but shows no requirement for an activating upstream kinase, since p35 already interacts tightly with the T-loop of CDK5.37,42 Both CDK5 activators carry an N-terminal myristoylation motif that is required for their membrane targeting.58 As expected for a membrane proximal signaling complex, the activity of the p35/CDK5 complex is transient, tightly regulated and influenced by the composition of the surrounding membrane.59 Membrane-tethered p35 is unphosphorylated and has high affinity for CDK5. After binding to and activating CDK5, p35 becomes phosphorylated by CDK5, causing p35 to dissociate again from CDK5. Subsequently, phospho-p35 either remains membrane-tethered60 or becomes rapidly degraded in the cytoplasm.61 Thus, CDK5 is regulated by an ultra-short negative feedback loop.
Until recently, CDK5 was the only known “membrane-associated” CDK, but PCTAIREs and PFTAIREs can also be targeted to the plasma membrane by their activator cyclin Y (CCNY).14,22,23,58 CCNY and p35/39 are the only known cyclin and cyclin-like proteins with a myristoylation signal that exert their function at the plasma membrane. As discussed in detail below, the interaction between CDK16 and CCNY is also regulated by phosphorylation. In summary, there are at least three different types of cyclin-CDK pairs with very different interaction half-lives. The long-lived complexes, e.g., cell cycle-regulatory cyclin-CDKs, are controlled by cell cycle phase-specific degradation of cyclins. In shorter lived complexes, a constitutively unstable component is stabilized by incorporation into multiprotein complexes, which makes CDK activity dependent on complex assembly. Another way to generate short-lived complexes is to condition the ability to interact on a posttranscriptional modification, e.g., protein phosphorylation, of one or both of the partners, as seen in the CDK5 and CDK16 complexes.
PCTAIREs Interact with CCNY
Bacterially expressed recombinant CDK16 shows no activity against MBP or histone H1 unless incubated with tissue extracts.14,62 Consistent with the requirement for an activator, murine CDK16 (496 amino acid residues) eluted from gel-filtration columns of brain or testis lysates with an apparent mass of > 80 kDa.62 There have been many attempts to track down a cyclin partner for CDK16, but most of the identified interactors failed to activate CDK16, at least toward myelin basic protein (MBP) or histone H1 as substrates.15,63,64 By using full-length human CDK16 as a bait in a yeast two-hybrid interaction screen, we have identified several novel interactors, of which CCNY was found to robustly activate CDK16 against MBP.9 The interaction of CDK16 with CCNY was confirmed in vivo, and the kinase complex isolated from murine testis and brain was found to be active.9 Similarly, the Caenorhabditis elegans (C.elegans) homolog PCT-1 interacts with CYY-1, the nematode CCNY ortholog.65
CCNY is well conserved among eumetazoa, but in contrast to conventional cyclins contains only a single cyclin fold.66 Because conventional cyclins require two such folds for activity, it is unknown how the single cyclin fold protein CCNY binds to and activates its partner kinases. For CCNY and CDK16 it is known that conserved residues in the cyclin fold as well as the PCTAIRE helix are essential.9 It is, therefore, conceivable that the structure of CCNY is similar to that observed for p25, a proteolytic fragment of p3542 or its yeast ortholog Pho80,47 which adopt a cyclin fold-like structure. Similar to CDK5, there is also no evidence that CDK16 requires T-loop phosphorylation, suggesting that CCNY, like p25, might tightly interact with the activation loop, alleviating the need for an activating phosphorylation.
The available structural data of a CDK16 fragment encompassing residues 163–473 (PDB ID: 3MTL) shows overall similarity to CDK2 (Fig. 1) but contains more unresolved regions, including the “PCTAIRE” helix. In contrast to CDK2, the kinase domains of CDK14 and 16 are not sufficient for CCNY binding, but require also the N-terminal extensions.9,23 In CDK16, the N-terminal domain has another important regulatory function, which was first identified by showing that bacterially expressed recombinant CDK16 harboring a non-phosphorylatable alanine at position 153 had higher kinase activity after incubation with brain extracts than wild-type CDK16.62 We found that phosphorylation of S153, which is embedded within a PKA consensus motif, prevents CCNY binding. In addition to this residue, a second domain encompassing residues 112–121, which, like S153, is also conserved among PCTAIRE proteins, is essential for CCNY binding.9 The S153 kinase has not been identified yet, but the sensitivity of S153 phosphorylation to forskolin (a stimulator of adenylate cyclase) and the PKA kinase inhibitor H89 strongly imply a regulatory role for PKA. In many human cell lines, CDK16 was found to be co-expressed with CCNY but inactive due to S153 phosphorylation. In contrast, S153 phosphorylation was found to be low in murine testis, in which CDK16 interacted with CCNY and was found to be active.9
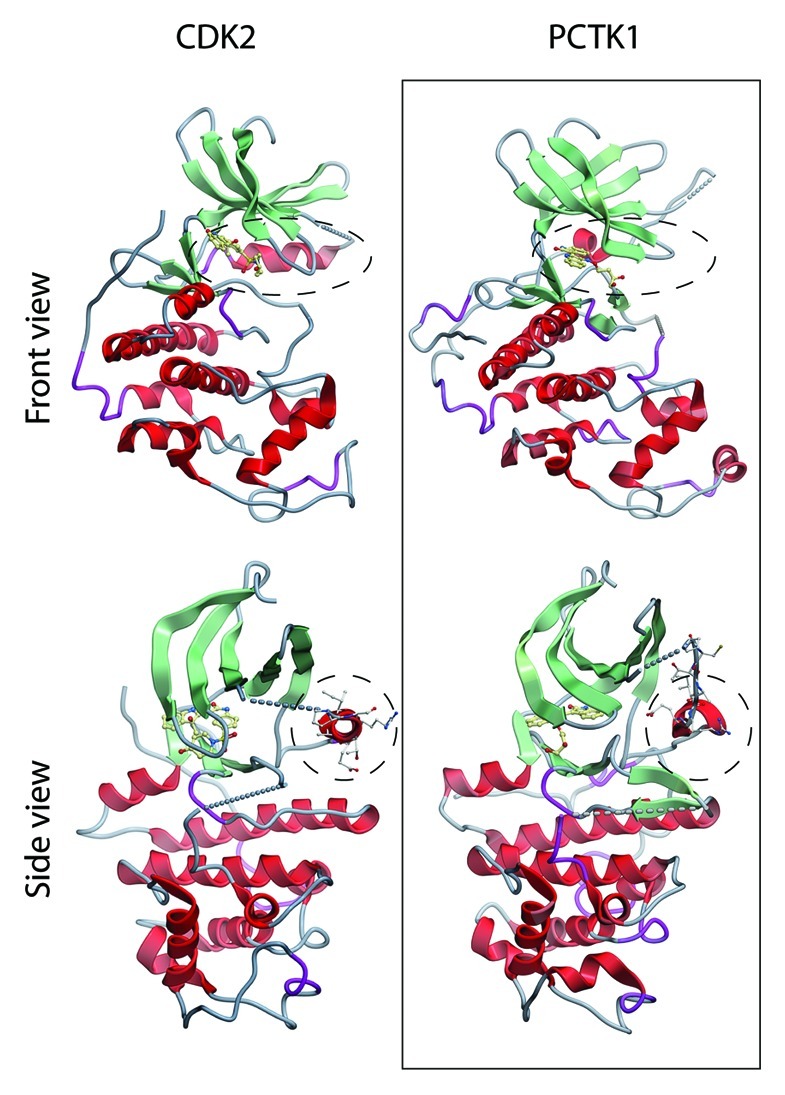
Figure 1. Structural comparison between PCTK1/CDK16 and CDK2. Structural data obtained from PDB files 1JST and 3MTL were visualized using Molsoft (Molsoft L.L.C). The PS/CTAIRE helices are highlighted by a dashed circle line. CDK16 (NΔ163, CΔ23) shows overall similarity to CDK2 but large parts including the PS/CTAIRE helix are unstructured.
