Abstract
Two genes coding for S-adenosyl-l-methionine synthase (SAMS, EC 2.5.1.6) were previously isolated from pea (Pisum sativum) ovaries. Both SAMS genes were highly homologous throughout their coding regions but showed a certain degree of sequence divergence within the 5′ and the 3′ untranslated regions. These regions have been used as gene-specific probes to analyze the differential expression of SAMS1 and SAMS2 genes in pea plants. The ribonuclease protection assay revealed different expression patterns for each individual gene. SAMS1 was strongly expressed in nearly all tissues, especially in roots. SAMS2 expression was weaker, reaching its highest level at the apex. Following pollination, SAMS1 was specifically up-regulated, whereas SAMS2 was expressed constitutively. The up-regulation of SAMS1 during ovary development was also observed in unpollinated ovaries treated with auxins. In unpollinated ovaries an increase in SAMS1 expression was observed as a consequence of ethylene production associated with the emasculation process. In senescing ovaries both SAMS1 and SAMS2 genes showed increased expression. Ethylene treatment of unpollinated ovaries led to an increase in the SAMS1 mRNA level. However, SAMS2 expression remained unchangeable after ethylene treatment, indicating that SAMS2 induction during ovary senescence was not ethylene dependent. SAMS mRNAs were localized by in situ hybridization at the endocarp of developing fruits and in the ovules of senescing ovaries. Our results indicate that the transcriptional regulation of SAMS genes is developmentally controlled in a specific way for each gene.
SAMS (EC 2.5.1.6) is a key enzyme in plant metabolism, catalyzing the biosynthesis of SAM from Met and ATP. SAM is a precursor for the biosynthesis of ethylene (Yang and Hoffman, 1984) and polyamines (Heby and Persson, 1990) and is involved in methylation reactions (Tabor and Tabor, 1984). SAMS cDNA clones have been isolated from several plants (Izhaki et al., 1995). Recently, two genes from pea (Pisum sativum), SAMS1 and SAMS2, were cloned in our laboratory (Gómez-Gómez and Carrasco, 1996). In all of the characterized systems, a multigene family encodes SAMS enzymes. The existence of different SAMS genes has generally been ascribed to the metabolic importance of SAM (Thomas and Surdin-Kerjan, 1987; Peleman et al., 1989b). Moreover, it has been suggested that some of the SAMS genes would be expressed constitutively, whereas others would be specifically regulated by developmental and/or environmental factors strictly controlled according to the requirement for SAM (Boerjan et al., 1994).
Marked variations in the levels of SAMS mRNA have been observed at different stages of plant development (Woodson et al., 1992; Boerjan et al., 1994; Izhaki et al., 1995) and in different plant tissues (Peleman et al., 1989a; Dekeysen et al., 1990). SAMS expression is also increased by a variety of environmental factors, including salt stress (Espartero et al., 1994; Van Breusegem et al., 1994), fungal and bacterial elicitors (Somssich et al., 1989; Gowri et al., 1991; Kawalleck et al., 1992), ozone exposure (Tuomainen et al., 1996), and mechanical stimuli (Espartero et al., 1994; Kim et al., 1994), all of which are also known to induce ethylene biosynthesis (Abeles et al., 1992). Hormonal regulation of SAMS has also been reported in wheat embryos (Mathur et al., 1992) and dwarf mutants of pea (Mathur et al., 1993), in which treatment with GA3 induced two additional isoenzymes of SAMS, and in tomato roots, in which treatment with ABA induced the accumulation of SAMS transcripts (Espartero et al., 1994).
In this paper we report a detailed study of the individual expression of SAMS genes, their developmental regulation, and the patterns of their response to auxins and ethylene in unpollinated pea ovaries, a convenient system with which to study both the regulation of senescence and the induction of fruit set by plant-growth regulators. Removing stamens 2 d before anthesis avoids self-pollination. Intact, unpollinated ovaries start to senesce naturally about 3 DPA, but treatment of unpollinated ovaries with different plant-growth regulators on the equivalent to the day of anthesis (d 0) prevents senescence and promotes fruit development (García-Martínez and Carbonell, 1980). The pea SAMS gene family consists of two genes. Analysis of the expression of the SAMS genes during ovary senescence and fruit development showed that SAMS transcript levels were up-regulated by auxins during fruit setting and by ethylene during ovary senescence (Gómez-Gómez and Carrasco, 1996). However, the high degree of identity made it impossible to differentiate the expression of each individual SAMS gene.
Using RPA and gene-specific probes, we now show that the SAMS1 gene is highly expressed during fruit development and senescence and is also expressed in pea ovaries following treatment with ethylene. Furthermore, we report that SAMS2 gene expression is reduced during auxin-induced parthenocarpic fruit development and that its expression during ovary senescence is not induced by ethylene. Spatial localization by in situ hybridization showed that SAMS mRNAs are differentially accumulated in the endocarp of developing ovaries, suggesting a role for SAM in pod wall development. We conclude that SAMS genes in pea are differentially regulated during ovary development and senescence.
MATERIALS AND METHODS
Pea (Pisum sativum L. cv Alaska) plants were grown as previously described (Carbonell and García-Martinez, 1985). Unpollinated ovaries were obtained by removing petals and stamens from flowers 2 d before anthesis. Only the first and second flowers of each plant were used for the experiments. Samples were obtained from (a) pollinated ovaries; (b) untreated, unpollinated ovaries from emasculated flowers (including presenescent and senescent ovaries); (c) parthenocarpic ovaries, which were unpollinated and derived from emasculated flowers treated on d 0 with a 2,4-D solution (20 μL of 100 μg mL−1 2,4-D in 0.1% Tween-80); and (d) unpollinated ovaries from emasculated flowers treated on d 0 with 10 μL L−1 ethylene. After the plants were harvested, samples to be used for RNA isolation were frozen in liquid N and stored at −80°C until used.
Ethylene production was measured by placing ovaries at different stages of development in 5-mL gas-tight containers for 0.5 h, after which time the headspace gas was assayed for ethylene by GC. The gas chromatograph (model GC-14BPFSC, Shimadzu, Columbia, MD) was equipped with an activated alumina column and a flame-ionization detector. An ethylene standard was used for calibration of concentration and retention time.
