Abstract
The DNA damage checkpoint controls cell cycle arrest in response to DNA damage, and activation of this checkpoint is in turn cell cycle-regulated. Rad9, the ortholog of mammalian 53BP1, is essential for this checkpoint response and is phosphorylated by the cyclin-dependent kinase (CDK) in the yeast Saccharomyces cerevisiae. Previous studies suggested that the CDK consensus sites of Rad9 are important for its checkpoint activity. However, the precise CDK sites of Rad9 involved have not been determined. Here we show that CDK consensus sites of Rad9 function in parallel to its BRCT domain toward checkpoint activation, analogous to its fission yeast ortholog Crb2. Unlike Crb2, however, mutation of multiple rather than any individual CDK site of Rad9 is required to completely eliminate its checkpoint activity in vivo. Although Dpb11 interacts with CDK-phosphorylated Rad9, we provide evidence showing that elimination of this interaction does not affect DNA damage checkpoint activation in vivo, suggesting that additional pathway(s) exist. Taken together, these findings suggest that the regulation of Rad9 by CDK and the role of Dpb11 in DNA damage checkpoint activation are more complex than previously suggested. We propose that multiple phosphorylation of Rad9 by CDK may provide a more robust system to allow Rad9 to control cell cycle-dependent DNA damage checkpoint activation.
Keywords: BRCT, CDK, Dpb11, Mec1, Rad53, Rad9
Introduction
The DNA damage checkpoint has an evolutionarily conserved role in controlling DNA damage-induced cell cycle arrest.1-3 RAD9 was the first DNA damage checkpoint gene identified in the yeast Saccharomyces cerevisiae and was found to control ionizing radiation-induced G2/M cell cycle arrest.4 Remarkably, a single DNA double-stranded break (DSB) in yeast is sufficient to activate the DNA damage checkpoint and cause a G2/M arrest,5 but how cells achieve these tasks is not fully understood. 53BP1, the mammalian ortholog of Rad9, mediates ATM-dependent DNA damage response in mammalian cells.6,7 Both Rad9 and 53BP1 are phosphorylated extensively by CDK.8-11 Mutation to 18 SP/TP sites, i.e., the consensus phosphorylation motif of CDK, of Rad9 was found to cause a defect in DNA damage-induced Rad53 phosphorylation, a hallmark of DNA damage checkpoint activation.12 On the other hand, mutation of Rad9 to Ser-11, a CDK phosphorylation site, was shown to act synergistically with mutations of histone methylation to reduce the chromatin association of Rad9.13 More recently, Dpb11 was found to interact with CDK-phosphorylated Rad9 via its N-terminal BRCT domains and two CDK phosphorylation sties of Rad9, i.e., S462 and T474.14 However, there is a lack of adequate in vivo evidence for the checkpoint functions of Dpb11 and the CDK phosphorylation sites of Rad9 involved in Dpb11-binding. The precise CDK sites of Rad9 that are responsible for DNA damage checkpoint activation remain undetermined. Studies of Crb2, the Rad9 ortholog in fission yeast, suggested that its C-terminal Tudor and BRCT domains aid in its recruitment to the site of DNA damage via histone modifications.15-17 Interestingly, phosphorylation of Thr-215, a CDK phosphorylation site of Crb2, acts synergistically with its BRCT domain to control the activation of Chk1, the fission yeast ortholog of Rad53, suggesting a redundant role between them.16,18 Similarly, Cut5, the fission yeast ortholog of Dpb11, has also been shown to interact with Crb2,19,20 although the biochemical basis of this interaction has not been examined. In light of these studies, it appears that Dpb11/Cut5 may be critical in recognizing CDK phosphorylated Rad9/Crb2 to mediate DNA damage checkpoint activation. To examine the underlying mechanism further, we sought the in vivo evidence for the roles of Dpb11 and CDK phosphorylation of Rad9 in DNA damage checkpoint activation.