How S153 phosphorylation disrupts the ability of CDK16 to interact with CCNY is not understood but could be due to phosphorylation-induced structural changes incompatible with CCNY binding. This hypothesis is supported by the observation that mutation of the nearby glycine 158 into alanine, which might reduce the flexibility of this domain, also prevents CCNY binding to CDK16.9 Another possibility is mutually exclusive or competitive binding of CCNY and 14-3-3 proteins, which were previously shown to bind to unphosphorylated and phosphorylated CDK16,9,62 respectively.
CCNY is a Promiscuous Cyclin
CCNY does not only interact with PCTAIREs but also with PFTAIRE proteins, and these interactions are evolutionarily conserved. Whether CCNY interacts with still other proteins, or has additional CDK independent functions, remains to be investigated. Human CDK1423 as well as the Drosophila ortholog L63 interacted with CG14939, the fly version of CCNY.8 This interaction involves the typical CDK-cyclin domains, such as the cyclin fold domain in CCNY and the “PFTAIRE” motif in CDK14, but also, additional regions outside the CDK domain.23 CCNY activates and recruits CDK14 to the cell membrane to phosphorylate the WNT co-receptor LRP5/6.8,23 Thus, PCTAIREs and PFTAIREs share the same activator, and their co-expression in many murine and human tissues suggests functional redundancy. In contrast to PCTAIRE kinases, there is no evidence that CCNY binding to PFTAIRE kinases is regulated by phosphorylation,23 although serine153 is conserved. One explanation for this difference could be that Ser119, which is responsible for 14-3-3 binding, is not conserved between the two kinases. Thus, although PFTAIREs and PCTAIREs are very similar, are co-expressed in many cell types, are both activated by CCNY and can exert similar functions, they can be differentially regulated by upstream kinases, suggesting that they also may have unique functions.
Functions of PCTAIREs and PFTAIREs
Over the past couple of years, the functions of PCTAIREs and PFTAIREs have begun to emerge by using genetic approaches in the fruit fly, in nematodes as well as in mice. Drosophila has only one PFTAIRE ortholog, L63, which is essential for development,7 but lacks a CDK16 homolog. L63/CDK14 interacts with and is activated by CG14939, the fly CCNY homolog, which is an essential gene in flies.66 The similarity of developmental phenotypes of the PFTK/L63 and CCNY/CG14939 mutants suggest that there are no CDK-independent functions of CCNY, and that CCNY is the only activator of PFTK in Drosophila.7 In cultured Drosophila cells, CCNY and L63 were shown to participate in WNT signaling by phosphorylating and activating the WNT co-receptor Arrow/LRP6.8 L63/CCNY activity was found to be cell cycle-regulated and to peak in mitosis, suggesting an essential function for WNT signaling in cell division,67 such as orientation of the mitotic spindle.68 In vertebrates, CCNY and CCNY-activated kinases are also involved in WNT signaling and are essential for development.8
C.elegans contains one CCNY gene, CYY-1, and one copy each of a PCTAIRE and a PFTAIRE kinase, PCT-1 (47% identical to PCTAIRE-2, Table 1) and the uncharacterized gene ZC123.4 (40% identical to human PFTAIRE (CDK14, Table 1), respectively. Worms lacking CYY-1 or PCT-1 have a mild phenotype, but additional deletion of CDK5 causes paralysis, pointing to a WNT-signaling independent function of PCTAIRE kinases. Although PCT-1 and CDK5 have different subcellular localizations, both regulate presynaptic vesicle transport and synapse turnover.65,69 In contrast to flies and vertebrates, CCNY is not essential in nematodes, probably because WNT signaling in nematodes does not depend on LRP6.70
In vertebrates, functional analysis of CCNY, PCTAIRE and PFTAIRE genes is more complicated due to the presence of multiple genes. Functional overlap of these kinases was demonstrated by knockdown experiments in Xenopus embryos, which revealed that depletion of the individual kinases failed to produce a phenotype, while depletion of CCNY and its highly related homolog CCNY-like1 resulted in a WNT loss-of-function phenotype.8 To investigate whether PCTAIRE kinases may have individual functions, we generated a conditional CDK16-knockout mouse, which revealed that CDK16 is essential for the completion of spermatogenesis.9 Analysis of human CDK16 in cell lines has suggested that this kinase has a role in intracellular vesicle formation64 and transport,71,72 as well as regulation of neurite outgrowth.62,73 Whether PCTAIRE signaling is important for neuronal function in vertebrates will have to await the construction and analysis of additional PCTAIRE knockout mice.
