Abstract
We observed that mouse neural progenitor cells (NPCs) have a secretory protein profile distinct from other brain cells and modulate microglial activation, proliferation, and phagocytosis in culture and in vivo. NPC-derived vascular endothelial growth factor was necessary and sufficient to exert at least some of these effects in mice. Neural stem or precursor cells may thus not only be shaped by microglia but regulate in turn microglia functions and activity.
Neural stem cells (NSCs) in the adult mammalian brain can give rise to rapidly dividing neural progenitor cells (NPCs) to produce neurons, astrocytes and oligodendrocytes and functionally contribute to cognition and repair processes after injury1. Cultured stem cells derived from adult or neonatal brains typically contain few NSCs and are made up mostly of NPCs instead2. Transplanted NPCs, even in small numbers, have beneficial effects on recovery from CNS trauma or disease3, 4, but the mechanisms of action remain unclear. And while increasing evidence shows how local environmental cues, including microglia and secreted proteins5, influence NPC functionality, only few studies have investigated whether NPCs may secrete signaling factors and thus influence their own environment6. In support of this idea, tissue specific stem cells outside the brain have been shown to secrete various cytokines, chemokines, and other intercellular signaling proteins7, and systemically administered NPCs taking residence in draining lymph nodes can modulate dendritic cell activity in models of multiple sclerosis8.
In investigating the interactions of NPCs with other cells types in vivo, we observed that microglia are more densely populated and proliferative in neurogenic niches and appear closely associated with NPCs, suggesting that endogenous microglia and NPCs are well-positioned to interact with each other (Fig. 1). To gain a broader view of the communication between cell types within the neurogenic niche and the potential of NPCs to modulate their environment, we characterized the profile of well-known secreted signaling proteins. We measured 58 secreted signaling proteins available on multiplex antibody-based immunoassays in cell culture supernatants collected from mouse primary microglia, astrocytes, neurons, and NPCs. Of the 46 factors detectable, each cell type secreted a distinct profile of signaling factors, as visualized by unbiased cluster analysis, and some factors could classify the four cell types (Fig. 2a). We identified several factors prominently secreted by NPCs, including tissue inhibitor of metalloproteinase type-1 (TIMP-1), vascular endothelial cell growth factor (VEGF), and haptoglobin (Fig. 2b). Notably, several of these factors are already known to be immunomodulatory proteins and in particular are prominent regulators of microglia9, 10. Given these observations, we hypothesized that NPCs may not only be regulated by microglia as suggested by others5, but that they may in turn be capable of regulating microglia as well. We thus tested whether NPC-derived factors can modulate major cellular functions of microglia.
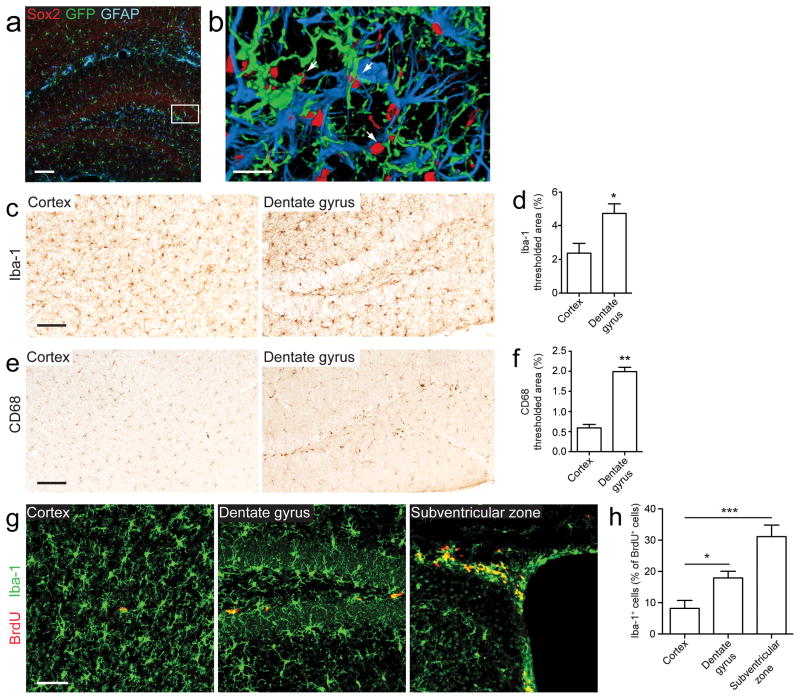
Figure 1. Microglia and neural progenitor cells are well-positioned to interact in vivo.