Results
The interaction between Dpb11 and Rad9 is not required for Rad53 activation in the G2/M phase
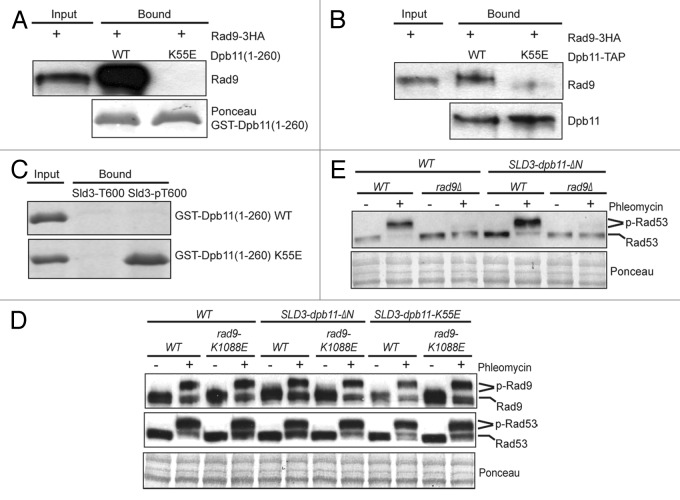
We first confirmed the interaction between the N-terminal BRCT domains of Dpb11 and Rad9 as reported previously (Fig. 1A).13,14 Further, a mutation to the conserved K55 residue within the first BRCT domain of Dpb11 abrogates its binding to endogenous Rad9–3HA derived from G2/M phase-arrested cells (Fig. 1A). The same K55E mutation of Dpb11 also eliminates the binding between full-length Dpb11 and Rad9 (Fig. 1B). Thus, Dpb11 interacts with Rad9 via its N-terminal BRCT domains as reported previously.14 We found that the same K55E mutation of Dpb11 also eliminates its interaction with a phosphopeptide that contains phosphorylated Thr-600 of Sld3, indicating that K55 is the critical residue involved in the phospho-recognition of Dpb11 (Fig. 1C). Next, we examined whether the interaction between Rad9 and Dpb11 is necessary for DNA damage checkpoint activation in vivo. Because Dpb11 is essential for cell viability, which interacts with Sld3 during DNA replication,21 we constructed a SLD3-dpb11 fusion mutant by fusing dpb11-K55E to endogenous SLD3 and then deleting DPB11 from its chromosomal locus. As reported previously,21 this fusion of Dpb11 to Sld3 bypasses the essential function of the N-terminal BRCT domains of Dpb11, allowing us to examine its role outside of DNA replication. Next, we analyzed DNA damage-induced Rad9 and Rad53 phosphorylation using their gel mobility shifts, which are the hallmark of DNA damage checkpoint activation.22,23 Unexpectedly, Rad9 and Rad53 are still hyper-phosphorylated in the SLD3-dpb11-K55E dpb11Δ mutant, similar to WT cells following phleomycin treatment, which causes DNA DSBs (Fig. 1D). To more rigorously test the role of the N-terminal BRCT domains of Dpb11, we also generated the SLD3-dpb11-ΔΝ mutant, in which an N-terminal truncated Dpb11(253–764) is fused to SLD3 with a concurrent deletion of endogenous DPB11. Once again, there is little defect in phleomycin-induced Rad9 and Rad53 phosphorylation in this SLD3-dpb11ΔN mutant (Fig. 1D). In both cases, an additional K1088E mutation in the Rad9 BRCT domain causes little synergistic effect when combined with the dpb11 mutations. To ask whether the SLD3-dpb11ΔN mutant could bypass the need for Rad9 to activate the DNA damage checkpoint, we examined the effect of rad9Δ. As shown in Figure 1E, the SLD3-dpb11ΔN rad9Δ fails to activate the checkpoint in the presence of phleomycin during G2/M phase. Therefore, despite that the Rad9 binds specifically to Dpb11 via its N-terminal BRCT domains, eliminating this interaction causes little effect on Rad9-dependent checkpoint activation in the G2/M phase.
Figure 1. The interaction between Rad9 and the N-terminal BRCT domains of Dpb11 is dispensable for DNA damage checkpoint activation. (A) Recombinant GST-Dpb11(1–260) (WT or K55E) was bound to glutathione beads and used to bind to Rad9–3HA in cell lysates derived from G2/M-arrested cells. Bound protein was detected with anti-HA antibody. (B) Effect of the K55E mutation of full-length Dpb11-TAP on its binding to endogenous Rad9–3HA from G2/M-arrested cells. The TAP tag consists of a 6xHIS-3xFLAG-Protein-A tag.24 Dpb11-TAP was first bound to the IgG beads and then used to bind to Rad9. (C) An N-terminally biotinylated Sld3 peptide corresponding to amino acids 588–612 (RVDSEENVQVQAT600PAVKKRTVTPNK) with the T600 residue unphosphorylated or phosphorylated was immobilized on straptavidin beads and used to pull-down recombinant GST-Dpb11(1–260) fragments (WT or K55E mutant). Bound proteins were detected by Coomassie blue staining. (D) Western blot analysis of Rad9–3HA and Rad53 gel mobility shift in G2/M-arrested WT, SLD3-dpb11-ΔN and SLD3-dpb11-K55E cells with or without rad9-K1088E. Cells were treated with or without phleomycin. Ponceau stain was used as a loading control. SLD3-dpb11-ΔN has an N-terminal truncated Dpb11 (amino acids 253–764). (E) western blot analysis of Rad53 gel mobility shift in G2/M-arrested WT, rad9Δ, SLD3-dpb11-ΔN and SLD3-dpb11-ΔN rad9Δ cells with or without phleomycin treatment.