PCTAIREs and PFTAIREs: Eumetazoan-Specific Genes
“PCTAIRE” sequence containing kinases have been identified in the genomes of many organisms including unicellular eukaryotes, such as trypanosomes, choanoflagellates and slime molds, but are missing in yeasts, plants and, curiously, insects.74 This peculiar pattern prompted us to re-investigate the evolutionary relationship of “PCTAIRE”-signature containing kinase genes in different model organisms. In “higher” eukaryotes, PCTAIREs, which are about 500 amino acid residues long, consist of a highly conserved CDK-like kinase domain (~50% identical to CDK1) and long N- and shorter C-terminal extensions. Most of the differences between the various PCTAIRE variants are located in these extensions, which are only 37% (N-terminal extension) and 17% (C-terminal extension) identical between human CDKs 16, 17 and 18, while the kinase core is more similar (73% identity) among human PCTAIREs.10
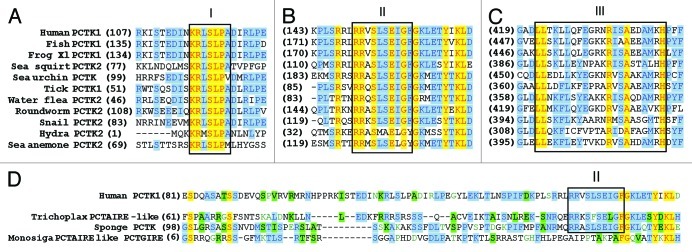
Within the N-terminal extension of CDKs 16–18, there are two sequence clusters that show higher sequence conservation between PCTAIRE genes of various species, suggesting important structural or regulatory functions. One domain comprises residues 115–123 [domain I, KRL(M/Y)S119LPA(M/V), numbering according to human CDK16), which is found in all vertebrate PCTAIREs (Fig. 2A). The second conserved region (domain II (150–158), RRXS153LSE(I/L)G] is not only present in all PCTAIREs (Fig. 2B), but also in the closely related PFTAIREs. A short constant region present in the C-terminal extension spans through residues 422–443 (Fig. 2C), followed by less well-conserved amino acid residues.9 Based on the findings that the conserved domains in the N-terminal extension are important for CCNY binding,9 we propose that only proteins harbouring domains I and II should be classified as true PCTK1/CDK16-like protein kinases.
Figure 2. Alignment of primary sequences of the N- and C-terminal extensions of PCTAIRE kinases from various organisms. CDK16 shows three major clusters of conservation in the N-terminal extension. Domain I is confined to PCTAIREs (A), while domain II (B) is also present in PFTAIRE kinases. Species most closely related to real animals such as that parazoan Trichoplax and sponges, and the choanoflagellate Monosiga, have proteins similar to PCTAIREs, but lack the trademark KRL(M/Y)S119LPA(M/V) domain I. Nevertheless, Trichoplax and sponges do contain a conserved domain II. (D) The C-terminal domain also contains a conserved domain (III) that might be important for proper folding of the catalytic domain.
By applying these criteria to bioinformatic database searches, we found that CDK16-like kinases are only present in eumetazoa (Table 1), while the “PCTAIRE”-sequence containing kinases present in more “primitive” eukaryotes are more closely related to CDK5 than to CDK16. Thus, the parasitic kinetoplastids (Trypanosoma brucei and Leishmania braziliensis) and amoebozoa (Dictyostelium discoideum75 and Polysphondylium pallidum) lack true CDK16 homologs. This is in agreement with the evolutionary position of kinetoplastids and amoebozoa, believed to have diverged around the time of origin of plants,76 which also lack PCTAIRE kinases.
The assignment of PCTAIRE proteins as CDK16 homologs in parazoans, such as the demosponge Amphimedon queenslandica, the placozoan Trichoplax adhaerens or the unicellular choanoflagellate Monosiga brevicollis, was more difficult. The collared flagellates are believed to be the closest living relatives of metazoans, and the Monosiga brevicollis genome contains a gene encoding a protein kinase harboring a “PCTGIRE” sequence kinase that exhibits 40% identity to the kinase domain of CDK16. This protein also contains long N- and C-terminal extensions, which, however, lack the above mentioned conserved domains I and II. Therefore, we conclude that collared flagellates lack a true CDK16 homolog. The genomes of Amphimedon queenslandica and Trichoplax adhaerens also have PCTAIRE candidate protein kinase genes that contain conserved domain II surrounding S153 (Fig. 2D), but domain I, positioned around S119, is missing. Thus, parazoans most likely also lack a true CDK16 homolog. Although both organisms contain most genes found in eumetazoa, they lack neurons, suggesting that they are secondarily simplified life forms that had a more sophisticated ancestor.77-79 We conclude from these analyses that true CDK16 homologs only arose along with the development of a nervous system and can only be found in animals, with the notable exception of insects.
Similar to the CDK16-like kinases, CDK14-like proteins are also found in all eumetazoa (including insects) as well as in Amphimedon. Thus, all eumetazoans appear to have at least one member of the PCTAIRE and the PFTAIRE families (Table 1 and Fig. 4C). Interestingly, although crustaceans, such as Daphnia pulex, contain both a CDK14 and a CDK16 homolog, other branches of arthropods lack one of these related kinases. Insects, e.g., Drosophila species, lack a CDK16 homolog, while arachnids, such as the tick Ixodes scapularis, lack CDK14. This suggests that in arthropods, one of these kinases is sufficient to carry out the essential functions of this kinase family.