(a) A hippocampal section from a 2-month-old CX3CR1GFP/wt mouse brain stained for the stem cell marker Sox2 (red), GFP+ microglia (green), and the astrocyte marker GFAP (blue) demonstrates that microglia and NPCs within the neurogenic niche are closely associated. (b) A high-magnification three-dimensional reconstruction of the area in the white box in panel (a); arrows point to NPCs that are encircled by microglia. (c–g) Microglial numbers and activity are increased in neurogenic niches. Microglia are more densely populated (c, d), activated (e, f), and proliferative (g, h) in brain regions with endogenous NPCs. Brain sections from the cortex, dentate gyrus, and subventricular zone were stained and compared for expression of Iba-1 (a general macrophage marker that labels all microglia; c, d), CD68 (a microglia activation marker; e, f), and Iba-1/BrdU (to label proliferating microglia; g, h). Scale bars, 100 μm for (a, c, e), 20 μm for (b), and 50 μm for (g) (n = 3–5 mice/group). Data are presented as mean + s.e.m. *, P < 0.05; **, P < 0.001; ***, P < 0.0001 by two-tailed t test.
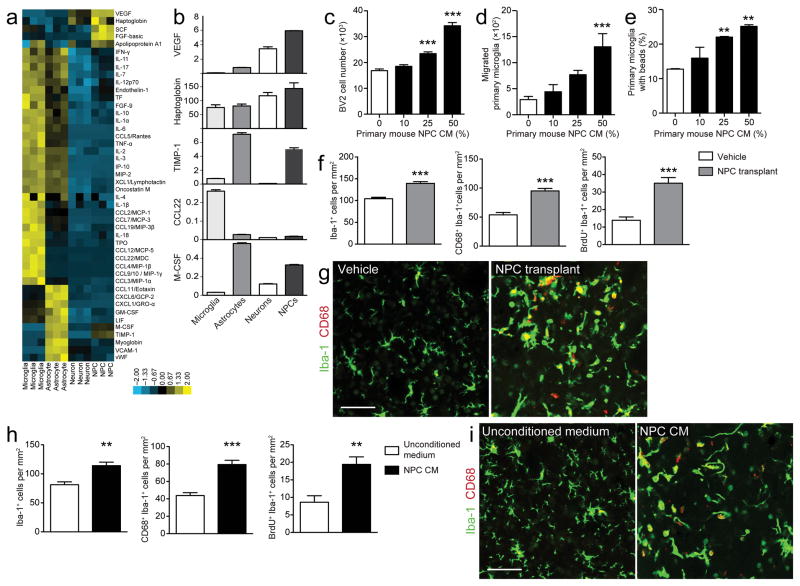
Figure 2. Neural progenitor cells secrete a distinct profile of signaling proteins and regulate microglial functions.
(a) Heat map of the secretory profiles of primary NPCs, neurons, astrocytes, and microglia, based on unsupervised clustering of immunoassay measurements of the listed proteins. Yellow shades represent increased expression of proteins relative to other cell types; blue shades, decreased expression. (b) Bar graphs of representative factors with differences in the levels of secreted proteins between cell types (in ng ml−1; mean of triplicates). (c–e) NPC-secreted factors regulate microglia in vitro. Conditioned medium from primary mouse NPCs (NPC CM) induces microglial proliferation (c), chemotaxis across a filter (d), and phagocytosis of fluorescent latex beads (e) (n = 3–6 wells/condition). (f–i) NPCs regulate microglia in vivo. Mice receiving intrastriatal transplants of NPCs (f, g) or NPC CM (h, i) demonstrate significant increases in the number of Iba-1+, Iba-1+/CD68+, and Iba-1+/BrdU+ cells compared to vehicle-treated controls (n = 10 mice/group). (g, i) Representative merged confocal images show brain sections from mice injected with vehicle and NPCs (h) or unconditioned medium and NPC CM (i) stained for Iba-1 (green) and CD68 (red); scale bar, 50 μm for all images (see Supplementary Fig. 4a, c for representative images of Iba-1/BrdU staining). Data are presented as mean + s.e.m. *, P < 0.05; **, P < 0.001; ***, P < 0.0001 by one-way ANOVA with Dunnett’s Multiple Comparison Test (c–d, compared to control, white columns, 0% NPC CM) or two-tailed t test (f, h).