N-terminal CDK consensus sites of Rad9 are involved in checkpoint activation
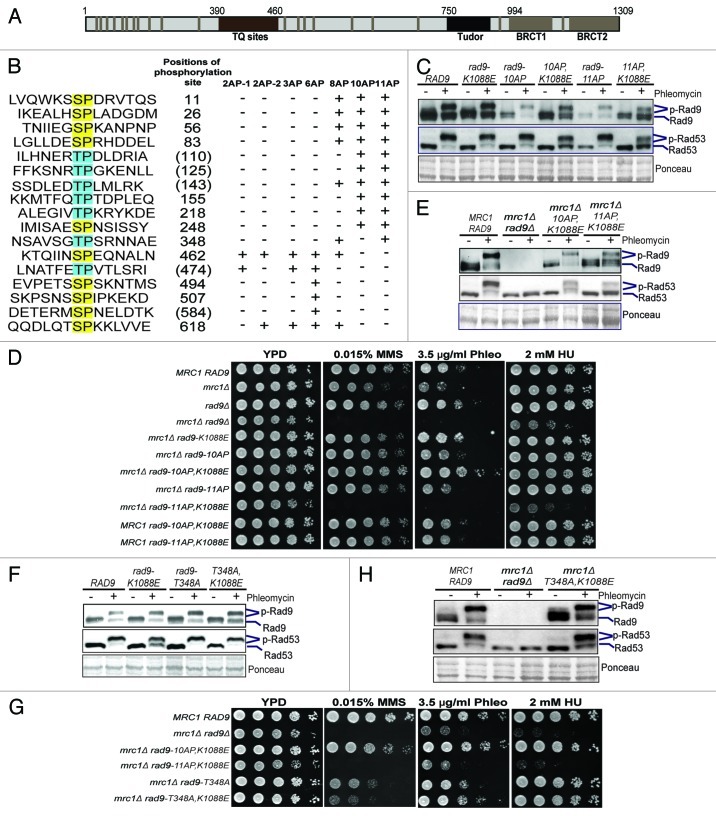
Rad9 contains C-terminal Tudor and BRCT domains, a cluster of TQ sites that are phosphorylated by Mec1 and Tel122 and 20 SP/TP sites that are targets of CDK (Fig. 2A). Previous studies have identified S462 and T474 of Rad9 to be involved in binding to Dpb11;14 however, there is a lack of significant checkpoint defect when these Ser/Thr residues are mutated into Ala. To further understand the interaction between Dpb11 and Rad9 and, more importantly, how CDK phosphorylation of Rad9 controls its checkpoint function, we introduced a number of Ser/Thr-to-Ala mutations to eliminate various CDK sites of Rad9 in its chromosomal locus (Fig. 2B). First, we examined the role of N-terminal 11 SP/TP sites of Rad9. As shown in Figure 2C, neither rad9–10AP nor rad9–11AP shows a noticeable defect in phleomycin-induced Rad53 and Rad9 phosphorylation in the G2/M phase. However, there is a small but appreciable reduction in phleomycin-induced Rad9 and Rad53 phosphorylation in the rad9–11AP,K1088E mutant, indicating a synergistic role between CDK phosphorylation and the BRCT domain of Rad9. This synergistic effect is not surprising considering that CDK phosphorylation of Crb2 in fission yeast also functions synergistically with its CDK phosphorylation of T215.15,16,18 Since Rad9 and Mrc1 function redundantly to control Rad53 activation during the cell cycle,24-26 we next analyzed these rad9 mutants in the mrc1Δ sml1Δ background. As expected, the mrc1Δ rad9Δ mutant is hypersensitive to genotoxic agents, including as little as 2 mM hydroxyurea (HU) (Fig. 2D). Among the rad9 mutants analyzed, only the rad9–11AP,K1088E mrc1Δ mutant shows a sensitivity comparable to that of mrc1Δ rad9Δ (Fig. 2D), suggesting a defective DNA damage checkpoint. Consistent with this notion, phleomycin-induced Rad9 and Rad53 phosphorylation in asynchronous cells is greatly impaired in the rad9–11AP,K1088E mrc1Δ mutant, whereas it is much less affected in the rad9–10AP,K1088E mrc1Δ mutant (Fig. 2E).
Figure 2. The N-terminal SP/TP sites of Rad9 are involved in DNA damage checkpoint activation. (A) Schematic of Rad9 domain structure and the consensus CDK phosphorylation sites (gray bars). (B) Summary of the Rad9 SP/TP phosphorylation sites, which are mutated as indicated in each rad9-AP mutant. Positions of the Ser/Thr residues are indicated to their right. Parentheses indicate phosphorylation not detected by MS.8 “+” indicates Ser/Thr-to-Ala mutations. “-“ indicates no mutation. Gel mobility shift assays of Rad9-3HA and Rad53 were performed in G2/M-arrested cells as indicated in (C and F). Plate sensitivity assays of MMS, phleomycin (Phleo) and HU with WT, mrc1Δ, rad9 and various double mutants of mrc1Δ rad9 were performed in (D and G). Gel mobility shift assays were performed using asynchronous cells, comparing WT and various double mutants of mrc1Δ rad9 as indicated in (E and H).
Considering that a single CDK phosphorylation site, i.e., T215, of Crb2 is involved in its checkpoint activity,15,16,18 we asked whether this difference between the rad9–10AP and rad9–11AP mutants is attributed to T348 of Rad9 specifically. As shown in Figure 2F and H, the rad9-T348A single mutant does not have a significant defect in Rad53 activation when combined with rad9-K1088E, mrc1Δ or both. Interestingly, the rad9-T348A mutation does cause an elevated sensitivity to methyl methane sulfonate (MMS) when it is combined with mrc1Δ and/or rad9-K1088E, indicating that T348 of Rad9 has an unspecified role in the DNA damage response (Fig. 2G). Taken together, these results show that the N-terminal SP/TP sites of Rad9 act redundantly to regulate its checkpoint activity.