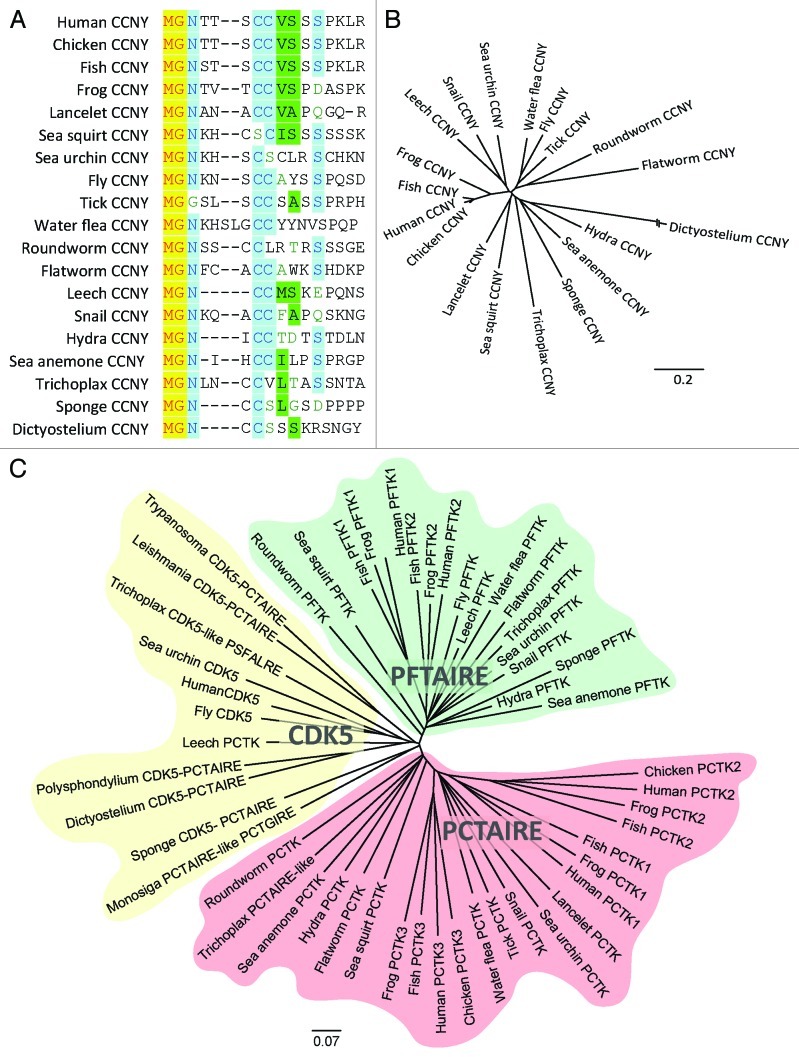
Figure 4. CCNY family and membrane-targeted CDKs. (A) CCNYs of eumetazoa contain a conserved N-terminal myristoylation signal. (B). Phylogenetic tree of all the animal CCNY primary sequences shows high conservation of this protein with exception of Dictyostelium CCNY due to its AT-enriched genome. (C) Phylogentic tree depicting the membrane-associated subfamily of CDKs.
Evolution of CCNY
CCNY is a single-cyclin fold protein that is highly conserved among metazoans, but proteins harboring a Y-type cyclin fold can also be found in plants and yeast,66 species which lack obvious PCTAIRE/PFTAIRE orthologs. The closest homolog to CCNY in yeasts is Pcl1, one of the many cyclins that activates Pho85, a kinase involved in environmental stress responses, cell cycle progression, morphogenesis and cell polarity.80 In contrast to yeast CCNY-like proteins, the role of the plant CCNY-like proteins, like the P-cyclins in Arabidopsis thaliana,81 is only poorly defined. Because these cyclins interact with the CDK1/2 homolog CDK-A, they are likely to be involved in cell cycle regulation. The conservation of single-cyclin fold proteins suggests important and possibly conserved functions, but as argued below, we don’t consider these “ancient,” single-cyclin fold proteins to be CCNY homologs.
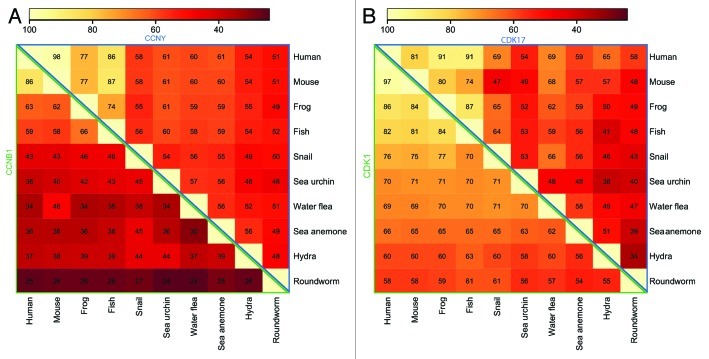
In metazoans, CCNY is considerably conserved over its entire length, which is remarkable, because sequence conservation among other cyclins is usually confined to the cyclin box, i.e., the first cyclin fold. The only other cyclin, whose sequence conservation involves the entire protein, is cyclin C.82 For example, the B-type cyclins, which are essential activators of the main mitosis regulator CDK1, show a sequence identity between 24–86%, while the CCNY orthologs are 38–98% identical (Fig. 3A and Table 1). Dictyostelium CCNY sequence homology is somewhat lower (Fig. 4B), but that can be attributed to the fact that the Dictyostelium genome is AT-rich76 resulting in long His and Asp repeats that are also found in the N-terminal part of CCNY. Removal of these stretches before sequence alignments increased the identity between the Dictyostelium and human proteins to 33% (Table 1).
Figure 3. Cyclin Y and PCTK2/CDK17 are highly conserved. Identity (in percentages) between the primary sequences of cyclin Y (CCNY) and cyclin B1 (CCNB1) of various species were converted to greyscale intensity according to standard heat map scale depicted above the matrices. CCNY is evolutionarily better conserved than cyclin B1 (CCNB1) (A). Similarly, CDK17 shows similar conservation as CDK1 (B).
CCNY seems to have emerged before PCTAIRE and/or PFTAIRE proteins, which are restricted to eumetazoans (Table 1 and Fig. 4B and C). In nematodes, the fruit fly and vertebrates, CCNY targets PCTAIRE and PFTAIRE kinases to the plasma membrane.8,9,66 We therefore analyzed CCNY homologs for the presence of an N-terminal myristoylation signal, and found that in all organisms that contain PCTAIRE/PFTAIRE homologs, CCNY harbors a myristoylation signal (Fig. 4A). In contrast, such a signal is missing in CCNY-like proteins of yeasts, plants and fungi, which lack a PCTAIRE/PFTAIRE homolog. CCNY homologs containing an N-terminal lipidation signal are, however, found in slime molds, such as Dictyostelium (Table 1),76 but their function and kinase partner are not defined yet.
