Abstract
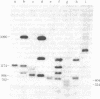
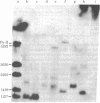
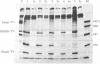
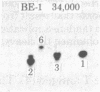
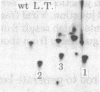
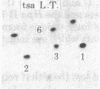
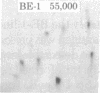
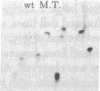
Rat-1 cells were transfected with the restriction enzyme fragment of polyoma virus DNA that extends clockwise from the Bcl I site ((65.4 map units) to the EcoRI site (0/100 map units). Six transformed cell lines were obtained and one of them (BE-1) has been investigated in detail. The viral DNA that is integrated into host DNA in this line appeared to consist of two fragments arranged in a "head-to-tail" tandem with no detectable intervening host sequences. BE-1 cells contained polyoma virus small and middle tumor antigens that were indistinguishable from the corresponding tumor antigens from lytically infected cells. No large tumor antigen was detected but a "new" Mr 34,000 protein, which proved to be a truncated version of large tumor antigen, was immunoprecipitated by anti-tumor-antigen antiserum. After injection of 10(6) BE-1 cells into young syngeneic Fischer rats, tumors appeared within 3--4 weeks. Thus, the coding capacity of the Bcl I/EcoRI fragment of polyoma virus DNA is sufficient to enable the cells to produce all of small and middle tumor antigens and about a third of large tumor antigen, to transform cells stably in culture, and to produce tumors in vivo.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Basilico C., Gattoni S., Zouzias D., Valle G. D. Loss of integrated viral DNA sequences in polyomatransformed cells is associated with an active viral A function. Cell. 1979 Jul;17(3):645–659. doi: 10.1016/0092-8674(79)90272-1. [DOI] [PubMed] [Google Scholar]
- Birg F., Dulbecco R., Fried M., Kamen R. State and organization of polyoma virus DNA sequences in transformed rat cell lines. J Virol. 1979 Feb;29(2):633–648. doi: 10.1128/jvi.29.2.633-648.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhart W. Complementation between temperature-sensitive (ts) and host range nontransforming (hr-t) mutants of polyoma virus. Virology. 1977 Apr;77(2):589–597. doi: 10.1016/0042-6822(77)90484-6. [DOI] [PubMed] [Google Scholar]
- Feunteun J., Sompayrac L., Fluck M., Benjamin T. Localization of gene functions in polyoma virus DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4169–4173. doi: 10.1073/pnas.73.11.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluck M. M., Staneloni R. J., Benjamin T. L. Hr-t and ts-a: two early gene functions of polyoma virus. Virology. 1977 Apr;77(2):610–624. doi: 10.1016/0042-6822(77)90486-x. [DOI] [PubMed] [Google Scholar]
- Francke B., Eckhart W. Polyoma gene function required for viral DNA synthesis. Virology. 1973 Sep;55(1):127–135. doi: 10.1016/s0042-6822(73)81014-1. [DOI] [PubMed] [Google Scholar]
- Freeman A. E., Gilden R. V., Vernon M. L., Wolford R. G., Hugunin P. E., Huebner R. J. 5-Bromo-2'-deoxyuridine potentiation of transformation of rat-embryo cells induced in vitro by 3-methylcholanthrene: induction of rat leukemia virus gs antigen in transformed cells. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2415–2419. doi: 10.1073/pnas.70.8.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M. Characterization of a temperature-sensitive mutant of polyoma virus. Virology. 1970 Mar;40(3):605–617. doi: 10.1016/0042-6822(70)90205-9. [DOI] [PubMed] [Google Scholar]
- Fried M., Griffin B. E., Lund E., Robberson D. L. Polyoma virus--a study of wild-type, mutant and defective DNAs. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):45–52. doi: 10.1101/sqb.1974.039.01.009. [DOI] [PubMed] [Google Scholar]