Microglia retain the ability to proliferate throughout adulthood, particularly in response to brain injury11. Conditioned medium from primary NPCs induced an increase in the number of BV2 mouse microglia cells over two days, consistent with increased proliferation (Fig. 2c). Secondly, microglia can migrate to injury sites within the brain upon detection of small homeostatic disturbances9. Using standard migration assays we found that primary mouse microglia and BV2 cell chemotaxis increased several fold in response to conditioned medium from primary NPCs, relative to control unconditioned medium (Fig. 2d and Supplementary Fig. 1a, respectively). Thirdly, microglia can take up extracellular material, including entire cells, and function as phagocytes9. Using flow cytometry to quantify bead uptake of fluorescent latex beads by microglia (Supplementary Fig. 1b), we found that conditioned medium from primary mouse NPCs increased phagocytosis by both primary microglia and BV2s in a concentration-dependent manner (Fig. 2e and Supplementary Fig. 1c, respectively). Additionally, BV2 cells engulfed a greater number of beads per individual cell with increasingly higher concentrations of conditioned medium (Supplementary Fig. 1d). We obtained similar results with conditioned medium collected from a rat NPC line (Supplementary Fig. 1e–g). Lastly, microglia modulate their environment and respond to injury by secreting cytokines and chemokines9. We thus added conditioned medium from NPCs to primary microglia and measured secreted signaling proteins 48 h later in the supernatant (Supplementary Fig. 2a). This resulted in a remarkable change in the overall secretory profile of microglia (Supplementary Fig. 2b, c). While the net impact of these changes is not known at this time, it is interesting to note that NPC conditioned medium does not simply result in an increase or decrease in factors classically linked with pro-inflammatory activities (e.g. chemokine CXCL1, interleukin (IL)-α, IL-1β, IL-6). Rather, some of these factors are increased and others are decreased by treatment with NPC conditioned medium. Together, the above findings demonstrate that NPCs can provide secreted signals that control major microglial functions.
To investigate the interaction of NPCs and microglia in vivo, we stereotactically injected primary mouse NPCs (analyzed for their purity and differentiation potential, Supplementary Fig. 3) or vehicle into the striatum of 2-month-old C57BL/6 mice. Consistent with our cell culture data, injection of NPCs resulted in increases in the number of Iba-1+ microglia, with more of these cells being activated and expressing the lysosomal marker CD68 (Iba-1+/CD68+), and more microglia proliferating (Iba-1+/BrdU+) (Fig. 2f, g, Supplementary Fig. 4a). Additionally, we analyzed microglial activation by morphological characterization of Iba-1+ cells, in which resting microglia were morphologically defined as cells with many, highly ramified branching processes and activated microglia were classified as cells with a larger soma and “bushy” appearance or as rounded/amoeboid macrophage-like cells. Consistent with our CD68 and BrdU data, we found significantly higher numbers of microglia with activated morphology in striata receiving NPC transplants compared to vehicle controls (Supplementary Fig. 4b). We also observed that the density of Iba-1+ cells was highest at the center of the NPC injection site and progressively less dense in intermediate and peripheral regions of analysis (data not shown), a trend that was not apparent for vehicle-treated animals. Importantly, conditioned medium from primary mouse NPCs injected into the striatum was sufficient to cause a similar increase in absolute number, activation, and proliferation of microglia (Fig. 2h, i, Supplementary Fig. 4c). These data suggest that NPCs, and moreover, NPC-secreted factors, can regulate microglia activation in vivo.
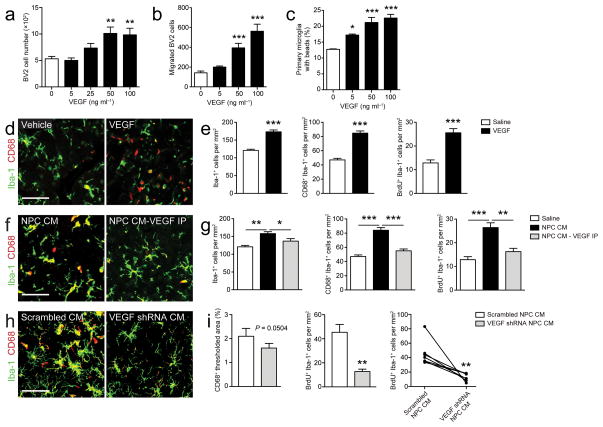
Having found that proteins secreted by NPCs can regulate microglial activity both in vitro and in vivo, we next sought to identify a specific NPC-secreted immune factor that is involved in this regulation. VEGF is highly secreted by NPCs relative to microglia or other CNS cells (Fig. 2b), confirming earlier findings12, and has been shown to induce proliferation and chemotaxis in microglia10. We thus tested whether this factor is sufficient and necessary to mimic part of the effects of NPC-derived factors in our in vitro and in vivo paradigms. Indeed, treatment of cultured microglia with recombinant VEGF protein induced proliferation, migration, and phagocytosis (Fig. 3a–c, respectively). We also injected recombinant VEGF protein into the right striatum and, as a control, saline into the left striatum of mice and again examined microglia close to the injection sites. Injection of VEGF protein resulted in increases in the number of Iba-1+ cells, Iba-1+/CD68+ cells, and Iba-1+/BrdU+ cells compared to saline injection (Fig. 3d, e, Supplementary Fig. 4d). On the other hand, depletion of VEGF from NPC conditioned medium with immunoaffinity beads almost completely reversed the effects of NPC conditioned medium on microglia (Fig. 3f, g, Supplementary Fig. 4e). Finally, in order to further address the necessity of NPC-derived VEGF in the regulation of microglia, we generated lentiviral vectors expressing shRNA constructs targeted against VEGF. Infection of cultured primary NPCs with the shRNAs reduced secreted VEGF levels in NPC conditioned medium compared to that of NPCs infected with a control (“Scrambled”) shRNA (Supplementary Fig. 4f). We injected conditioned medium from the most effective VEGF knockdown (i.e., VEGF shRNA I) and control conditioned medium (“Scrambled NPC CM”) into the right and left striatum, respectively. Knocking down VEGF attenuated the effects of NPC conditioned medium on microglia (Fig. 3h, i, Supplementary Fig. 4g). Thus our findings indicate that VEGF, which is secreted by NPCs in significant amounts, is sufficient and, at least in part, necessary to regulate microglial number, activation, and proliferation.