Outlook
The PCTAIRE- and PFTAIRE-related kinases are highly expressed in neurons and in the testis. They are targeted to the plasma membrane via myristoylated CCNY, which is essential for WNT signaling.8 These kinases are, however, likely to have additional functions, which remain to be fully characterized. In nematodes PCT-1 and CCNY are required in neurons,65,69 while in vertebrates CDK16 is essential for spermatogenesis.9 The precise functions of CCNY-dependent kinases in these processes are, however, still unclear, and it will be important to define the specific substrates of these kinases to understand their mode of action.
PCTAIREs and PFTAIREs seem to have some overlapping functions, e.g., during vertebrate development.8 In Drosophila, which lacks a PCTAIRE homolog, deletion of PFTAIRE/CDK14 or CCNY is lethal,7,66 probably due to their essential role in regulating the WNT co-receptor LRP5/6/Arrow during mitosis.8 In human cells, CDK16 activity is prevented in mitosis due to the phosphorylation of S153, and it will be important to define the contribution of these kinases for WNT signaling in interphase and mitosis. Redundancy can also be seen between PCT-1 and CDK5 in the motor neurons of C.elegans. Given the different subcellular localization of these kinases65 and the different phenotypes of the mutants,69 it seems likely, however, that these two kinases exert different functions in the same pathway, and it will be important to find out whether PCTAIRE kinases do cooperate with CDK5 in vertebrate neurons as well.
CCNY and its dependent kinases are evolutionarily very well conserved; in fact they are better conserved than mitotic cyclins and CDK1 of the same organisms4,83-85 (Fig. 3). This might reflect the importance of their overall structure and not only catalytic activity, suggesting that these kinases are functioning in larger protein complexes, which remain to be characterized.
CCNY is present in social amoeboid organisms, which show motility, a wide range of membrane-dependent signaling pathways76 and have a CDK5 homolog containing a “PCTAIRE” sequence.86 PFTAIREs emerged in metazoans and are present in species that lack a nervous system, e.g., in demosponges. In contrast, true PCTAIRE kinases (“CDK16”-homologs) are only present in animals, again supporting the hypothesis that these kinases might have an important role in neurons.87 In addition to their redundant functions, PFTAIRE and PCTAIRE kinases might have specialized roles in other cellular processes, as evidenced by the critical role of CDK16 in mammalian spermatogenesis.9 The ongoing genetic and biochemical analysis of PCTAIRE and PFTAIRE kinases will certainly yield a few more interesting insights into the function of these membrane-CDKs, which might have great impact on human health, ranging from fertility to mental disorders.
Acknowledgments
This work was supported by funding from the Austrian Science Funds (FWF) P20860-B12 (S.G.), SFB021 “Cell proliferation and cell death in tumors,” the Austrian National Bank Jubiläumsstiftung grant #12787, a research grant from the Austrian Society for Endocrinology and Metabolism (ÖGES), as well as the Tiroler Zukunftsstiftung (SG), Österreichische Krebshilfe Tirol and assets of Eva von Lachmüller, Brixen, Italy.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21592
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–4. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 3.Gao CY, Zelenka PS. Cyclins, cyclin-dependent kinases and differentiation. Bioessays. 1997;19:307–15. doi: 10.1002/bies.950190408. [DOI] [PubMed] [Google Scholar]
- 4.Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci. 2005;30:630–41. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Doonan JH, Kitsios G. Functional evolution of cyclin-dependent kinases. Mol Biotechnol. 2009;42:14–29. doi: 10.1007/s12033-008-9126-8. [DOI] [PubMed] [Google Scholar]
- 6.Malumbres M, Harlow E, Hunt T, Hunter T, Lahti JM, Manning G, et al. Cyclin-dependent kinases: a family portrait. Nat Cell Biol. 2009;11:1275–6. doi: 10.1038/ncb1109-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stowers RS, Garza D, Rascle A, Hogness DS. The L63 gene is necessary for the ecdysone-induced 63E late puff and encodes CDK proteins required for Drosophila development. Dev Biol. 2000;221:23–40. doi: 10.1006/dbio.2000.9685. [DOI] [PubMed] [Google Scholar]
- 8.Davidson G, Shen J, Huang YL, Su Y, Karaulanov E, Bartscherer K, et al. Cell cycle control of wnt receptor activation. Dev Cell. 2009;17:788–99. doi: 10.1016/j.devcel.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Mikolcevic P, Sigl R, Rauch V, Hess MW, Pfaller K, Barisic M, et al. Cyclin-dependent kinase 16/PCTAIRE kinase 1 is activated by cyclin Y and is essential for spermatogenesis. Mol Cell Biol. 2012;32:868–79. doi: 10.1128/MCB.06261-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirose T, Tamaru T, Okumura N, Nagai K, Okada M. PCTAIRE 2, a Cdc2-related serine/threonine kinase, is predominantly expressed in terminally differentiated neurons. Eur J Biochem. 1997;249:481–8. doi: 10.1111/j.1432-1033.1997.t01-1-00481.x. [DOI] [PubMed] [Google Scholar]
- 11.Besset V, Rhee K, Wolgemuth DJ. The cellular distribution and kinase activity of the Cdk family member Pctaire1 in the adult mouse brain and testis suggest functions in differentiation. Cell Growth Differ. 1999;10:173–81. [PubMed] [Google Scholar]