RNA Isolation and RPA
RNA was prepared from frozen plant material following the method of Prescott and Martin (1987) and quantified by measuring the A260. The integrity of rRNA was verified by electrophoresis through 1% (w/v) agarose gels containing 2.2 m formaldehyde. RPA (Lee and Costlow, 1987) was performed using [α-32P]UTP strand-specific radiolabeled probes generated from a 124-bp fragment of the 5′ portion of the SAMS1 cDNA and a 423-bp fragment containing the 3′ end of the SAMS2 cDNA (Gómez-Gómez and Carrasco, 1996). The sequences selected to generate the probes were different enough in both pea SAMS genes to ensure probe specificity. To synthesize the SAMS1 and SAMS2 antisense RNA probes, both SAMS cDNAs were cloned in pGEM-1 (Promega), and the plasmids were linearized with PstI and PvuII and transcribed in vitro with T7 and Sp6 RNA polymerase, respectively. Ten micrograms of total RNA was hybridized with 55,000 dpm, 760 and 250 attomoles of SAMS1- and SAMS2-labeled riboprobes, respectively.
Following hybridization, digestion buffer was added and unhybridized RNA was removed by digestion with a mixture of RNase-A and RNase-T1 at 30°C for 30 min following the directions of the manufacturer (Boehringer Mannheim). Adding 20% SDS and proteinase K and incubating at 37°C for 15 min stopped the digestion. After phenol extraction and ethanol precipitation, RNAs were separated on a 6% polyacrylamide-sequencing gel. An aliquot of undigested RNA probe was run as a marker for the size of the transcript. A quantitative measure of transcript abundance was obtained using a radioanalytical imaging system (InstantImager 2024, Packard Instruments, Meriden, CT). Radioactivity was directly measured on the gel. The data were corrected for the number of uracil residues within each probe, and the specific activity of the probes was calculated to determine the attomoles of mRNA according to the following equation:
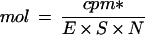 |
where cpm* is the counts per minute counted by the imager and corrected according to the reference date of the [32P]UTP, E is the efficiency of the counting, S is the specific activity of the [α-32P]UTP (800 Ci mmol−1), and N is the number of uracil residues in the probe. All values are the means of three independent experiments. Autoradiography of the gel was independently obtained on radiographic film (XAR-5, Kodak) using an intensifying screen at −80°C.
In Situ RNA Hybridization
Ovaries were harvested from flowers at various stages of development, fixed, embedded in paraffin, and tissue-sectioned as described previously (Vercher et al., 1984). In situ hybridization was carried out essentially as described by Duck (1994). Tissue samples were cut into 10-μm sections and placed on replicated slides coated with poly-l-Lys. The sections were dewaxed, hydrated, and treated with HCl, proteinase K, and acetic anhydride. Digoxigenin-labeled sense and antisense riboprobes were generated by in vitro transcription of a pGEM-1 template containing a 400-bp insert with the 5′ region of the SAMS1 cDNA (Gómez-Gómez and Carrasco, 1996). Plasmids were linearized, and digoxigenin-11-UTP was incorporated using either T7 or SP6 polymerase according to the instructions of the manufacturer (Boehringer Mannheim).
Slides were hybridized for 30 h at 45°C in 50% formamide, 30 mm NaCl, 10 mm Tris-HCl pH 7.5, 1 mm EDTA, 1× Denhardt's solution, 10% dextran sulfate, 100 mm DTT, 500 μg/mL denatured salmon-sperm DNA, 150 μg mL−1 yeast tRNA, and 0.2 μg mL−1 digoxigenin-labeled probe. Following hybridization, slides were treated with RNase-A to remove nonhybridized probe and washed once at room temperature in 2× SSC for 30 min, once at room temperature in 1× SSC for 30 min, and once at 37°C in 0.5× SSC for 30 min. Hybridization of the riboprobes was detected with anti-digoxigenin antibody conjugated to alkaline phosphatase and visualized by color development following the manufacturer's directions (Boehringer Mannheim).
RESULTS
Differential Expression of SAMS Genes in Pea Plants
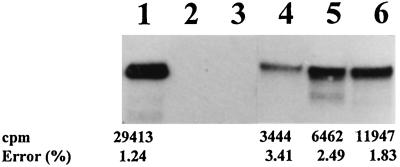
Previous work in our laboratory showed that SAMS transcripts were present in vegetative and reproductive tissues. SAMS mRNA was detected at higher levels in roots and stems than in leaves when the SAMS1-coding sequence was used as a probe for northern analysis (Gómez-Gómez and Carrasco, 1996). To determine whether both SAMS genes contributed to this expression pattern, RPA (Lee and Costlow, 1987) was performed with RNA samples from different tissues. RPA was carried out using 10 μg of total RNA, as described in Methods. Antisense RNA probes of 124 and 436 bp, respectively, were synthesized from cDNA clones for SAMS1 and SAMS2 and labeled with [α-32P]UTP. No RNase protection was observed after hybridizing the SAMS gene-specific probes to yeast tRNA or human RNA (Fig. 1).
Figure 1.
RPA of SAMS1 transcripts. Control lanes for the SAMS1 RNA probe are: lane 1, undigested probe; lane 2, SAMS1 RNA probe hybridized to yeast tRNA; lane 3, SAMS1 RNA probe hybridized to total human RNA; and lanes 4 to 6, RNA (5, 10, and 15 μg, respectively) from pollinated pea ovaries 5 DPA. All were hybridized to the SAMS1 gene-specific RNA probe, and RPA was performed as described in Methods. The counts per minute detected in the samples are shown under each band. Radioactivity was measured using a radioanalytical imaging system (see Methods).