Figure 3. Neural progenitor cell-secreted VEGF is necessary and sufficient for the regulation of microglia.
(a–b) VEGF regulates microglia in vitro. Microglia treated with recombinant VEGF show increased proliferation (a), chemotaxis (b), and phagocytosis (c) (n = 3–6 wells/condition). (d, e) Similarly, intrastriatal injection of VEGF induces an increase in cell number, activation and proliferation of microglia around the injection site (n = 5 mice/group). (f–i) NPC-derived VEGF is necessary for the regulation of microglia by NPCs. Immunodepletion of VEGF from NPC CM (f, g) (n = 10 mice/group) or depletion of VEGF from NPC CM by lentiviral shRNA-mediated knockdown in cultured NPCs (h, i) (n = 7 mice/group) attenuates the effects of NPC CM on microglia in vivo. For the representative confocal images of each in vivo experiment (d, f, h), brain sections were stained for Iba-1 (green) and CD68 (red); scale bar, 50 μm for all images (see Supplementary Fig. 4d, e, g for representative images of Iba-1/BrdU staining). Data are presented as mean + s.e.m. *, P < 0.05; **, P < 0.001; ***, P < 0.0001 by one-way ANOVA with Dunnett’s Multiple Comparison Test (a–c, compared to control, white columns, no VEGF), paired two-tailed t test (e, i), or unpaired two-tailed t test (g).
In summary, we show that NPC-derived secreted factors and transplanted NPCs are powerful regulators of their environment, specifically of microglia. We have not yet tested if resident NPCs in the brain have similar functions, as such experiments will require genetic deletion of each factor individually from NPCs. Nevertheless, we show here that isolated and transplanted NPCs have the capacity to secrete a multitude of signaling proteins, many of which have been linked with injury responses or immune cell functions. In particular, we find that NPC-derived VEGF represents one mediator in the regulation of microglia by NPCs. We expect that other NPC-secreted immune factors are also likely to act on microglia, and furthermore may regulate other cell types within the NPC environment. Moreover, NPCs may also utilize such secreted proteins to self-regulate; for example stem cell factor (SCF), which is also highly expressed by NPCs relative to the other cell types, is known to regulate hematopoietic stem cells13. Together with findings from our lab showing that NPCs express several major components of the B lymphocyte receptor14 and the observation that transplanted NPCs tend to exert repair functions in injury models3, 4, or are capable of phagocytosis15, it is tempting to speculate that NPCs assume a function in maintaining tissue integrity and immune function in the CNS apart from their purpose to produce neural cells.
METHODS
Cell Culture
We isolated and cultured primary mouse neural progenitor cells (NPCs) from cortices of P0 C57Bl/6 mouse pups as previously described16. Briefly, brains were dissected and incubated in DMEM (Gibco) containing 2.5 U ml−1 papain (Worthington Biochemicals), 250 U ml−1 DNase I (Worthington Biochemicals) and 1 U ml−1 dispase II (Boehringer Mannheim) at 37°C for 45 min. Tissue pieces were washed twice in DMEM with 10% fetal bovine serum (FBS; Gibco), and cellular pellets were resuspended in Neurobasal A supplemented with B27 without Vitamin A (Gibco), EGF (20 ng ml−1, Peprotech), FGF-2 (20 ng ml−1, Peprotech), and 1% penicillin-streptomycin (Gibco). To passage cells, neurospheres were harvested by centrifugation, dissociated in Hanks-based cell dissociation buffer (Gibco), and grown in the same medium as above. A rat NPC line was obtained from Dr. T. Palmer, Stanford University, and cultured as previously described5.