C-terminal CDK consensus sites of Rad9 also act redundantly to control checkpoint activation
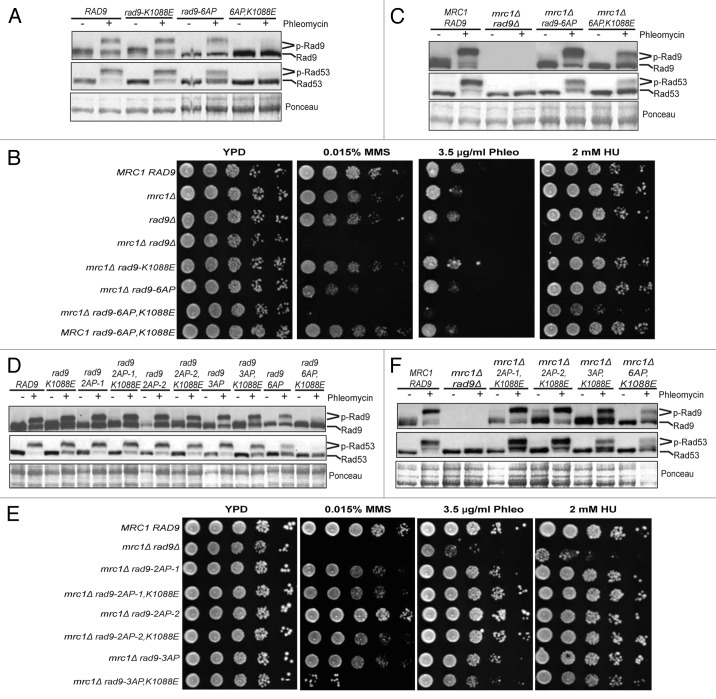
Next we examined whether the C-terminal six SP/TP sites of Rad9 regulate its checkpoint function (Fig. 2B). Mutation of all six SP/TP sites of Rad9 compromises but does not eliminate phleomycin-induced Rad9 and Rad53 phosphorylation in the G2/M phase (Fig. 3A). Interestingly, the rad9–6AP,K1088E mutant is almost completely defective in phleomycin-induced Rad53 and Rad9 phosphorylation in the G2/M phase (Fig. 3A), indicating a loss of checkpoint activity. Consistent with this notion, the rad9–6AP,K1088E mrc1Δ mutant is hypersensitive to various genotoxic agents, similar to the rad9Δ mrc1Δ mutant, whereas rad9–6AP mrc1Δ or rad9-K1088E mrc1Δ shows only modest sensitivity (Fig. 3B). Moreover, phleomycin-induced Rad53 phosphorylation in asynchronous cells is greatly reduced but not eliminated in the rad9–6AP,K1088E mrc1Δ mutant (Fig. 3C), although the phosphorylation of Rad9 is reduced to a lesser extent. Further mutagenesis of Rad9 shows that neither rad9–2AP-1 nor rad9–2AP-2 has a defect in phleomycin-induced Rad53 and Rad9 phosphorylation, regardless of the rad9-K1088E mutation (Fig. 3D). The rad9–2AP-1 mutant contains the same mutations to S462 and T474 of Rad9, as reported previously.14 Interestingly, the rad9–3AP,K1088E mutant does show a slight reduction of phleomycin-induced Rad53 phosphorylation, albeit to a lesser extent compared with rad9–6AP,K1088E (Fig. 3D). Additionally, neither rad9–2AP-1 nor rad9–2AP-2 shows an elevated sensitivity when combined with rad9-K1088E and mrc1Δ (Fig. 3E). In contrast, the rad9–3AP,K1088E mrc1Δ mutant shows a slightly higher sensitivity to MMS, although it is less than that of rad9Δ mrc1Δ and rad9–6AP,K1088E mrc1Δ. Phleomycin-induced Rad9 and Rad53 phosphorylation in asynchronous cells is essentially unaffected in the rad9–2AP-1 and rad9–2AP-2 mutants; however, it is moderately reduced in the rad9–3AP,K1088E mrc1Δ mutant (Fig. 3F). Taken together, multiple SP/TP sites in this region of Rad9 are also involved in the regulation of its checkpoint activity.
Figure 3. The C-terminal SP/TP sites of Rad9 are also involved in DNA damage checkpoint activation. Gel mobility shift assay of Rad9–3HA and Rad53 was performed using G2/M-arrested cells as indicated in (A and D) and asynchronous cells as indicated in (C and F). Plate sensitivity assay of WT, mrc1Δ, rad9 and various double mutants of mrc1Δ rad9 was performed in (B and E).