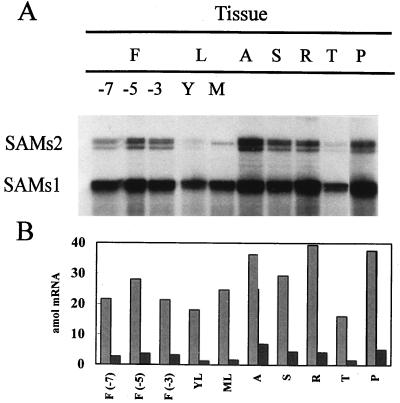
Quantification of RPA of increasing RNA amounts showed the linearity of the method (Fig. 1). As shown in Figure 2, expression of SAMS1 mRNA was strong in most tissues, especially in roots, apices, and petals; a little lower in stems, flower buds, and mature leaves; and lowest in young leaves and tendrils. By contrast, SAMS2 was expressed preferentially in the apex and was almost absent from vegetative tissues such as young and mature leaves and tendrils (Fig. 2). In general, changes in SAMS1 mRNA level were more significant than changes in SAMS2 mRNA. RPA of SAMS2 transcripts gave rise to two bands, because the local denaturation of the 3′ end of the probe located at the poly(A+) tail of the mRNA. Quantification of the counts and correction by the specific activity of the probes showed that in all tissues SAMS2 transcripts were expressed at lower levels than SAMS1 transcripts (Fig. 2B).
Figure 2.
RPA of SAMS transcripts in pea tissues. A, Ten micrograms of total RNA extracted from different tissues of pea plants was hybridized to SAMS gene-specific RNA probes, and RPA was performed. F, Floral buds (−7, −5, and −3, d 7, 5, and 3 before anthesis, respectively); L, leaves (Y, young; M, mature); A, apex; S, stem; R, root; T, tendrils; and P, petals. B, Quantification of the SAMS mRNAs at the stages shown in A. SAMS transcript levels were quantified using a radioanalytical imaging system. Light gray bars, SAMS1; dark gray bars, SAMS2. The data were corrected for the number of uracil residues within each probe.
Expression of SAMS Genes during Fruit Development
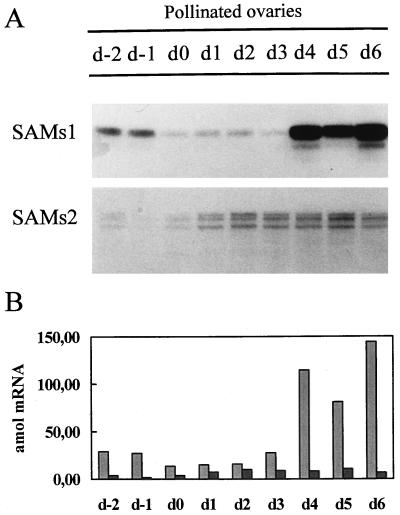
Pea ovaries ranging from 2 d before to 6 DPA were used to analyze the differential expression of the SAMS genes during fruit development. RPA analysis showed different expression patterns for SAMS1 and SAMS2 genes during fruit development (Fig. 3). Before d 0, SAMS1 was expressed at a higher level than SAMS2 mRNA, which was almost undetectable (Fig. 3). Immediately after pollination, SAMS1 transcript level decreased and remained invariable up to 3 DPA, then increased suddenly on 4 DPA, and stayed high through 6 DPA (Fig. 3). On the contrary, SAMS2 level increased after anthesis and reached its maximum after 24 h, which was maintained for the following 3 d (Fig. 3).
Figure 3.
Temporal expression of SAMS genes during fruit setting. A, Ten micrograms of total RNA extracted from pea ovaries at different stages of development were hybridized to SAMS gene-specific RNA probes, and RPA was performed. d−2 to d6, Days with respect to anthesis. B, Quantification of the SAMS mRNAs at the stages shown in A. SAMS transcript levels were quantified using a radioanalytical imaging system. Light gray bars, SAMS1; dark gray bars, SAMS2. The data were corrected for the number of uracil residues within each probe.
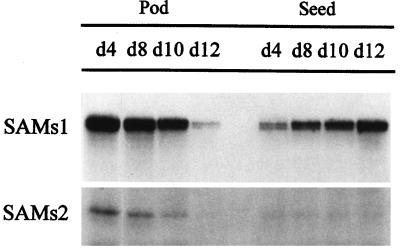
Analysis of SAMS expression in pods and seeds during fruit development showed opposite gradients of SAMS1 mRNA expression. SAMS1 mRNA accumulated during seed development (Fig. 4). On the contrary, SAMS1 mRNA level in the pod decreased from 4 to 10 DPA, when the pod began to dry (Fig. 4). In the meantime, SAMS2 levels varied in pods in a way similar to SAMS1 but were present at almost undetectable levels in seeds during fruit development (Fig. 4).
Figure 4.
Analysis of SAMS transcripts in pea pods and ovules during fruit development. Ten micrograms of total RNA extracted from pea pods and seeds from pollinated pea ovaries at different stages of development was hybridized to SAMS gene-specific RNA probes, and RPA was performed. d4 to d12, Days with respect to anthesis.
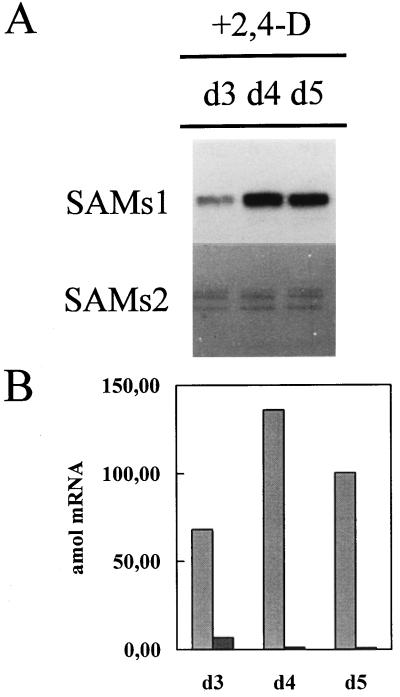
Treatment of emasculated ovaries with auxins induced parthenocarpic development, generating fruits without seeds. Previous work in our laboratory showed that SAMS transcripts accumulate only in parthenocarpic ovaries generated by auxin application in a way similar to that produced by pollination (Gómez-Gómez and Carrasco, 1996). We analyzed the expression of the individual SAMS genes during this process. RPA using SAMS1 and SAMS2 antisense RNA probes showed a differential expression pattern for both genes during auxin-induced parthenocarpy (Fig. 5). In parthenocarpic ovaries, SAMS1 mRNA levels accumulate after 3 DPA (Fig. 5) in a way similar to that observed after pollination (Fig. 3). On the contrary, SAMS2 mRNA levels were lower than those observed in pollinated ovaries (Fig. 5).
Figure 5.