We obtained primary microglia and astrocytes from P0–P3 C57Bl/6 mouse pups using a previously described protocol17, but with some modifications. Briefly, cortices were isolated and stripped of meninges and dissociated with a 25-gauge needle. The cell suspension was seeded into poly-L-lysine-coated (Sigma-Aldrich) T75 tissue culture flasks and maintained in DMEM/F12 with 10% FBS and 1% penicillin-streptomycin for two weeks to grow a mixed astrocyte/microglia population. We then applied an antigen-antibody-mediated magnetic cell-sorting assay to isolate high-purity microglia and astrocytes (adapted from ref. 18 and Miltenyi Biotech). The mixed glial population was resuspended in MACS buffer (PBS with 0.5% bovine serum albumin and 2 mM EDTA, pH 7.2) and incubated with CD11b MicroBeads (Miltenyi Biotech). The cell suspension was passed through a magnetic cell separator column (Miltenyi Biotech) fitted into a QuadroMACS cell separator and washed to isolate the unlabeled, purified astrocyte population. The column was then removed from the magnetic separator and flushed with MACS buffer to collect the purified microglia population. The purity of each primary cell type was assessed by immunocytochemistry (data not shown).
The microglia BV2 cell line was obtained from Dr. Elisabetta Blasi and maintained in DMEM with 10% FBS and 1% penicillin-streptomycin19.
We isolated primary hippocampal neurons from 16-day-old CF1 mouse embryos, as described previously20. Briefly, single-cell suspensions obtained from hippocampi were plated on 24-well culture plates were coated with poly-L-lysine at a density of 3×104 cells/well in DMEM/F-12 with 10% FBS and 1% penicillin-streptomycin and subsequently maintained in Neurobasal medium containing 2% B27 supplement plus retinoic acid (Gibco).
Cell Supernatant Collection (Conditioned Media)
We seeded 1×105 mouse primary NPCs at passage 5–7 in 1 ml of culture medium in 12-well plates and collected conditioned medium (CM) 48 h later. 1×105 rat NPCs were cultured in 0.5 ml of medium, and CM was collected 24 h later.
For the proteomic analysis of secreted proteins from primary neural cells, we plated 1×106 cells/well in 1 ml of Neurobasal A with B27 on coated 12-well plates and collected supernatants 48 h later. To examine the effects of NPC CM on the levels of proteins secreted from microglia, we followed the experimental scheme outlined in Supplementary Fig. 2a. The supernatants from microglia treated with or without 10% NPC CM (diluted in Neurobasal A) were collected after 48 h.
Proteomic Analyses
The relative concentrations of signaling molecules were measured in supernatants described above using antibody-based multiplex immunoassays (Luminex) by Rules Based Medicine Inc., a fee-for-service provider. Measurements were obtained from at least three independent samples.
Proliferation Assay
To assess microglia proliferation, we treated 2×103 BV2 cells/well (six wells/condition) in a 96-well plate with NPC CM or recombinant mouse VEGF 164 (R&D Systems) at varying concentrations, as indicated in Figures; Neurobasal A with B27 was used for all dilutions. After 48 h, we stained all cells with Hoechst 33342 (Molecular Probes) and counted the number of cells in each well using an automated imaging system (Cellavista Analyzer, Roche). Data are from multiple independent experiments.
Chemotaxis Assay
To test chemotactic activity, we added NPC CM or VEGF at varying concentrations to the bottom chamber of 96-well chemotaxis systems (ChemoTx chemotaxis system, 101-8sp, Neuroprobe Inc.), and BV2s or primary microglia (2×105 cells/well, five wells/condition) to the top compartment (Neurobasal A with B27 was used for all dilutions). After 3 h, we spun down cells that had migrated through the filter into the bottom chamber, as described by the manufacturer. We removed the filter and stained the cells in the bottom chamber with Hoechst 33342. We then counted the number of migrated cells in each well using the Cellavista imaging system. Data are from multiple independent experiments.
Phagocytosis Assay
We analyzed phagocytic function using a previously described assay21, 22. We treated BV2s or primary microglia (2×105 cells/well, three wells/condition) in 12-well plates with NPC CM or VEGF for 2 h. We then added 2×106 microspheres (flash-red, 6 μm; Bangs Laboratories) to each well and incubated the cells and microspheres for 1 h at 37°C on a rotating platform. Prior to being added, the microspheres were opsonized by incubation in 50% FBS/PBS for 30 min at 37°C. We stopped the assay by adding ice-cold PBS to the cultures, then collected, washed, and resuspended the samples in FACS buffer (PBS with 2% FBS and 0.1% sodium azide) in preparation for flow cytometry. We analyzed the samples for fluorescence (PerCP-Cy5, phagocytosed microspheres) and light scatter (cell size) to quantify microglia with microspheres. Data are from multiple independent experiments.