N-terminal and C-terminal CDK consensus sites of Rad9 can function together to control checkpoint activation
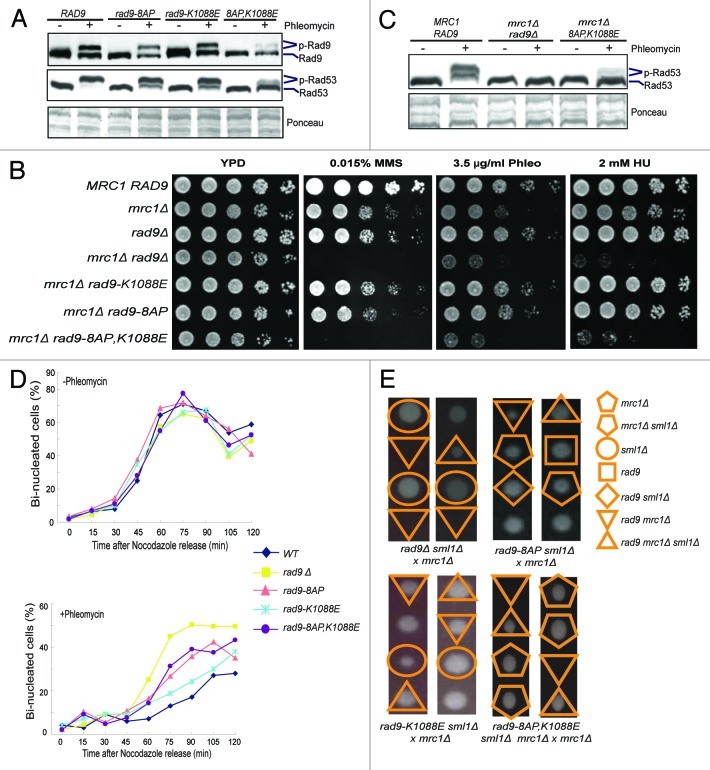
The above results demonstrate that the CDK sites in two distinct regions of Rad9 participate in checkpoint activation, which raises the possibility that they could also function redundantly. To test this, we generated the rad9–8AP mutant, which combines partial mutations to the SP/TP sites in both regions (Fig. 2B). As shown in Figure 4A, phleomycin-induced Rad9 and Rad53 phosphorylation is largely eliminated in the rad9–8AP,K1088E mutant, but not the rad9–8AP mutant. Moreover, the rad9–8AP,K1088E mrc1Δ mutant is hypersensitive to various genotoxic agents, similar to mrc1Δ rad9Δ (Fig. 4B). Phleomycin-induced Rad53 phosphorylation in asynchronous cells is also mostly eliminated in the rad9–8AP,K1088E mrc1Δ mutant (Fig. 4C), consistent with a loss of Rad9 checkpoint activity. Because Rad9 is known to control and induce cell cycle arrest in response to DNA damage,4 we next examined the nuclear division phenotypes of various rad9 mutants following a transient phleomycin treatment. As shown in Figure 4D, the percentages of bi-nucleated cells are indistinguishable between WT and the various rad9 mutants without phleomycin treatment. However, following a transient phleomycin treatment, wild-type cells show a strong delay in the formation of bi-nucleated cells, whereas rad9Δ cells display a minimal such delay, as expected. Both the rad9–8AP,K1088E and rad9–8AP cells show a mild delay, like rad9Δ, while the rad9-K1088E cells exhibit a stronger delay, similar to WT cells. Deletion of both RAD9 and MRC1 is lethal,24,27 which is suppressed by deleting SML1. Deletion of SML1 also suppresses the lethality of rad53Δ and mec1Δ.28 This genetic interaction profile provides a useful test of the ability of Rad9 to activate Rad53. Tetrad dissection analysis showed that neither rad9–8AP nor rad9-K1088E is lethal in the mrc1Δ background (Fig. 4E). Interestingly, rad9–8AP,K1088E is synthetically lethal with mrc1Δ, which is rescued by sml1Δ. Collectively, these findings show that the ability of Rad9 to promote Rad53 activation is essentially eliminated in the rad9–8AP,K1088E mutant.
Figure 4. N-terminal and C-terminal SP/TP sites of Rad9 act redundantly to control DNA damage checkpoint activation. Gel mobility shift assays of Rad9–3HA and Rad53 were performed using G2/M-arrested cells and asynchronous cells as indicated in (A and C), respectively. (B) Plate sensitivity assay of WT, mrc1Δ, rad9Δ and various double mutants of mrc1Δ rad9. (D) Percentages of bi-nucleated cells after release from phleomycin treatment in the G2/M phase, comparing WT, rad9Δ, rad9–8AP, rad9-K1088E and rad9–8AP,K1088E mutants. (E) Tetrad dissection analysis of the genetic interactions between various rad9 mutants, mrc1Δ and sml1Δ. Synthetic lethality between mrc1Δ and either rad9Δ or rad9–8AP,K1088E is rescued by sml1Δ. No synthetic lethality was detected between mrc1Δ and rad9–8AP or rad9-K1088E. Representative tetrads are shown here for simplicity. Unmarked spores contain RAD9 MRC1 SML1.