Effect of 2,4-D treatment on d 0 on SAMS expression in unpollinated pea ovaries. A, Ten micrograms of total RNA extracted from 2,4-D-treated pea ovaries was hybridized to SAMS gene-specific RNA probes, and RPA was performed. d3 to d5, Days with respect to anthesis. B, Quantification of the SAMS mRNAs at the stages shown in A. SAMS transcript levels were quantified using a radioanalytical imaging system. Light gray bars, SAMS1; dark gray bars, SAMS2. The data were corrected for the number of uracil residues within each probe.
Expression of SAMS Genes in Senescing Pea Ovaries
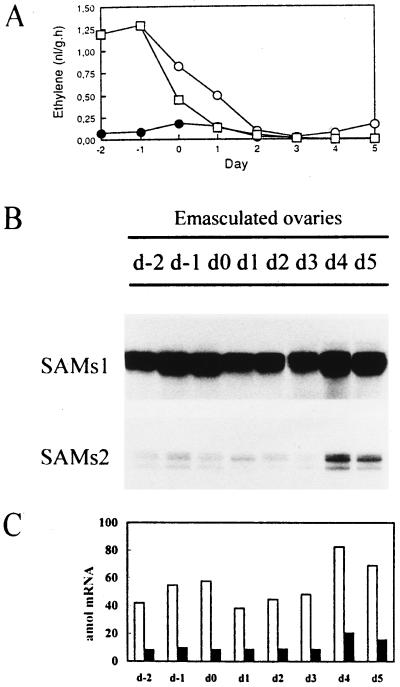
Since ethylene levels are related to the senescence of plant organs (Davies and Grierson, 1989) and because SAM is involved in ethylene biosynthesis, we determined ethylene production in pea ovaries (Fig. 6A) and compared it with SAMS expression in ovaries that had been pollinated (Fig. 3), 2,4-D-treated (Fig. 5), or emasculated (Fig. 6B). To induce senescence, flowers were emasculated 2 d before anthesis. Unpollinated ovaries grow slightly until the equivalent of 3 DPA, and then enter a degenerative process that ends with the death of the organ. A strong increase in ethylene production was immediately detected in ovaries emasculated 2 d before anthesis (Fig. 6A).
Figure 6.
Ethylene levels and temporal expression of SAMS genes during ovary senescence. A, Ethylene levels were determined in emasculated ovaries from 2 d before anthesis (at the time of emasculation) to 6 DPA. •, Ovaries allowed to be pollinated; ○, emasculated ovaries; and □, emasculated ovaries treated with 2,4-D on d 0. d-2 to d5, Days with respect to anthesis. B, Ten micrograms of total RNA extracted from the same samples used to measure ethylene was hybridized to SAMS gene-specific RNA probes, and RPA was performed. C, Quantification of the SAMS mRNAs at the stages shown in A. SAMS transcript levels were quantified using a radioanalytical imaging system. White bars, SAMS1; black bars, SAMS2. The data were corrected for the number of uracil residues within each probe.
Because of the wounding caused by the emasculation process, this ethylene increase was accompanied by an increase in the level of SAMS1 mRNA with no change in the expression of SAMS2 (Fig. 6B). Later, on the equivalent of 4 DPA, there was a slight increase in ethylene production that could be associated with ovary senescence (Fig. 6A). There was a transcript accumulation of SAMS1 mRNA at 4 DPA that was associated with the onset of senescence (Fig. 6, B and C). Thereafter, a decline in SAMS1 mRNA abundance was associated with the advanced stages of ovary senescence. The SAMS2 mRNA level remained almost unchanged in emasculated ovaries but was slightly induced at the onset of ovary senescence (Fig. 6B).
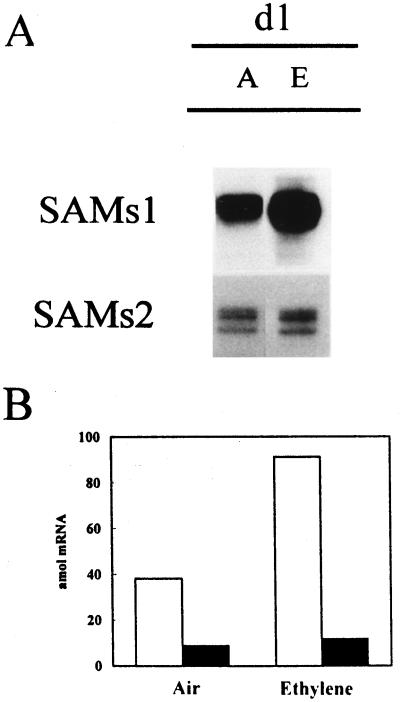
We examined the effect of exogenous ethylene on SAMS1 and SAMS2 gene expression in emasculated pea ovaries on d 0. As shown in Figure 7, the levels of SAMS1 mRNA increased in the ovaries in response to ethylene treatment. The level of SAMS1 mRNA after 24 h of treatment was the same as the level in senescing ovaries (Figs. 5 and 6). In contrast to SAMS1 mRNA, SAMS2 mRNA remained almost constant, which is consistent with the lack of stimulation of this gene during the emasculation process.
Figure 7.
Effect of ethylene treatment on SAMS expression in unpollinated pea ovaries. Treatment was made on d 0, and ovaries were collected 24 h later. A, Ten micrograms of total RNA extracted from ethylene-treated pea ovaries was hybridized to SAMS gene-specific RNA probes, and RPA was performed. Lane A, Air; lane E, after ethylene treatment. B, Quantification of the SAMS mRNAs at the stages shown in A. SAMS transcript levels were quantified using a radioanalytical imaging system. White bars, SAMS1; black bars, SAMS2. The data were corrected for the number of uracil residues within each probe.