Mice
Wildtype C57BL/6J mice were used for stereotactic injections and primary cell cultures (The Jackson Laboratory). CX3CR1GFP/wt mice (The Jackson Laboratory) were used to analyze the neurogenic niche. Mice were kept on a 12 h light/dark cycle and allowed free access to food and water. All animal care and use complied with the Animal Welfare Act and was in accordance with institutional guidelines and approved by the VA Palo Alto Committee on Animal Research and the institutional administrative panel of laboratory animal care at Stanford University.
Regional Microglial Expression Analyses
To study microglia expression patterns, we administered 5′-bromo-2′-deoxyuridine (BrdU, 100 mg kg−1 body weight; dissolved in 0.9% NaCl to 25 mg ml−1; Sigma-Aldrich) by intraperitoneal injection once a day for 3 days to two-month-old male wildtype C57/BL6J mice and sacrificed the mice one day after injections. The brains were processed and analyzed as described below (“Immunohistochemistry”). Two independent experiments, n = 3–5 mice/group.
Stereotactic Injections
NPC Transplantation
We dissociated neurospheres into single cells and resuspended the cells at a density of 1×106 cells/ml in culture medium. We performed intrastriatal transplantation in 2-month-old male C57/BL6J mice by infusing of 2×105 viable cells in 2 μl of saline or, in control animals, 2 μl of saline alone into the right hemisphere. Briefly, anesthesia was induced and maintained with 2% isoflurane in N2O:Air (80:20). Body temperature was monitored and maintained at 37°C. After fixation of the head in a stereotactic frame (Stoelting Instruments), a midline incision was made to expose the cranium. A frontal burr hole was drilled and a cannula (30G) was stereotactically inserted into the striatum (anterior, +0.1; lateral, +/−2.5; ventral −3.5 mm; relative to bregma23). The cell suspension was slowly infused at a rate of 0.5 μl/min using a microinfusion system (UltraMicroPump III, World Precision Instruments). The cannula was left in place for additional 5 min and slowly withdrawn. The wound was sutured and the animal allowed to recover. We administered BrdU 24 h before stereotactic injections and immediately afterwards and sacrificed the mice 24 h after surgery. Two independent experiments, n = 10 mice/group.
NPC Conditioned Medium Injections
We injected 5 μl of conditioned medium from primary mouse NPC cultures, or, in control animals, unconditioned medium, into the right striatum, as described above. Two independent experiments, n = 10 mice/group.
VEGF Protein and VEGF Immunodepleted Conditioned Medium Injections
For injection of VEGF protein, we stereotactically infused 2 μl of recombinant VEGF protein (diluted to 200 ng μl−1 in saline; R&D Systems) into the right striatum, as described above. As a control, injections of 2 μl vehicle were performed on the left side. n = 5 mice/group.
For VEGF immunodepletion of NPC CM, a protein G immunoprecipitation kit (Roche Applied Science) was used according to the manufacturer’s instructions. Briefly, we incubated 5 μg of a monoclonal anti-VEGF antibody (Millipore, 05-443) with 1 ml of NPC CM for 60 min at 4°C and then incubated the mixture with a protein G/agarose suspension for 3 h. Agarose beads with the immunoprecipitate were cleared by centrifugation and the supernatant removed. As a control, conditioned medium was processed in an identical way, but without addition of the antibody. Immunodepleted and undepleted media were intrastriatally infused in the left and right striatum, respectively, as described above. Two independent experiments, n = 10 mice/group.
VEGF shRNA NPC Conditioned Medium Injections
VEGF knockdown in primary NPCs was performed using a lentiviral shRNA expression system (pSIH-H1 shRNA Cloning and Lentivector Expression system; System Biosciences) according to the manufacturer’s instructions. Three VEGF-targeted sequences were cloned into the pSIH1-H1-copGFP vector (VEGF I: 5′-ACGAGATATTCCGTAGTACATA-3′; VEGF II: 5′-CCGAGATAGAGTACATCTTCAA-3′; VEGF III: 5′-AGGTACTTATTTAATAGCCCTT-3′). The constructs were packaged into viral particles by the Stanford Neuroscience Gene Vector and Virus Core. Conditioned media were collected as described above from primary NPCs infected with the shRNA viruses or a control scrambled sequence (5′-GAATCATCAGGCGTACAGA-3′). The efficacy of the VEGF-shRNA knockdown in NPC CM was analyzed by ELISA (Mouse VEGF DuoSet; R&D Systems), according to the manufacturer’s instructions. VEGF shRNA I was used for all subsequent experiments.