Multiple CDK sites of Rad9 mediate its interaction with Dpb11
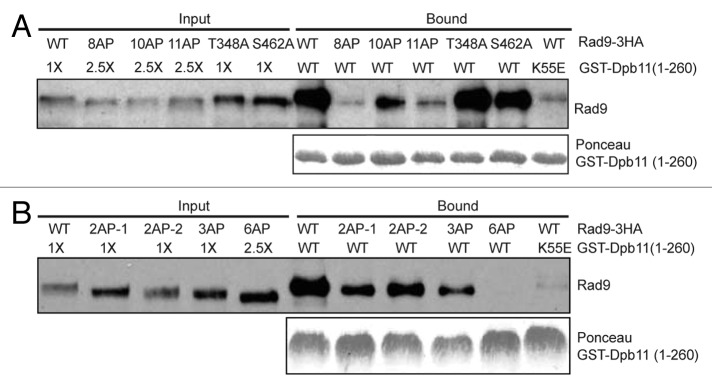
Previous studies showed that Rad9 interacts with Dpb11 via Ser-11, Ser-462 and Thr-474.13,14 Since mutation of these residues of Rad9 does not eliminate its ability to mediate Rad53 activation, we chose to examine the interaction between Dpb11 and Rad9 further. Because Dpb11 binds to Rad9 via its N-terminal BRCT domains only (Fig. 1A and B), we used Dpb11 BRCT-pull-down assay to examine its interaction with endogenous Rad9 in various rad9 mutant backgrounds. As shown in Figure 5A, while Rad9–10AP retains a weaker interaction with Dpb11, both Rad9–11AP and Rad9–8AP fail to show a significant binding to Dpb11. The T348A or S462A single mutation of Rad9 has little effect on this interaction. We also examined the effect of mutation to the C-terminal CDK consensus sites of Rad9. As shown in Figure 5B, mutation of either two or three SP/TP sites of Rad9 causes only a gradual reduction in their interactions with Dpb11, a loss of binding to Dpb11 is only observed for Rad9–6AP. This observation strongly suggests that multiple CDK consensus sites of Rad9 mediate its interaction with Dpb11.
Figure 5. Multiple CDK consensus sites of Rad9 are involved in binding to Dpb11. GST-Dpb11(1–260) was used to pull-down WT Rad9–3HA and various Rad9-AP mutant proteins in (A and B), as described in Figure 1A. 2.5-fold more cell lysates of rad9–6AP, rad9–8AP, rad9–10AP or rad9–11AP were used in the binding reactions to compensate for their reduced band intensities. GST-Dpb11(1–260) was detected by Ponceau staining.
Discussion
Rad9 has a critical role in DNA damage checkpoint activation in yeast, particularly during the G2/M phase when CDK activity is high.4,5 Previous studies have shown that a single DNA DSB in yeast is sufficient to activate the DNA damage checkpoint in a Rad9-dependent manner,3 indicating a highly sensitive signal transduction system. The key question is how Rad9 mediates this robust and sensitive checkpoint activation. Rad9 and its orthologs are known CDK targets, and they contain multiple CDK consensus sites.8-11 The observations that CDK-phosphorylated Rad9 interacts with Dpb11 and Dpb11 can activate Mec1 have led to a model that Dpb11 may be a critical regulator of Rad9.14 However, there has been a lack of in vivo evidence of such a role for Dpb11. Additionally, mutation of the SP/TP sites of Rad9 involved in binding to Dpb11 fails to cause a strong defect in Rad53 activation,14 raising the question on how CDK phosphorylation of Rad9 is involved. Here we present multiple lines of evidence, which suggest that multiple phosphorylation of Rad9 by CDK is responsible for its ability to control DNA damage checkpoint activation. Several new observations were made and summarized here.
First, despite that Dpb11 interacts with Rad9 via its N-terminal BRCT domains,13,14 elimination of this Rad9-Dpb11 interaction does not cause any appreciable defects in DNA damage-induced Rad9 and Rad53 phosphorylation (Fig. 1D). One explanation of this surprising finding is that alternative pathway(s) may exist to mediate the function of CDK-phosphorylated Rad9, which remain to be identified in further studies. Our observation also does not exclude that Dpb11 may regulate DNA damage checkpoint activation via alternative mechanism(s). For example, the C-terminal BRCT domains of Dpb11 may be involved, considering its interaction with the Mec3-Rad17-Ddc1 complex.29-31 Since Rad9 specifically interacts with the N-terminal BRCT domains of Dpb11 (Fig. 1B),14 Dpb11 cannot be the only protein that recognizes CDK-phosphorylated Rad9, regardless of its other roles.
Second, previous studies on the binding specificity of Dpb11 BRCT domains showed that they bind to CDK-phosphorylated Sld2 and Sld3 in a highly sequence specific manner.21,32 Indeed, a phosphopeptide of Sld3 containing phosphorylated T600 of Sld3 shows a robust binding to the N-terminal BRCT domains of Dpb11 (Fig. 1C).33 Our analysis of the binding between Rad9 and the same BRCT domains of Dpb11 reveals a quite different mode of interaction, despite that Rad9 also binds to Dpb11 specifically (Figs. 1A and B).14 Using the same phosphopeptide pull-down assay, we could not detect any appreciable binding between any individual phosphopeptide of Rad9, which contains each of the phosphorylated SP/TP sites, with the N-terminal BRCT domains of Dpb11 (results not shown). This promoted us to examine the binding between Dpb11 BRCT domains and endogenous Rad9 instead.
