Abstract
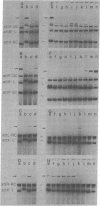
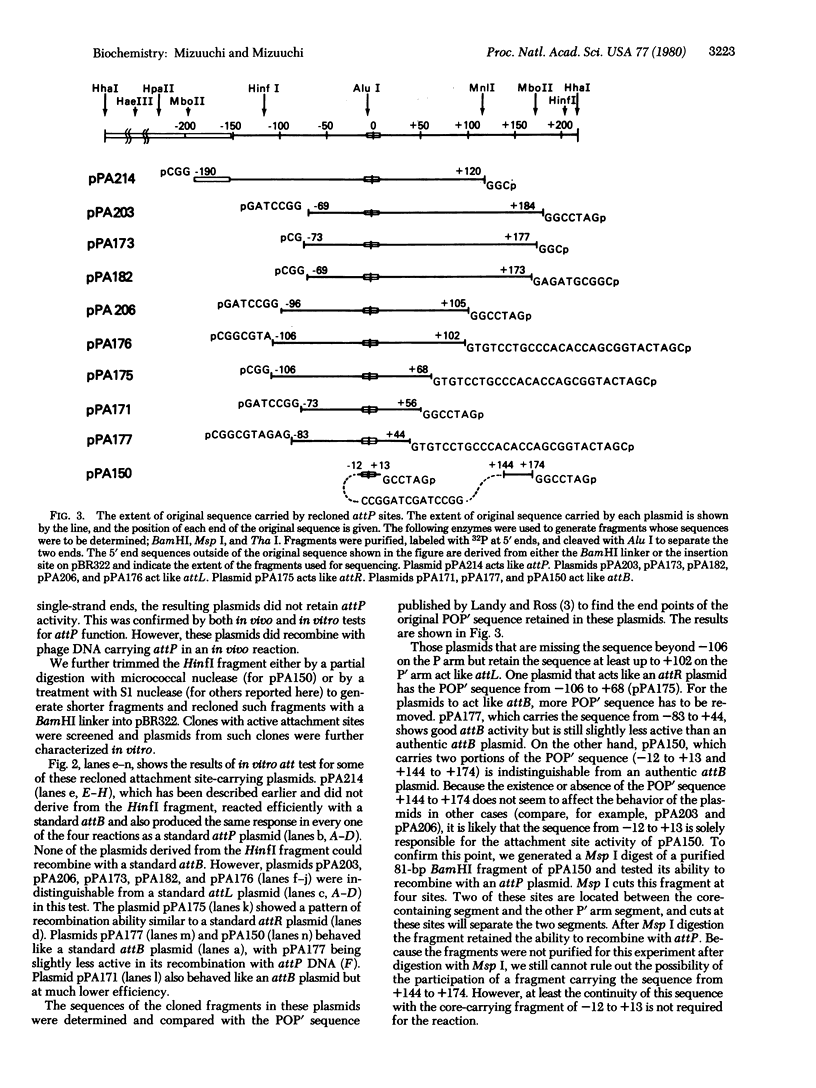
We have investigated the minimum extent of DNA sequence required for the attachment site of bacteriophage lambda to function in integrative recombination. A DNA fragment carrying the phage attachment site (attP) of bacteriophage lambda was trimmed, recloned, and tested for recombination proficiency. In order to recombine with the bacterial attachment site (attB), the phage attachment site must retain about 250 base pairs of its original sequence. On the left side, the essential sequence extends beyond 106 base pairs from the center of the 15-base-pair common core sequence but not beyond 152 base pairs. On the right side the required sequence extends beyond 68 base pairs but not beyond 99 base pairs from the center of the core. A trimmed site that has lost part of the sequence mentioned above cannot function as the phage attachment site. However, depending on which part of the sequence is present, such a site can still act in reactions normally requiring one of the prophage attachment sites or the bacterial attachment site. The results also suggest that the essential suquence of the bacterial attachment site consists only of the sequence common to the phage and bacterial attachment sites.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoess R. H., Landy A. Structure of the lambda att sites generated by int-dependent deletions. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5437–5441. doi: 10.1073/pnas.75.11.5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigman A., Hu S. L., Szybalski W. Regulation of integration by coliphage lambda: activation of int transcription by the cII and cIII proteins. Virology. 1979 Jan 30;92(2):542–556. doi: 10.1016/0042-6822(79)90156-9. [DOI] [PubMed] [Google Scholar]
- Hsu P. L., Ross W., Landy A. The lambda phage att site: functional limits and interaction with Int protein. Nature. 1980 May 8;285(5760):85–91. doi: 10.1038/285085a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., Nash H. A. Nicking-closing activity associated with bacteriophage lambda int gene product. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3760–3764. doi: 10.1073/pnas.76.8.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., Nash H. Integrative recombination of bacteriophage lambda: requirement for supertwisted DNA in vivo and characterization of int. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1099–1109. doi: 10.1101/sqb.1979.043.01.122. [DOI] [PubMed] [Google Scholar]
- Landy A., Ross W. Viral integration and excision: structure of the lambda att sites. Science. 1977 Sep 16;197(4309):1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Mizuuchi M. Integrative recombination of bacteriophage lambda: in vitro study of the intermolecular reaction. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1111–1114. doi: 10.1101/sqb.1979.043.01.123. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Nash H. A. Restriction assay for integrative recombination of bacteriophage lambda DNA in vitro: requirement for closed circular DNA substrate. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3524–3528. doi: 10.1073/pnas.73.10.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash H. A. Integration and excision of bacteriophage lambda. Curr Top Microbiol Immunol. 1977;78:171–199. doi: 10.1007/978-3-642-66800-5_6. [DOI] [PubMed] [Google Scholar]
- Nash H. A. Integrative recombination in bacteriophage lambda: analysis of recombinant DNA. J Mol Biol. 1975 Feb 5;91(4):501–514. doi: 10.1016/0022-2836(75)90276-4. [DOI] [PubMed] [Google Scholar]
- Panet A., van de Sande J. H., Loewen P. C., Khorana H. G., Raae A. J., Lillehaug J. R., Kleppe K. Physical characterization and simultaneous purification of bacteriophage T4 induced polynucleotide kinase, polynucleotide ligase, and deoxyribonucleic acid polymerase. Biochemistry. 1973 Dec 4;12(25):5045–5050. doi: 10.1021/bi00749a003. [DOI] [PubMed] [Google Scholar]
- Ross W., Landy A., Kikuchi Y., Nash H. Interaction of int protein with specific sites on lambda att DNA. Cell. 1979 Oct;18(2):297–307. doi: 10.1016/0092-8674(79)90049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk T. E., Rhodes C., Rigby P. W., Berg P. Biochemical method for mapping mutational alterations in DNA with S1 nuclease: the location of deletions and temperature-sensitive mutations in simian virus 40. Proc Natl Acad Sci U S A. 1975 Mar;72(3):989–993. doi: 10.1073/pnas.72.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman M. J., Mizuuchi K., Gottesman M. M. New att mutants of phage lambda. Virology. 1976 Jul 1;72(1):13–22. doi: 10.1016/0042-6822(76)90307-x. [DOI] [PubMed] [Google Scholar]