Spatial Distribution of SAMS Genes in Pea Ovaries
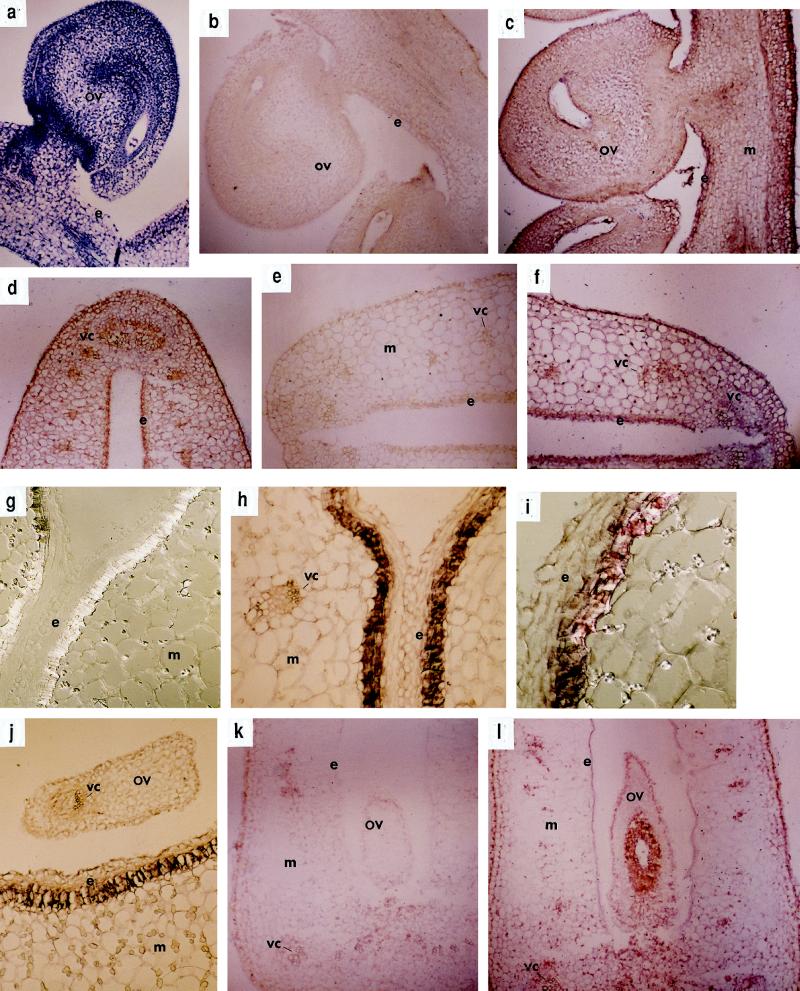
Previous reports have described the expression of SAMS-GUS reporter genes in roots, stems, and leaves of Arabidopsis (Peleman et al., 1989a) and tobacco (Peleman et al., 1989b) and in leaves of rice (Dekeysen et al., 1990). We determined the spatial distribution of SAMS genes at different stages of ovary development and senescence. In situ hybridization was performed using SAMS1 (Gómez-Gómez and Carrasco, 1996) sense and antisense RNA probes. Hybridization of longitudinal (Fig. 8c) and transverse (Fig. 8d) sections of pollinated ovaries on d 0 with an antisense SAMS probe revealed a general pattern of expression in all living cells, especially in vascular tissue, the ovule, and ovary epidermal cells. Control for cell density, which was accomplished by staining a longitudinal section with toluidine blue, showed that higher SAMS expression was not correlated with greater cell density (Fig. 8a).
Figure 8.
(Figure appears on facing page.)
In situ localization of SAMS mRNAs in pea ovaries. Pollinated and unpollinated ovaries at different stages of development were sectioned and hybridized to an antisense SAMS1 probe. a, b, and c, Longitudinal sections of a pollinated pea ovary at anthesis stained with toluidine blue (a) or hybridized with a SAMS sense probe (b) or a SAMS antisense probe (c). d to h, Transverse sections of a pollinated pea ovary at anthesis hybridized with a SAMS antisense probe (d), at 3 DPA hybridized with a SAMS sense probe (e) or a SAMS antisense probe (f), and at 5 DPA hybridized with a SAMS sense probe (g) or a SAMS antisense probe (h). i, Detail of a transverse section of a pollinated pea ovary 5 DPA hybridized with a SAMS antisense probe. j, Transverse section of a pollinated pea ovary at 5 DPA, including an ovule, hybridized with a SAMS antisense probe. k and l, Transverse sections of an emasculated pea ovary at the equivalent to 4 DPA hybridized with a SAMS sense probe (k) and a SAMS antisense probe (l). Bar in i = 200 μm; bars in all other panels = 100 μm. e, Endocarp; m, mesocarp; ov, ovule; and vc, vascular cell. {/ANNT;152064n;;11968n;1056n}
In pollinated ovaries on 3 DPA, SAMS was expressed mostly in the endocarp and vascular bundles (Fig. 8f). This pattern of expression was reinforced 5 DPA (Fig. 8, h–j), when the transcripts were present in cells associated with the vascular tissue of developing seed and pod mesocarp. Moreover, at this stage of development, SAMS mRNAs were particularly abundant in endocarp cells. In senescing, unpollinated ovaries 4 DPA, SAMS mRNAs were localized in vascular tissues and also in the aborted ovules (Fig. 8l). No signal was detected when a sense probe was used to hybridize sections at different stages (Fig. 8, b, e, g, and k).
DISCUSSION
SAMS genes have been considered to be “housekeeping genes” that are expected to be expressed constitutively in all tissues. Previous work showed that SAMS transcripts were induced in developing pea ovaries in response to pollination and auxins and also during ovary senescence (Gómez-Gómez and Carrasco, 1996). However, northern analysis was not able to determine whether the genes were biologically equivalent or whether they were differentially regulated. In this work we used RPA with gene-specific probes to distinguish the expression pattern of each SAMS gene. The steady-state levels of mRNA, corresponding to both SAMS1 and SAMS2 genes, were monitored for different pea tissues at different developmental stages and in response to different plant hormones.
Our studies demonstrate that SAMS1 and SAMS2 genes are individually regulated, with their transcripts accumulating to varying degrees among different tissues and at different stages of ovary development or senescence. SAMS1 mRNA was found to be expressed at higher levels than SAMS2 in all tissues. In fact, the SAMS1 expression pattern was found to be the same as that obtained by northern analysis using a probe able to hybridize both SAMS1 and SAMS2 genes (Gómez-Gómez and Carrasco, 1996). Nonetheless, the expression pattern for SAMS2 was quite different, although its contribution to the pool of SAMS mRNAs was much lower. A similar relationship between SAMS genes has been found in tomato, in which transcripts corresponding to the sam1 gene were the most abundant, although the expression pattern for each gene was different in each tissue (Espartero et al., 1994).