As shown in Figure 5, we observed a complex interaction profile between Dpb11 and Rad9. Mutation of the first 10 SP/TP sites of Rad9 reduces this interaction considerably. Despite a further reduction of this interaction observed for Rad9–11AP, mutation of T348 alone has relatively little effect on this interaction. On the other hand, mutation of the six C-terminal SP/TP sites of Rad9 completely eliminates its binding to Dpb11, suggesting that these residues of Rad9 have a strong role. Once again, mutation of S462 and T474 of Rad9 partially reduces its interaction with Dpb11, suggesting that these residues are partially involved in their binding to Dpb11.14 These observations raise the question whether there is any sequence specificity of Dpb11 N-terminal BRCT domains toward binding to Rad9. We reason that Rad9 and Sld3 represent two types of ligands, which are not necessarily contradictory to each other. While Sld3 has one high-affinity binding site for Dpb11, Rad9 appears to have multiple lower affinity binding sites to interact with Dpb11. There also exists a partial sequence-specificity among the CDK phosphorylation sites of Rad9 for Dpb11-binding. For example, T348, S462 and T474 of Rad9 appear to have a relatively stronger role than others. We emphasize here that one potential caveat is that mutation of a subset of SP/TP sites of Rad9 could lead to unintended effects on the phosphorylation of the other SP/TP sites of Rad9. Moreover, we cannot exclude the possibility that a residual binding between Rad9 and Dpb11 may exist for some Rad9-AP proteins, which is below the detection limit of the binding assay used here. Nevertheless, our results here are consistent with the notion that multiple CDK sites, rather than any one or two sites, of Rad9 mediate its interaction with Dpb11 to ensure a specific protein-protein interaction.
Third, consistent with that not all SP/TP sites of Rad9 have equivalent roles, our analysis of the rad9 mutant phenotypes indicated that they exhibit varying degrees of checkpoint defects. For example, the six C-terminal SP/TP sites of Rad9 appear to have a stronger role than its 11 N-terminal SP/TP sites toward Rad53 activation in the G2/M phase (comparing Figs. 2C and 3A). On the other hand, partial Rad53 activation in asynchronous cells was still detected in either the rad9–11AP,K1088E mrc1Δ or the rad9–6AP,K1088E mrc1Δ mutant (Figs. 2E and 3C). We also found that CDK sites in these two regions of Rad9 can function redundantly. The rad9–8AP mutant contains partial mutations to the SP/TP sites in both regions of Rad9. We observed a near complete failure of rad9–8AP to activate Rad53 when it is combined with rad9-K1088E and mrc1Δ. The synergistic defect in checkpoint activation when the rad9-AP mutations are combined with the K1088E mutation in the BRCT domain of Rad9 also suggests that the checkpoint defects of these rad9-AP mutants are less likely due to a defect in protein stability caused by mutations to these CDK consensus sites of Rad9. Instead, the CDK phosphorylation and the BRCT domain of Rad9 act redundantly to control its DNA damage checkpoint activity, which is reminiscent of the observations of Crb2 in fission yeast,15,16,18 although the situation with CDK phosphorylation regulation of Rad9 appears to be far more complex.
Finally, considering that Rad9 and its eukaryotic orthologs invariably have a large number of SP/TP sites that are targeted by CDK, the mechanism uncovered in the yeast Saccharomyces cerevisiae may be conserved in other eukaryotes. By employing multivalent interactions, rather than a single CDK phosphorylation event, this may enable cells to achieve the robustness in signaling as well as an ultra-sensitive response to DNA damages during the cell cycle, analogous to the multiple phosphorylation of Sic1 by CDK in controlling the cell cycle.34
Materials and Methods
Yeast strains
Standard yeast genetic methods were used to generate mutants. All rad9 mutants were introduced into the endogenous locus of RAD9 using PCR-based transformation method and confirmed via DNA sequencing.35 The details are available upon request. All strains used in this study are listed in Table S1.
Analysis of bi-nucleated cells
Log-phase cells were arrested in the G2/M phase with 15 µg/ml nocodazole for 3 h at 30°C and were then split into two sets and shifted to room temperature. One set was treated with 50 µg/ml phleomycin for 1 h. Then the cells were washed and released into fresh YPD medium. Samples were taken every 15 min, fixed with 70% ethanol, treated with RNase and pepsin and stained with propidium iodide. The bi-nucleated cells were counted blindly. Three independent experiments were performed for quantification.
Western blot analysis of Rad9 and Rad53
Cells were grown and arrested in the G2/M phase as described above or kept as asynchronous and treated with 50 µg/ml phleomycin. Protein extracts were prepared using trichloroacetic acid precipitation. Rad9–3HA and Rad53 were probed using anti-HA (3F10) and anti-Rad53 antibodies, respectively.
Pull-down assays
C-terminally TAP-tagged, full-length Dpb11 was overexpressed from the yeast strain SCY249 and was bound to the IgG resins. The GST-tagged N-terminal BRCT domains of Dpb11 (amino acids 1–260) was overexpressed from E. coli and bound to glutathione sepharose beads. The Dpb11-bound beads were then incubated with an equal amount of cell lysate prepared from yeast cells arrested in the G2/M phase. The binding was performed in TBSN buffer (50 mM Tris pH 7.2, 50 mM NaCl, 0.5% NP-40, 50 mM β-glycerophosphate, 10 mM NaF, 10 mM EDTA, 2 mM PMSF, 2X protease inhibitor mix and 2 mM DTT) at 4°C for 2 h, and the beads were washed with TBSN (concentration of NaCl was increased to 100 mM during washing) three times. The bound proteins were eluted and then detected by western blot using anti-HA antibody or by Ponceau staining.
Plate sensitivity assays
10-fold serial dilutions of log-phase cells were spotted on YPD plates with or without MMS, hydroxyurea (HU) or phleomycin (Phleo) at the indicated concentrations and incubated at 30°C for 48 to 72 h.