- 12.Besset V, Rhee K, Wolgemuth DJ.The identification and characterization of expression of Pftaire-1, a novel Cdk family member, suggest its function in the mouse testis and nervous system Mol Reprod Dev 19985018–29.3>3.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 13.Rhee K, Wolgemuth DJ. Cdk family genes are expressed not only in dividing but also in terminally differentiated mouse germ cells, suggesting their possible function during both cell division and differentiation. Dev Dyn. 1995;204:406–20. doi: 10.1002/aja.1002040407. [DOI] [PubMed] [Google Scholar]
- 14.Charrasse S, Carena I, Hagmann J, Woods-Cook K, Ferrari S. PCTAIRE-1: characterization, subcellular distribution, and cell cycle-dependent kinase activity. Cell Growth Differ. 1999;10:611–20. [PubMed] [Google Scholar]
- 15.Le Bouffant F, Capdevielle J, Guillemot JC, Sladeczek F. Characterization of brain PCTAIRE-1 kinase immunoreactivity and its interactions with p11 and 14-3-3 proteins. Eur J Biochem. 1998;257:112–20. doi: 10.1046/j.1432-1327.1998.2570112.x. [DOI] [PubMed] [Google Scholar]
- 16.Hirose T, Kawabuchi M, Tamaru T, Okumura N, Nagai K, Okada M. Identification of tudor repeat associator with PCTAIRE 2 (Trap). A novel protein that interacts with the N-terminal domain of PCTAIRE 2 in rat brain. Eur J Biochem. 2000;267:2113–21. doi: 10.1046/j.1432-1327.2000.01218.x. [DOI] [PubMed] [Google Scholar]
- 17.Yamochi T, Nishimoto I, Okuda T, Matsuoka M. ik3-1/Cables is associated with Trap and Pctaire2. Biochem Biophys Res Commun. 2001;286:1045–50. doi: 10.1006/bbrc.2001.5493. [DOI] [PubMed] [Google Scholar]
- 18.Zukerberg LR, Patrick GN, Nikolic M, Humbert S, Wu CL, Lanier LM, et al. Cables links Cdk5 and c-Abl and facilitates Cdk5 tyrosine phosphorylation, kinase upregulation, and neurite outgrowth. Neuron. 2000;26:633–46. doi: 10.1016/S0896-6273(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 19.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–8. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 20.Lazzaro MA, Albert PR, Julien JP. A novel cdc2-related protein kinase expressed in the nervous system. J Neurochem. 1997;69:348–64. doi: 10.1046/j.1471-4159.1997.69010348.x. [DOI] [PubMed] [Google Scholar]
- 21.Le Bouffant F, Le Minter P, Traiffort E, Ruat M, Sladeczek F. Multiple subcellular localizations of PCTAIRE-1 in brain. Mol Cell Neurosci. 2000;16:388–95. doi: 10.1006/mcne.2000.0881. [DOI] [PubMed] [Google Scholar]
- 22.Yang T, Chen JY. Identification and cellular localization of human PFTAIRE1. Gene. 2001;267:165–72. doi: 10.1016/S0378-1119(01)00391-2. [DOI] [PubMed] [Google Scholar]
- 23.Jiang M, Gao Y, Yang T, Zhu X, Chen J. Cyclin Y, a novel membrane-associated cyclin, interacts with PFTK1. FEBS Lett. 2009;583:2171–8. doi: 10.1016/j.febslet.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massagué J, et al. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–20. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 25.Brown NR, Noble ME, Endicott JA, Johnson LN. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat Cell Biol. 1999;1:438–43. doi: 10.1038/15674. [DOI] [PubMed] [Google Scholar]
- 26.Pines J. Cyclins, CDKs and cancer. Semin Cancer Biol. 1995;6:63–72. doi: 10.1006/scbi.1995.0009. [DOI] [PubMed] [Google Scholar]
- 27.Pines J. Cyclins and cyclin-dependent kinases: a biochemical view. Biochem J. 1995;308:697–711. doi: 10.1042/bj3080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brandeis M, Hunt T. The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S phase. EMBO J. 1996;15:5280–9. [PMC free article] [PubMed] [Google Scholar]
- 29.Evans T, Rosenthal ET, Youngblom J, Distel D, Hunt T. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983;33:389–96. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- 30.Lew DJ, Dulić V, Reed SI. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell. 1991;66:1197–206. doi: 10.1016/0092-8674(91)90042-W. [DOI] [PubMed] [Google Scholar]
- 31.Noble ME, Endicott JA, Brown NR, Johnson LN. The cyclin box fold: protein recognition in cell-cycle and transcription control. Trends Biochem Sci. 1997;22:482–7. doi: 10.1016/S0968-0004(97)01144-4. [DOI] [PubMed] [Google Scholar]
- 32.Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–91. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 33.Gibson TJ, Thompson JD, Blocker A, Kouzarides T. Evidence for a protein domain superfamily shared by the cyclins, TFIIB and RB/p107. Nucleic Acids Res. 1994;22:946–52. doi: 10.1093/nar/22.6.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuoka M, Matsuura Y, Semba K, Nishimoto I. Molecular cloning of a cyclin-like protein associated with cyclin-dependent kinase 3 (cdk 3) in vivo. Biochem Biophys Res Commun. 2000;273:442–7. doi: 10.1006/bbrc.2000.2965. [DOI] [PubMed] [Google Scholar]
- 35.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–9. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersen G, Busso D, Poterszman A, Hwang JR, Wurtz JM, Ripp R, et al. The structure of cyclin H: common mode of kinase activation and specific features. EMBO J. 1997;16:958–67. doi: 10.1093/emboj/16.5.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarricone C, Dhavan R, Peng J, Areces LB, Tsai LH, Musacchio A. Structure and regulation of the CDK5-p25(nck5a) complex. Mol Cell. 2001;8:657–69. doi: 10.1016/S1097-2765(01)00343-4. [DOI] [PubMed] [Google Scholar]
- 38.Lolli G, Lowe ED, Brown NR, Johnson LN. The crystal structure of human CDK7 and its protein recognition properties. Structure. 2004;12:2067–79. doi: 10.1016/j.str.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 39.Hoeppner S, Baumli S, Cramer P. Structure of the mediator subunit cyclin C and its implications for CDK8 function. J Mol Biol. 2005;350:833–42. doi: 10.1016/j.jmb.2005.05.041. [DOI] [PubMed] [Google Scholar]