- Graham F. L., Abrahams P. J., Mulder C., Heijneker H. L., Warnaar S. O., De Vries F. A., Fiers W., Van Der Eb A. J. Studies on in vitro transformation by DNA and DNA fragments of human adenoviruses and simian virus 40. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):637–650. doi: 10.1101/sqb.1974.039.01.077. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. Transformation of rat cells by DNA of human adenovirus 5. Virology. 1973 Aug;54(2):536–539. doi: 10.1016/0042-6822(73)90163-3. [DOI] [PubMed] [Google Scholar]
- Griffin B. E., Fried M., Cowie A. Polyoma DNA: a physical map. Proc Natl Acad Sci U S A. 1974 May;71(5):2077–2081. doi: 10.1073/pnas.71.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B. E., Maddock C. New classes of viable deletion mutants in the early region of polyoma virus. J Virol. 1979 Sep;31(3):645–656. doi: 10.1128/jvi.31.3.645-656.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hutchinson M. A., Hunter T., Eckhart W. Characterization of T antigens in polyoma-infected and transformed cells. Cell. 1978 Sep;15(1):65–77. doi: 10.1016/0092-8674(78)90083-1. [DOI] [PubMed] [Google Scholar]
- Israel M. A., Simmons D. T., Hourihan S. L., Rowe W. P., Martin M. A. Interrupting the early region of polyoma virus DNA enhances tumorigenicity. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3713–3716. doi: 10.1073/pnas.76.8.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Brocklehurst J. R., Dulbecco R. Virus-specific proteins in the plasma membrane of cells lytically infected or transformed by pol-oma virus. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4666–4670. doi: 10.1073/pnas.74.10.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y. Polyoma virus-specific 55K protein isolated from plasma membrane of productively infected cells is virus-coded and important for cell transformation. Virology. 1979 Oct 15;98(1):261–266. doi: 10.1016/0042-6822(79)90545-2. [DOI] [PubMed] [Google Scholar]
- Kamen R., Lindstrom D. M., Shure H., Old R. W. Virus-specific RNA in cells productively infected or transformed by polyoma virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):187–198. doi: 10.1101/sqb.1974.039.01.025. [DOI] [PubMed] [Google Scholar]
- Lania L., Gandini-Attardi D., Griffiths M., Cooke B., De Cicco D., Fried M. The polyoma virus 100K large T-antigen is not required for the maintenance of transformation. Virology. 1980 Feb;101(1):217–232. doi: 10.1016/0042-6822(80)90497-3. [DOI] [PubMed] [Google Scholar]
- Miller L. K., Fried M. Construction of infectious polyoma hybrid genomes in vitro. Nature. 1976 Feb 19;259(5544):598–601. doi: 10.1038/259598a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Restriction endonucleases. CRC Crit Rev Biochem. 1976 Nov;4(2):123–164. doi: 10.3109/10409237609105456. [DOI] [PubMed] [Google Scholar]
- Seif R., Cuzin F. Temperature-sensitive growth regulation in one type of transformed rat cells induced by the tsa mutant of polyoma virus. J Virol. 1977 Dec;24(3):721–728. doi: 10.1128/jvi.24.3.721-728.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Smart J. E., Ito Y. Three species of polyoma virus tumor antigens share common peptides probably near the amino termini of the proteins. Cell. 1978 Dec;15(4):1427–1437. doi: 10.1016/0092-8674(78)90066-1. [DOI] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Griffin B. E. Sequence from early region of polyoma virus DNA containing viral replication origin and encoding small, middle and (part of) large T antigens. Cell. 1979 Jun;17(2):357–370. doi: 10.1016/0092-8674(79)90162-4. [DOI] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Walsh J. E., Griffin B. E. Coding potential and regulatory signals of the polyoma virus genome. Nature. 1980 Jan 31;283(5746):445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Zouzias D., Prasad I., Basilico C. State of the viral DNA in rat cells transformed by polyma virus. II. Identification of the cells containing nonintegrated viral DNA and the effect of viral mutations. J Virol. 1977 Oct;24(1):142–150. doi: 10.1128/jvi.24.1.142-150.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]