Supplementary Material
Acknowledgments
We would like to thank Dr. Marco Foiani for anti-Rad53 antibody and members of the Zhou lab for suggestions during the course of this study. This work was supported by NIGMS GM080469 and the Ludwig Institute for Cancer Research to H.Z.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental material may be found here: www.landesbioscience.com/journals/cc/article/21987
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21987
References
- 1.Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Toward maintaining the genome: DNA damage and replication checkpoints. Annu Rev Genet. 2002;36:617–56. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 2.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–9. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 3.Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet. 2006;40:209–35. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 4.Weinert TA, Hartwell LH. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–22. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 5.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–7. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298:1435–8. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- 7.Lee JH, Goodarzi AA, Jeggo PA, Paull TT. 53BP1 promotes ATM activity through direct interactions with the MRN complex. EMBO J. 2010;29:574–85. doi: 10.1038/emboj.2009.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics. 2008;7:1389–96. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vialard JE, Gilbert CS, Green CM, Lowndes NF. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 1998;17:5679–88. doi: 10.1093/emboj/17.19.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Vugt MA, Gardino AK, Linding R, Ostheimer GJ, Reinhardt HC, Ong SE, et al. A mitotic phosphorylation feedback network connects Cdk1, Plk1, 53BP1, and Chk2 to inactivate the G(2)/M DNA damage checkpoint. PLoS Biol. 2010;8:e1000287. doi: 10.1371/journal.pbio.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jørgensen C, Miron IM, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–26. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonilla CY, Melo JA, Toczyski DP. Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol Cell. 2008;30:267–76. doi: 10.1016/j.molcel.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granata M, Lazzaro F, Novarina D, Panigada D, Puddu F, Abreu CM, et al. Dynamics of Rad9 chromatin binding and checkpoint function are mediated by its dimerization and are cell cycle-regulated by CDK1 activity. PLoS Genet. 2010;6:6. doi: 10.1371/journal.pgen.1001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfander B, Diffley JF. Dpb11 coordinates Mec1 kinase activation with cell cycle-regulated Rad9 recruitment. EMBO J. 2011;30:4897–907. doi: 10.1038/emboj.2011.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du LL, Nakamura TM, Russell P. Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev. 2006;20:1583–96. doi: 10.1101/gad.1422606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sofueva S, Du LL, Limbo O, Williams JS, Russell P. BRCT domain interactions with phospho-histone H2A target Crb2 to chromatin at double-strand breaks and maintain the DNA damage checkpoint. Mol Cell Biol. 2010;30:4732–43. doi: 10.1128/MCB.00413-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura TM, Du LL, Redon C, Russell P. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol Cell Biol. 2004;24:6215–30. doi: 10.1128/MCB.24.14.6215-6230.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura TM, Moser BA, Du LL, Russell P. Cooperative control of Crb2 by ATM family and Cdc2 kinases is essential for the DNA damage checkpoint in fission yeast. Mol Cell Biol. 2005;25:10721–30. doi: 10.1128/MCB.25.24.10721-10730.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 1997;11:3387–400. doi: 10.1101/gad.11.24.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mochida S, Esashi F, Aono N, Tamai K, O’Connell MJ, Yanagida M. Regulation of checkpoint kinases through dynamic interaction with Crb2. EMBO J. 2004;23:418–28. doi: 10.1038/sj.emboj.7600018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–5. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz MF, Duong JK, Sun Z, Morrow JS, Pradhan D, Stern DF. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol Cell. 2002;9:1055–65. doi: 10.1016/S1097-2765(02)00532-4. [DOI] [PubMed] [Google Scholar]
- 23.Sun Z, Hsiao J, Fay DS, Stern DF. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–4. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- 24.Chen SH, Smolka MB, Zhou H. Mechanism of Dun1 activation by Rad53 phosphorylation in Saccharomyces cerevisiae. J Biol Chem. 2007;282:986–95. doi: 10.1074/jbc.M609322200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka K, Russell P. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat Cell Biol. 2001;3:966–72. doi: 10.1038/ncb1101-966. [DOI] [PubMed] [Google Scholar]
- 26.Osborn AJ, Elledge SJ. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 2003;17:1755–67. doi: 10.1101/gad.1098303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Pagé N, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–8. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 28.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–40. doi: 10.1016/S1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 29.Wang H, Elledge SJ. Genetic and physical interactions between DPB11 and DDC1 in the yeast DNA damage response pathway. Genetics. 2002;160:1295–304. doi: 10.1093/genetics/160.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puddu F, Granata M, Di Nola L, Balestrini A, Piergiovanni G, Lazzaro F, et al. Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol Cell Biol. 2008;28:4782–93. doi: 10.1128/MCB.00330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Germann SM, Oestergaard VH, Haas C, Salis P, Motegi A, Lisby M. Dpb11/TopBP1 plays distinct roles in DNA replication, checkpoint response and homologous recombination. DNA Repair (Amst) 2011;10:210–24. doi: 10.1016/j.dnarep.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 32.Masumoto H, Muramatsu S, Kamimura Y, Araki H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature. 2002;415:651–5. doi: 10.1038/nature713. [DOI] [PubMed] [Google Scholar]
- 33.Zegerman P, Diffley JF. Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature. 2010;467:474–8. doi: 10.1038/nature09373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–21. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 35.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–61. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.