In contrast, the two Arabidopsis SAMS genes show a similar expression pattern (Peleman et al., 1989b): SAMS1 was highly expressed in roots, apex, and petals and also accumulated to significant levels in the other tissues examined; preferential expression of SAMS2 was found in the apex. The expression of both genes in this tissue could be due to polyamine biosynthesis, which is required for cell proliferation in dividing tissues (Evans and Malmberg, 1989), and to the biosynthesis of macromolecules associated with the meristematic activity of this tissue.
In the apex SAM could be decarboxylated and used for the biosynthesis of spermidine and spermine. This would agree with the high levels of Arg decarboxylase mRNA and enzymatic activity detected in the pea apex (Pérez-Amador and Carbonell, 1995; Pérez-Amador et al., 1995). In floral buds both mRNAs change their levels coordinately according to the developmental stage of the organ. A developmental stage pattern has also been observed during corolla development in petunia (Izhaki et al., 1995) and has been related to the rapid growth of the tissue (Weiss and Halevy, 1989).
Expression of SAMS Genes in Fruits Is Developmentally Regulated
Before the day of anthesis, coinciding with a period of elevated cell division rate, SAMS1 was highly expressed, whereas SAMS2 mRNA was almost undetectable. At the time of anthesis, when cell division was about to conclude, the relationship between SAMS1 and SAMS2 expression changed. SAMS1 mRNA expression decreased, and SAMS2 mRNA expression increased slightly 24 h after anthesis and remained almost constant through 6 DPA. The analysis of SAMS2 expression in pods and seeds showed that the expression detected in fruits was almost completely restricted to the pod. In pollinated ovaries, the most dramatic increase in SAMS1 expression was detected 4 DPA. SAMS1 expression was mainly due to the level of SAMS1 mRNA in the pod and was localized primarily to cells of the presclerenchyma zone of the endocarp. At this time the differentiation of the endocarp cells into sclerenchyma fibers starts (Vercher et al., 1984).
The accumulation of SAMS1 mRNA in cells under lignification would suggest that SAMS1 is involved in the biosynthesis of the secondary cell wall. During this process SAM is needed for methylation of lignin monomers before polymerization (Higuchi, 1981). At 8 DPA, when the pod reached its maximum length, SAMS1 expression decreased. Then, as the pod began to dry out, the amount of dry matter declined. Simultaneously, the seeds enlarged rapidly and increased in fresh weight by accumulation of dry matter, partly at the expense of the pod wall (Eeuwens and Schwabe, 1975). During pea seed development the synthesis and amount of storage and metabolic proteins increases (Bewley and Black, 1994). SAMS1 mRNA also increased, suggesting the participation of SAM in macromolecule biosynthesis as a methyl group donor. Moreover, the presence of stored SAMS mRNA in wheat embryos (Mathur et al., 1991) indicates that SAMS1 mRNA could remain in the dry seed and then be reutilized upon subsequent hydration. mRNAs coding for enzymes essential for intermediary metabolism and proteins essential for the successful completion of germination have been found in the stored or conserved messages in dry seeds (Kermade, 1990).
SAMS Genes Are Differentially Regulated in Parthenocarpic Developing Ovaries
The expression of SAMS1 and SAMS2 genes was investigated in parthenocarpic pea ovaries induced by auxin treatment. The weight of these ovaries is lower than that of the pollinated ovaries, but at the histological and physiological levels the developmental pattern of the ovaries is the same (Vercher and Carbonell, 1991). Northern analysis showed that in pollinated and auxin-induced parthenocarpic ovaries the expression pattern of SAMS genes was the same (Gómez-Gómez and Carrasco, 1996). However, the individual analysis of SAMS transcripts showed that SAMS1 mRNA was induced in auxin-treated unpollinated ovaries as well as in pollinated ovaries, which suggests that similar mechanisms are involved in the transcriptional control of the SAMS1 gene.
On the contrary, SAMS2 expression was dramatically reduced in parthenocarpic fruits. This could be due to the lack of pollination and/or fertilization in the parthenocarpic ovaries that may provide the stimulus for SAMS2 mRNA induction, which began at anthesis in pollinated ovaries. In addition, since auxins induce the parthenocarpic development of the ovaries, generating fruits without seeds, the lack of induction of SAMS2 mRNA could be associated with factors originating in the seeds and translocated to the pod, where they induce SAMS2 transcription.
SAMS Gene Expression during Ovary Senescence
The involvement of ethylene in pea ovary senescence has already been reported (Gómez-Gómez and Carrasco, 1996). During senescence, the increase in SAMS gene expression was dependent on ethylene production. In the present study we showed that ethylene increases in unpollinated pea ovaries as a consequence of the emasculation process and during senescence. Both of these increases in ethylene production are accompanied by increases in SAMS expression. In both cases SAMS1 was expressed at higher levels, whereas the SAMS2 transcript accumulated only at the onset of ovary senescence and was less abundant than SAMS1 mRNA. Because only the SAMS1 gene was induced by ethylene, accumulation of SAMS1 mRNA after emasculation and during ovary senescence could be associated with an increase in ethylene biosynthesis.
Increases in SAMS gene expression in response to mechanical stimuli have been reported in wounded Arabidopsis leaves (Kim et al., 1994) and tomato roots, in which only one of the three SAMS genes was induced (Espartero et al., 1994). However, induction of SAMS2 mRNA at the onset of senescence was ethylene independent and could have been age related. Leaf senescence in Arabidopsis (Grbic and Bleecker, 1995) and leaf senescence and ripening in tomato (Picton et al., 1993) are developmental processes that are regulated by the interplay between putative age-related factors and ethylene. In unpollinated pea ovaries, these age-related factors could determine the sensitivity of ovary tissue to ethylene and the lack of sensitivity of ovary tissue to plant-growth substances 5 d after emasculation (García-Martínez and Carbonell, 1980).
The expression of SAMS genes in unpollinated pea ovaries at the equivalent of 4 DPA was localized specifically in cells of the embryo sac and in vascular cells, which were differentiating at this time (Vercher et al., 1989) and in which SAM is necessary for lignin biosynthesis. Expression of other genes related to the senescence process has also been observed during the differentiation of vascular cells (Granell et al., 1992; Nadeau et al., 1993; Tang et al., 1994), a process related to senescence (Jones and Dangl, 1996).