Five μl of conditioned medium from NPCs infected with VEGF shRNA (“VEGF shRNA NPC CM”) or control scrambled shRNA (“Scrambled NPC CM”) were stereotactically infused into the right and left striatum, respectively, as described above. n = 7 mice/group.
Immunohistochemistry
Immunostaining was performed as described previously24. Briefly, mouse brains were immersed in 4% paraformaldehyde overnight at 4°C and sectioned at 40 μm on a freezing microtome. Sections were incubated in 2N HCL at 37°C for 30 min for BrdU antigen retrieval, washed in TBST, blocked with 10% goat serum, and incubated overnight at 4°C with primary antibodies (rabbit anti-Iba-1, 1:1000, WAKO Chemicals, 019-19741; mouse anti-BrdU, 1:500, Abcam, ab6326; rat anti-CD68, 1:250, Serotec, MCA1957). For fluorescent stains, primary antibody binding was detected by incubating with corresponding secondary antibodies (Alexa Fluor 488, 546, and 633; 1:500; Molecular Probes, A-11304, A-11003, A-21094, respectively). Nuclei were counterstained with DAPI (1 mg ml−1; AnaSpec) and the slices mounted on glass slides with FluorSave reagent (Calbiochem). 3,3′-Diaminobenzidine (DAB; Sigma-Aldrich) stains (Fig. 1c, e) were performed with an ABC labeling kit (Vector Laboratories).
For regional analyses of microglia expression patterns in DAB stains, Metamorph imaging software (Universal Imaging Corporation) was used to quantify the percentage of pixels above background staining (“thresholded area”) within a region drawn around the dentate granule cell layer and subgranular zone or around a cortical region of equivalent size in five coronal sections. Volocity software (Perkin Elmer) was used for 3D reconstruction with a coarse algorithm and deconvolution of Z-stack fluorescence images.
For analyses of the number and distribution of microglia in the striatum and other co-localization studies, a region of interest on each hemisphere was defined as a circular field covering the striatal area around the injection site (2.2 mm2). In six sections (120 μm apart) centered on the level of the injection sites, six z-stacks per ROI were acquired at random positions using a confocal laser-scanning microscope (Zeiss). Cell counts and CD68/Iba-1 and BrdU/Iba-1 co-localization analyses were carried using a digital image processing system (ImageJ 1.4025 for Macintosh, National Institutes of Health). Cell densities per mm2 were calculated using the ImageJ software.
For analyzing microglial activation by morphology, an investigator blinded to group assignment counted and classified Iba-1+ cells. We defined resting microglia morphologically as cells with many, highly ramified branching processes. Activated microglia were either classified as cells with a larger soma and a “bushy” appearance of the processes or as rounded/amoeboid macrophage-like cells26, 27.
Data and Statistical Analyses
Statistical analyses were performed with GraphPad InStat 3.05 and Prism 5.0 software (GraphPad Software). Means between two groups were compared with two-tailed, paired or unpaired (where indicated in Figure Legends) Student’s t tests. Comparisons of means from multiple groups against one control group were analyzed with one-way ANOVA and post-hoc Dunnett’s Multiple Comparison Test. All experiments were conducted in a randomized and blinded fashion.
To generate proteomic heat maps, immunoassay measurements of the listed proteins were normalized and clustered using an unsupervised clustering algorithm (Gene Cluster 3.0; Michiel de Hoon, Human Genome Center, University of Tokyo, Japan)28 to group across sample types and proteins (any proteins that were reported to be undetectable in two out of three measurements for a sample were removed from the analysis prior to data processing). Java TreeView 1.0.13 (Alok J. Saldanha, Department of Genetics, Stanford University) was used to visualize the heat maps29
Supplementary Material
Acknowledgments
The authors wish to thank Saul Villeda and Raphael Wabl for assistance with the neural progenitor cell cultures and Michael Lochrie and the Stanford Neuroscience Gene Vector and Virus Core for generating the shRNA lentiviruses (supported by NINDS P30 core grant, NS069375-01A1). This work was supported by grants from the Department of Veterans Affairs (T.W.-C.), National Institutes of Health Institute on Aging (R01 AG027505, T.W.-C), a California Initiative for Regenerative Medicine Award (T.W.-C.), a National Science Foundation predoctoral fellowship (K.I.M.), the Swiss National Science Foundation (PBBEB-117034 and PASMP3-123221/1, R.H.A.), and the Evelyn L. Neizer Fund (R.H.A.).
Footnotes
Author Contributions. K.I.M., R.H.A., and T.F. contributed equally to this work. K.I.M. performed in vitro experiments and performed and analyzed in vivo experiments. R.H.A. performed and analyzed in vivo experiments. T.F. performed in vitro experiments. G.B. assisted with VEGF shRNA experiment. M.H.-M. and Y.H. provided conceptual advice and input. K.I.M. and T.W.-C. wrote the manuscript. R.G. and T.W.-C. supervised the project.