- 40.Petri ET, Errico A, Escobedo L, Hunt T, Basavappa R. The crystal structure of human cyclin B. Cell Cycle. 2007;6:1342–9. doi: 10.4161/cc.6.11.4297. [DOI] [PubMed] [Google Scholar]
- 41.Baumli S, Lolli G, Lowe ED, Troiani S, Rusconi L, Bullock AN, et al. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J. 2008;27:1907–18. doi: 10.1038/emboj.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Echalier A, Endicott JA, Noble ME. Recent developments in cyclin-dependent kinase biochemical and structural studies. Biochim Biophys Acta. 20101804:511–519. doi: 10.1016/j.bbapap.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 43.De Bondt HL, Rosenblatt J, Jancarik J, Jones HD, Morgan DO, Kim SH. Crystal structure of cyclin-dependent kinase 2. Nature. 1993;363:595–602. doi: 10.1038/363595a0. [DOI] [PubMed] [Google Scholar]
- 44.Day PJ, Cleasby A, Tickle IJ, O’Reilly M, Coyle JE, Holding FP, et al. Crystal structure of human CDK4 in complex with a D-type cyclin. Proc Natl Acad Sci USA. 2009;106:4166–70. doi: 10.1073/pnas.0809645106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Connell-Crowley L, Solomon MJ, Wei N, Harper JW. Phosphorylation independent activation of human cyclin-dependent kinase 2 by cyclin A in vitro. Mol Biol Cell. 1993;4:79–92. doi: 10.1091/mbc.4.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russo AA, Jeffrey PD, Pavletich NP. Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat Struct Biol. 1996;3:696–700. doi: 10.1038/nsb0896-696. [DOI] [PubMed] [Google Scholar]
- 47.Welburn JP, Tucker JA, Johnson T, Lindert L, Morgan M, Willis A, et al. How tyrosine 15 phosphorylation inhibits the activity of cyclin-dependent kinase 2-cyclin A. J Biol Chem. 2007;282:3173–81. doi: 10.1074/jbc.M609151200. [DOI] [PubMed] [Google Scholar]
- 48.Udvardy A. The role of controlled proteolysis in cell-cycle regulation. Eur J Biochem. 1996;240:307–13. doi: 10.1111/j.1432-1033.1996.0307h.x. [DOI] [PubMed] [Google Scholar]
- 49.Murray A, Hunt T. The Cell Cycle: An Introduction. 1993 [Google Scholar]
- 50.Trembley JH, Hu D, Slaughter CA, Lahti JM, Kidd VJ. Casein kinase 2 interacts with cyclin-dependent kinase 11 (CDK11) in vivo and phosphorylates both the RNA polymerase II carboxyl-terminal domain and CDK11 in vitro. J Biol Chem. 2003;278:2265–70. doi: 10.1074/jbc.M207518200. [DOI] [PubMed] [Google Scholar]
- 51.Loyer P, Trembley JH, Katona R, Kidd VJ, Lahti JM. Role of CDK/cyclin complexes in transcription and RNA splicing. Cell Signal. 2005;17:1033–51. doi: 10.1016/j.cellsig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 52.Galbraith MD, Donner AJ, Espinosa JM. CDK8: a positive regulator of transcription. Transcr. 2010;1:4–12. doi: 10.4161/trns.1.1.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garriga J, Bhattacharya S, Calbó J, Marshall RM, Truongcao M, Haines DS, et al. CDK9 is constitutively expressed throughout the cell cycle, and its steady-state expression is independent of SKP2. Mol Cell Biol. 2003;23:5165–73. doi: 10.1128/MCB.23.15.5165-5173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barette C, Jariel-Encontre I, Piechaczyk M, Piette J. Human cyclin C protein is stabilized by its associated kinase cdk8, independently of its catalytic activity. Oncogene. 2001;20:551–62. doi: 10.1038/sj.onc.1204129. [DOI] [PubMed] [Google Scholar]
- 55.Ramakrishnan R, Rice AP. Cdk9 T-loop phosphorylation is regulated by the calcium signaling pathway. J Cell Physiol. 2011;227:609–17. doi: 10.1002/jcp.22760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chi Y, Zhang C, Zong H, Hong Y, Kong X, Liu H, et al. Thr-370 is responsible for CDK11(p58) autophosphorylation, dimerization, and kinase activity. J Biol Chem. 2011;286:1748–57. doi: 10.1074/jbc.M110.107367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsai LH, Takahashi T, Caviness VS, Jr., Harlow E. Activity and expression pattern of cyclin-dependent kinase 5 in the embryonic mouse nervous system. Development. 1993;119:1029–40. doi: 10.1242/dev.119.4.1029. [DOI] [PubMed] [Google Scholar]
- 58.Asada A, Yamamoto N, Gohda M, Saito T, Hayashi N, Hisanaga S. Myristoylation of p39 and p35 is a determinant of cytoplasmic or nuclear localization of active cyclin-dependent kinase 5 complexes. J Neurochem. 2008;106:1325–36. doi: 10.1111/j.1471-4159.2008.05500.x. [DOI] [PubMed] [Google Scholar]
- 59.Zhu YS, Saito T, Asada A, Maekawa S, Hisanaga S. Activation of latent cyclin-dependent kinase 5 (Cdk5)-p35 complexes by membrane dissociation. J Neurochem. 2005;94:1535–45. doi: 10.1111/j.1471-4159.2005.03301.x. [DOI] [PubMed] [Google Scholar]
- 60.Sato K, Zhu YS, Saito T, Yotsumoto K, Asada A, Hasegawa M, et al. Regulation of membrane association and kinase activity of Cdk5-p35 by phosphorylation of p35. J Neurosci Res. 2007;85:3071–8. doi: 10.1002/jnr.21438. [DOI] [PubMed] [Google Scholar]
- 61.Patrick GN, Zhou P, Kwon YT, Howley PM, Tsai LH. p35, the neuronal-specific activator of cyclin-dependent kinase 5 (Cdk5) is degraded by the ubiquitin-proteasome pathway. J Biol Chem. 1998;273:24057–64. doi: 10.1074/jbc.273.37.24057. [DOI] [PubMed] [Google Scholar]
- 62.Graeser R, Gannon J, Poon RY, Dubois T, Aitken A, Hunt T. Regulation of the CDK-related protein kinase PCTAIRE-1 and its possible role in neurite outgrowth in Neuro-2A cells. J Cell Sci. 2002;115:3479–90. doi: 10.1242/jcs.115.17.3479. [DOI] [PubMed] [Google Scholar]
- 63.Cheng K, Li Z, Fu WY, Wang JH, Fu AK, Ip NY. Pctaire1 interacts with p35 and is a novel substrate for Cdk5/p35. J Biol Chem. 2002;277:31988–93. doi: 10.1074/jbc.M201161200. [DOI] [PubMed] [Google Scholar]