In conclusion, SAMS1 and SAMS2 mRNAs are differentially expressed during ovary development in pea. Ethylene induction of SAMS1 mRNA and the lack of induction of SAMS2 mRNA by ethylene-treated and parthenocarpic fruits suggest the existence of different factors controlling SAMS gene transcription. These findings raise the question of how this separate action is established and maintained. Future research will focus on the mechanism regulating SAMS1 and SAMS2 expression during ovary development in pea.
ACKNOWLEDGMENTS
The authors are grateful to Drs. A. Molowny and C. López-García for their assistance with tissue preparation for the in situ hybridization experiments and to R. Martínez-Pardo and A. Villar for their technical assistance at the greenhouse.
Abbreviations:
- DPA
days postanthesis
- RPA
RNase protection assay
- SAM
S-adenosyl-l-Met
- SAMS
S-adenosyl-l-Met synthase
Footnotes
This research was supported by grant no. PB92-0018-C02-02 from Dirección General de Investigación Científica y Tecnológica (Spain) to P.C. L.G.G. was funded by the Conselleria d'Educació i Ciència de la Generalitat Valenciana.
LITERATURE CITED
- Abeles FB, Morgan PW, Saltveit ME (1992) Regulation of ethylene production by internal, environmental, and stress factors. In FB Abeles, PW Morgan, ME Salveit, eds, Ethylene in Plant Biology, Ed 2. Academic Press, New York, pp 56–119
- Bewley JD, Black M (1994) Seed development and maturation. In JD Bewley, M Black, eds, Seed, Physiology of Development and Germination, Ed 2. Plenum Press, New York, pp 35–115
- Boerjan W, Bauw G, Van Montagu M, Inzé D. Distinct phenotypes generated by overexpression and suppression of S-adenosyl-l-methionine reveal developmental patterns of gene silencing in tobacco. Plant Cell. 1994;6:1401–1414. doi: 10.1105/tpc.6.10.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell J, García-Martinez JL. Ribulose-1,5-bisphosphate carboxylase and fruit set or degeneration of unpollinated ovaries of Pisum sativum. Planta. 1985;164:534–539. doi: 10.1007/BF00395972. [DOI] [PubMed] [Google Scholar]
- Davies KM, Grierson D. Identification of cDNA clones for tomato (Lycopersicom esculentum Mill). mRNAs that accumulate during ripening and leaf senescence. Planta. 1989;179:73–80. doi: 10.1007/BF00395773. [DOI] [PubMed] [Google Scholar]
- Dekeysen RA, Claes B, De Rycke R, Mabets M, Van Montagu M, Caplan A. Transient gene expression in intact and organized tissues. Plant Cell. 1990;2:591–602. doi: 10.1105/tpc.2.7.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duck NB. RNA in situ hybridisation in plants. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual, Ed 2. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 1–13. [Google Scholar]
- Eeuwens CJ, Schwabe WW. Seed and pod wall development in Pisum sativum, L. in relation to extracted and applied hormones. J Exp Bot. 1975;26:1–14. [Google Scholar]
- Espartero J, Pintor-Toro JA, Pardo JM. Differential accumulation of S-adenosylmethionine synthetase transcripts in response to salt stress. Plant Mol Biol. 1994;25:217–227. doi: 10.1007/BF00023239. [DOI] [PubMed] [Google Scholar]
- Evans TP, Malmberg RL. Do polyamines have roles in plant development? Annu Rev Plant Physiol Plant Mol Biol. 1989;40:235–269. [Google Scholar]
- García-Martínez JL, Carbonell J. Fruit-set of unpollinated ovaries of Pisum sativum L. Influence of plant growth regulators. Planta. 1980;147:451–456. doi: 10.1007/BF00380187. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Carrasco P. Hormonal regulation of S-adenosylmethionine synthetase transcripts in pea ovaries. Plant Mol Biol. 1996;30:821–832. doi: 10.1007/BF00019014. [DOI] [PubMed] [Google Scholar]
- Gowri G, Bugos R, Campbell W, Maxwell C, Dixon R. Stress responses in alfalfa (Medicago sativa) Plant Physiol. 1991;97:7–14. doi: 10.1104/pp.97.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granell A, Harris N, Pisabarro AG, Carbonell J. Temporal and spatial expression of a thiol protease gene during pea ovary senescence, and its regulation by gibberellin. Plant J. 1992;2:907–915. doi: 10.1046/j.1365-313x.1992.t01-5-00999.x. [DOI] [PubMed] [Google Scholar]
- Grbic V, Bleecker A. Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 1995;8:595–602. [Google Scholar]
- Heby O, Persson L. Molecular genetics of polyamine synthesis in eukaryotic cells. Trends Biochem Sci. 1990;15:153–158. doi: 10.1016/0968-0004(90)90216-x. [DOI] [PubMed] [Google Scholar]
- Higuchi T. Biosynthesis of lignin. In: Tanner W, Loewurs FA, editors. Encyclopedia of Plant Physiology, Vol 138, New Series, Plant Carbohydrates II. Berlin: Springer-Verlag; 1981. pp. 194–224. [Google Scholar]
- Izhaki A, Shoseyov O, Weiss D. A petunia cDNA encoding S-adenosyl-methionine synthase. Plant Physiol. 1995;108:841–842. doi: 10.1104/pp.108.2.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Dangl JL. Logjam at the styx: programmed cell death in plants. Trends Plant Sci. 1996;1:114–119. [Google Scholar]
- Kawalleck P, Plesch G, Halhbrock K, Somssich IE. Induction of S-adenosyl-l-methionine synthetase and S-adenosyl-l-homocysteine hydrolase mRNAs in cultured cells and leaves of Petroselium crispum. Proc Natl Acad Sci USA. 1992;89:4713–4717. doi: 10.1073/pnas.89.10.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermade AR. Regulatory mechanisms involved in the transition from seed development to germination. CRC Crit Rev Plant Sci. 1990;9:155–195. [Google Scholar]