References
- 1.Ming GL, Song H. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golmohammadi MG, et al. Stem Cells. 2008;26:979–987. doi: 10.1634/stemcells.2007-0919. [DOI] [PubMed] [Google Scholar]
- 3.Pluchino S, et al. Nature. 2003;422:688–694. doi: 10.1038/nature01552. [DOI] [PubMed] [Google Scholar]
- 4.Ziv Y, Avidan H, Pluchino S, Martino G, Schwartz M. Proc Natl Acad Sci USA. 2006;103:13174–13179. doi: 10.1073/pnas.0603747103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monje ML, Toda H, Palmer TD. Science. 2003;302:1760–1765. doi: 10.1126/science.1088417. [DOI] [PubMed] [Google Scholar]
- 6.Klassen HJ, et al. Cytokine. 2003;22:101–106. doi: 10.1016/s1043-4666(03)00120-0. [DOI] [PubMed] [Google Scholar]
- 7.Skalnikova H, Motlik J, Gadher SJ, Kovarova H. Proteomics. 2011;11:691–708. doi: 10.1002/pmic.201000402. [DOI] [PubMed] [Google Scholar]
- 8.Pluchino S, et al. PLoS ONE. 2009;4:e5959. doi: 10.1371/journal.pone.0005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanisch UK, Kettenmann H. Nat Neurosci. 2007;10:1387–1394. doi: 10.1038/nn1997. [DOI] [PubMed] [Google Scholar]
- 10.Forstreuter F, Lucius R, Mentlein R. J Neuroimmunol. 2002;132:93–98. doi: 10.1016/s0165-5728(02)00315-6. [DOI] [PubMed] [Google Scholar]
- 11.Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FMV. Nat Neurosci. 2007;10:1538–1543. doi: 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- 12.Schänzer A, et al. Brain Pathol. 2004;14:237–248. doi: 10.1111/j.1750-3639.2004.tb00060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zsebo KM, et al. Cell. 1990;63:195–201. doi: 10.1016/0092-8674(90)90300-4. [DOI] [PubMed] [Google Scholar]
- 14.Moriyama M, et al. J Neurosci. 2011;31:3981–3989. doi: 10.1523/JNEUROSCI.3617-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Z, et al. Nature Cell Biology. 2011;13:1–9. doi: 10.1038/ncb0111-1. [DOI] [PubMed] [Google Scholar]
- 16.Reynolds BA, Rietze RL. Nat Meth. 2005;2:333–336. doi: 10.1038/nmeth758. [DOI] [PubMed] [Google Scholar]
- 17.Giulian D, Baker T. J of Neurosci. 1986;6:2163–2178. doi: 10.1523/JNEUROSCI.06-08-02163.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marek R, Caruso M, Rostami A, Grinspan JB, Das Sarma J. J Neurosci Methods. 2008;175:108–118. doi: 10.1016/j.jneumeth.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 19.Blasi E, Barluzzi R, Bocchini V, Mazzolla R, Bistoni F. J Neuroimmunol. 1990;27:229–237. doi: 10.1016/0165-5728(90)90073-v. [DOI] [PubMed] [Google Scholar]
- 20.Brionne TC, Tesseur I, Masliah E, Wyss-Coray T. Neuron. 2003;40:1133–1145. doi: 10.1016/s0896-6273(03)00766-9. [DOI] [PubMed] [Google Scholar]
- 21.Steinkamp J, Wilson J, Saunders G, Stewart C. Science. 1982;215:64–66. doi: 10.1126/science.7053559. [DOI] [PubMed] [Google Scholar]
- 22.Giulian D, Chen J, Ingeman J, George J, Noponen M. J Neurosci. 1989;9:4416–4429. doi: 10.1523/JNEUROSCI.09-12-04416.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. Academic Press; 2001. [Google Scholar]
- 24.Buckwalter MS, et al. Am J Pathol. 2006;169:154–164. doi: 10.2353/ajpath.2006.051272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider CA, Rasband WS, Eliceiri KW. Nat Meth. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soltys Z, Ziaja M, Pawlínski R, Setkowicz Z, Janeczko K. J Neurosci Res. 2001;63:90–97. doi: 10.1002/1097-4547(20010101)63:1<90::AID-JNR11>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 27.Streit WJ, Walter SA, Pennell NA. Prog Neurobiol. 1999;57:563–581. doi: 10.1016/s0301-0082(98)00069-0. [DOI] [PubMed] [Google Scholar]
- 28.De Hoon MJL, Imoto S, Nolan J, Miyano S. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 29.Saldanha AJ. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.