- 64.Palmer KJ, Konkel JE, Stephens DJ. PCTAIRE protein kinases interact directly with the COPII complex and modulate secretory cargo transport. J Cell Sci. 2005;118:3839–47. doi: 10.1242/jcs.02496. [DOI] [PubMed] [Google Scholar]
- 65.Ou CY, Poon VY, Maeder CI, Watanabe S, Lehrman EK, Fu AK, et al. Two cyclin-dependent kinase pathways are essential for polarized trafficking of presynaptic components. Cell. 2010;141:846–58. doi: 10.1016/j.cell.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu D, Guest S, Finley RL., Jr. Why cyclin Y? A highly conserved cyclin with essential functions. Fly (Austin) 2010;4:278–82. doi: 10.4161/fly.4.4.12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davidson G, Niehrs C. Emerging links between CDK cell cycle regulators and Wnt signaling. Trends Cell Biol. 2010;20:453–60. doi: 10.1016/j.tcb.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 68.Segalen M, Bellaïche Y. Cell division orientation and planar cell polarity pathways. Semin Cell Dev Biol. 2009;20:972–7. doi: 10.1016/j.semcdb.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 69.Park M, Watanabe S, Poon VY, Ou CY, Jorgensen EM, Shen K. CYY-1/cyclin Y and CDK-5 differentially regulate synapse elimination and formation for rewiring neural circuits. Neuron. 2011;70:742–57. doi: 10.1016/j.neuron.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eisenmann DM. Wnt signaling. WormBook. 2005:1–17. doi: 10.1895/wormbook.1.7.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Y, Cheng K, Gong K, Fu AK, Ip NY. Pctaire1 phosphorylates N-ethylmaleimide-sensitive fusion protein: implications in the regulation of its hexamerization and exocytosis. J Biol Chem. 2006;281:9852–8. doi: 10.1074/jbc.M513496200. [DOI] [PubMed] [Google Scholar]
- 72.Tang X, Guilherme A, Chakladar A, Powelka AM, Konda S, Virbasius JV, et al. An RNA interference-based screen identifies MAP4K4/NIK as a negative regulator of PPARgamma, adipogenesis, and insulin-responsive hexose transport. Proc Natl Acad Sci USA. 2006;103:2087–92. doi: 10.1073/pnas.0507660103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fu WY, Cheng K, Fu AK, Ip NY. Cyclin-dependent kinase 5-dependent phosphorylation of Pctaire1 regulates dendrite development. Neuroscience. 2011;180:353–9. doi: 10.1016/j.neuroscience.2011.02.024. [DOI] [PubMed] [Google Scholar]
- 74.Joubès J, Chevalier C, Dudits D, Heberle-Bors E, Inzé D, Umeda M, et al. CDK-related protein kinases in plants. Plant Mol Biol. 2000;43:607–20. doi: 10.1023/A:1006470301554. [DOI] [PubMed] [Google Scholar]
- 75.Michaelis C, Weeks G. The isolation from a unicellular organism, Dictyostelium discoideum, of a highly-related cdc2 gene with characteristics of the PCTAIRE subfamily. Biochim Biophys Acta. 1993;1179:117–24. doi: 10.1016/0167-4889(93)90132-9. [DOI] [PubMed] [Google Scholar]
- 76.Insall R. The Dictyostelium genome: the private life of a social model revealed? Genome Biol. 2005;6:222. doi: 10.1186/gb-2005-6-6-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Signorovitch AY, Dellaporta SL, Buss LW. Molecular signatures for sex in the Placozoa. Proc Natl Acad Sci USA. 2005;102:15518–22. doi: 10.1073/pnas.0504031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miller DJ, Ball EE. Animal evolution: the enigmatic phylum placozoa revisited. Curr Biol. 2005;15:R26–8. doi: 10.1016/j.cub.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 79.Schierwater B, de Jong D, Desalle R. Placozoa and the evolution of Metazoa and intrasomatic cell differentiation. Int J Biochem Cell Biol. 2009;41:370–9. doi: 10.1016/j.biocel.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 80.Carroll AS, O’Shea EK. Pho85 and signaling environmental conditions. Trends Biochem Sci. 2002;27:87–93. doi: 10.1016/S0968-0004(01)02040-0. [DOI] [PubMed] [Google Scholar]
- 81.Torres Acosta JA, de Almeida Engler J, Raes J, Magyar Z, De Groodt R, Inzé D, et al. Molecular characterization of Arabidopsis PHO80-like proteins, a novel class of CDKA;1-interacting cyclins. Cell Mol Life Sci. 2004;61:1485–97. doi: 10.1007/s00018-004-4057-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Léopold P, O’Farrell PH. An evolutionarily conserved cyclin homolog from Drosophila rescues yeast deficient in G1 cyclins. Cell. 1991;66:1207–16. doi: 10.1016/0092-8674(91)90043-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hemerly AS, Ferreira PC, Van Montagu M, Engler G, Inzé D. Cell division events are essential for embryo patterning and morphogenesis: studies on dominant-negative cdc2aAt mutants of arabidopsis. Plant J. 2000;23:123–30. doi: 10.1046/j.1365-313x.2000.00800.x. [DOI] [PubMed] [Google Scholar]
- 84.Eloy NB, Coppens F, Beemster GT, Hemerly AS, Ferreira PC. The Arabidopsis anaphase promoting complex (APC): regulation through subunit availability in plant tissues. Cell Cycle. 2006;5:1957–65. doi: 10.4161/cc.5.17.3125. [DOI] [PubMed] [Google Scholar]
- 85.Santamaría D, Barrière C, Cerqueira A, Hunt S, Tardy C, Newton K, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448:811–5. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- 86.Sharma SK, Brock DA, Ammann RR, DeShazo T, Khosla M, Gomer RH, et al. The cdk5 homologue, crp, regulates endocytosis and secretion in dictyostelium and is necessary for optimum growth and differentiation. Dev Biol. 2002;247:1–10. doi: 10.1006/dbio.2002.0684. [DOI] [PubMed] [Google Scholar]
- 87.Cole AR. PCTK proteins: the forgotten brain kinases? Neurosignals. 2009;17:288–97. doi: 10.1159/000231895. [DOI] [PubMed] [Google Scholar]