- Kim CS, Kwak JM, Nam HG, Kim K, Cho BH. Isolation and characterization of two cDNAs that are rapidly induced during the wound response of Arabidopsis thaliana. Plant Cell Rep. 1994;13:340–343. doi: 10.1007/BF00232633. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Costlow NA. A molecular titration assay to measure transcript prevalence levels. Methods Enzymol. 1987;152:633–648. doi: 10.1016/0076-6879(87)52070-5. [DOI] [PubMed] [Google Scholar]
- Mathur M, Saluja D, Sachar RC. Post-translational regulation of S-adenosylmethionine synthetase from its stored mRNA in germinated wheat embryos. Biochim Byophys Acta. 1991;1078:161–170. doi: 10.1016/0167-4838(91)99005-d. [DOI] [PubMed] [Google Scholar]
- Mathur M, Satpthy M, Sachar RC. Phytohormonal regulation of S-adenosylmethionine synthetase by gibberellic acid in wheat aleurones. Biochim Biophys Acta. 1992;1137:161–170. doi: 10.1016/0167-4889(92)90155-5. [DOI] [PubMed] [Google Scholar]
- Mathur M, Sharma N, Sachar RC. Differential regulation of S-adenosylmethionine synthetase isozymes by gibberellic acid in dwarf pea epicotyls. Biochim Biophys Acta. 1993;1162:283–290. doi: 10.1016/0167-4838(93)90292-y. [DOI] [PubMed] [Google Scholar]
- Nadeau JA, Zhang XS, Nair H, O′Neill SD. Temporal and spatial regulation of 1-aminocyclopropane-1-carboxylate oxidase in the pollination-induced senescence of orchid flowers. Plant Physiol. 1993;103:31–39. doi: 10.1104/pp.103.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleman J, Boerjan W, Engler G, Seurinck J, Botterman J, Alliotte T, Van Montagu M, Inzé D. Strong cellular preference in the expression of a housekeeping gene of Arabidopsis thaliana encoding S-adenosyl-methionine synthetase. Plant Cell. 1989a;1:81–93. doi: 10.1105/tpc.1.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleman J, Saito K, Cottyn B, Engler G, Seurinck J, Van Montagu M, Inzé D. Structure and expression analyses of the S-adenosylmethionine synthetase gene family in Arabidopsis thaliana. Gene. 1989b;84:359–369. doi: 10.1016/0378-1119(89)90510-6. [DOI] [PubMed] [Google Scholar]
- Pérez-Amador MA, Carbonell J. Arginine decarboxylase and putrescine oxidase in ovaries of Pisum sativum L. Plant Physiol. 1995;107:865–872. doi: 10.1104/pp.107.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Amador MA, Carbonell J, Granell A. Expression of arginine decarboxylase is induced during early fruit development and in young tissues of Pisum sativum L. Plant Mol Biol. 1995;28:997–1009. doi: 10.1007/BF00032662. [DOI] [PubMed] [Google Scholar]
- Picton S, Barton SL, Bouzayen M, Hamilton AJ, Grierson D. Altered fruit ripening and leaf senescence in tomatoes expressing an antisense ethylene-forming enzyme transgene. Plant J. 1993;3:469–481. [Google Scholar]
- Prescott A, Martin C. A rapid method for the quantitative assessment of levels of specific mRNAs in plants. Plant Mol Biol Rep. 1987;4:219–224. [Google Scholar]
- Somssich JE, Bollmann J, Hahlbrock L, Kombrink E, Schultz W. Differential early activation of defence-related genes in elicitor-treated parsley cells. Plant Mol Biol. 1989;12:227–234. doi: 10.1007/BF00020507. [DOI] [PubMed] [Google Scholar]
- Tabor CW, Tabor H. Methionine adenosyltransferase (S-adenosylmethionine synthetase) and S-adenosylmethionine decarboxylase. Adv Enzymol. 1984;56:251–282. doi: 10.1002/9780470123027.ch4. [DOI] [PubMed] [Google Scholar]
- Tang X, Gomes AMTR, Bhatia A, Woodson WR. Pistil-specific and ethylene-regulated expression of 1-aminocyclopropane-1-carboxylate oxidase genes in petunia flowers. Plant Cell. 1994;6:1227–1239. doi: 10.1105/tpc.6.9.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D, Surdin-Kerjan Y. SAMS1, the structural gene for one of the S-adenosylmethionine synthetases in Saccharomyces cerevisiae. J Biol Chem. 1987;262:16704–16709. [PubMed] [Google Scholar]
- Tuomainen J, Betz C, Ernst D, Langebartels C, Sandermann H Jr, Kangasjärvi J (1996) Ozone affects the ethylene biosynthetic pathway at both biochemical and mRNA levels. NATO Advanced Research Workshop. In Biology and Biotechnology of the Plant Hormone Ethylene. Chania, Greece, p 33
- Van Breusegem F, Dekeyser R, Gielen J, Van Montagu M, Kaplan A. Characterization of a S-adenosylmethionine synthase gene in rice. Plant Physiol. 1994;105:1463–1464. doi: 10.1104/pp.105.4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercher Y, Carbonell J. Changes in the structure of ovary tissues and in the ultrastructure of mesocarp cells during ovary senescence or fruit development induced by plan growth substances in Pisum sativum. Physiol Plant. 1991;81:518–526. [Google Scholar]
- Vercher Y, Carrasco P, Carbonell J. Biochemical and histochemical detection of endoproteolytic activities in ovary senescence or fruit development in Pisum sativum. Physiol Plant. 1989;76:405–411. [Google Scholar]
- Vercher Y, Molowny A, López C, García-Martínez JL, Carbonell J. Structural changes in the ovary of Pisum sativum L. induced by pollination and gibberellic acid. Plant Sci Lett. 1984;36:87–91. [Google Scholar]
- Weiss D, Halevy AH. Stamens and gibberellin in the regulation of corolla pigmentation and growth in Petunia hibrida. Planta. 1989;179:89–96. doi: 10.1007/BF00395775. [DOI] [PubMed] [Google Scholar]
- Woodson WR, Park KY, Drory A, Larsen PB, Wang H. Expression of ethylene biosynthetic pathway transcripts in senescing carnation flowers. Plant Physiol. 1992;99:526–532. doi: 10.1104/pp.99.2.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol. 1984;35:155–189. [Google Scholar]