Abstract
Craniosynostosis is the early fusion of one or more sutures of the infant skull and is a common defect occurring in approximately 1 of every 2,500 live births. Non-syndromic craniosynostosis accounts for approximately 80% of all cases and is thought to have strong genetic determinants that are yet to be identified. ALX4 is a homeodomain transcription factor with known involvement in osteoblast regulation. By direct sequencing of the ALX4 coding region in sagittal or sagittal-suture-involved non-syndromic craniosynostosis probands, we identified novel, nonsynonymous, familial variants in three of 203 individuals with NSC. Using dual-luciferase assay we show that two of these variants (V7F and K211E) confer a significant gain-of-function effect on ALX4. Our results suggest that ALX4 variants may have an impact on the genetic etiology of NSC.
Keywords: ALX4, sagittal, craniosynostosis, gain-of-function, dual-luciferase
The premature fusion of the various sutures in the human neurocranium (skull vault and base) is defined as craniosynostosis (CS). The clinical consequences of CS include abnormal head shape and increased intracranial pressure, which may result in neurologic symptoms, developmental delay, and hearing or vision problems [Kimonis et al., 2007]. About 20% of CS cases are caused by Mendelian mutations and are described as syndromic, as they are associated with extracranial anomalies and/or developmental delays [Passos-Bueno et al., 2008]. Gain-of-function (GOF) mutations in FGFR1, FGFR2 or FGFR3 leading to over-induced signaling of the MAPK/RAS pathway have been described in several autosomal dominant CS syndromes, including Apert, Crouzon, Muenke, Pfeiffer and Jackson-Weiss [Passos-Bueno et al., 2008]. Loss-of-function mutations in TWIST1, a repressor of FGFRs, lead to Saethre-Chotzen syndrome [Howard et al., 1997; Rice et al., 2000].
The remaining 80% of CS is classified as non-syndromic craniosynostosis (NSC), which presents with isolated suture fusion and no other associated anomalies. The etiology of this form is less understood. A few lines of evidence support genetic influence: observations of a discrepant male to female ratio [Lajeunie et al., 2005], the occurrence of multiple affected individuals in one family in approximately 6-8% of all families [Boyadjiev et al., 2007], and a high concordance rate in monozygotic as compared to dizygotic twins – 30% vs. 0% in sagittal, 43% vs. 5% in metopic [Lajeunie et al., 2005]. In addition, rare mutations in the FGFR genes or TWIST1 have been reported in some patients with NSC [Johnson et al., 2000; Seto et al., 2007]. Furthermore, three of 81 patients with coronal NSC were found to have heterozygous mutations in EFNA4 [Merrill et al., 2006]. However, these cases cumulatively account for a small percentage of NSC cases. Thus, more genetic factors are likely to contribute to NSC.
The GOF mutation P148H in MSX2 causes Boston type of craniosynostosis [Jabs et al., 1993] while loss-of function mutations in the same gene results in parietal foramina [Spruijt et al., 2005]. These results suggest that the regulation of the suture patency depends on precise dosage of certain developmental regulators. In this context, craniosynostosis (accelerated ossification) and parietal foramina (delayed ossification) phenotypes represent the opposite ends of a developmental spectrum. Similar effects have been observed from mutations in at least 3 other genes: TWIST1 [Howard et al., 1997; Stankiewicz et al., 2001], Nell1 [Desai et al., 2006; Zhang et al., 2003] and RUNX2 [Mundlos et al., 1997; Shevde et al., 2001], resulting in either craniosynostosis or delayed ossification of the skull.
ALX4 (NM_021926.3; MIM# 605420), a homeobox domain transcription factor, is known to play a role similar and additive to MSX2 in the development of the calvaria [Antonopoulou et al., 2004]. Indeed, heterozygous loss-of- function mutations of this gene result in a parietal foramina phenotype that is clinically indistinguishable from the one caused by MSX2 mutations. Furthermore, two ALX4 variants were identified in a cohort of 181 samples from patients with hot-spot negative non-syndromic or syndromic CS – a familial in frame deletion (c.314_325del encoding p.P105_Q108del) and c.605T>G encoding p.L202W [Mavrogiannis et al., 2006], though their functional significance was not determined. Thus, we suspected that GOF mutations in ALX4 may cause NSC or at least contribute in some way to its etiology. Here we report the analysis of ALX4 by direct sequencing and the functional effects of several missense variants identified in affected individuals.
We sequenced all four exons representing the entire coding region of ALX4 in 203 patients with NSC, of which 197 had single sagittal suture fusion and six had multiple suture fusion NSC with sagittal suture involvement. Sagittal suture fusion is the most common form of NSC, accounting for 40-58% of all NSC cases [Kimonis et al., 2007]. Thus, we chose to look primarily at sagittal NSC probands aiming to better elucidating the genetic etiology of this common disorder. The diagnoses were established by clinical geneticists on the basis of physical examination and review of medical records and head CT scans. Hot-spot mutation analysis of FGFR1-3 and TWIST1 [Boyadjiev et al., 2007] was performed on all probands with multiple suture NSC and 111 probands with single suture sagittal NSC. The remaining probands were not screened since no hot-spot mutations were identified in this initial cohort.
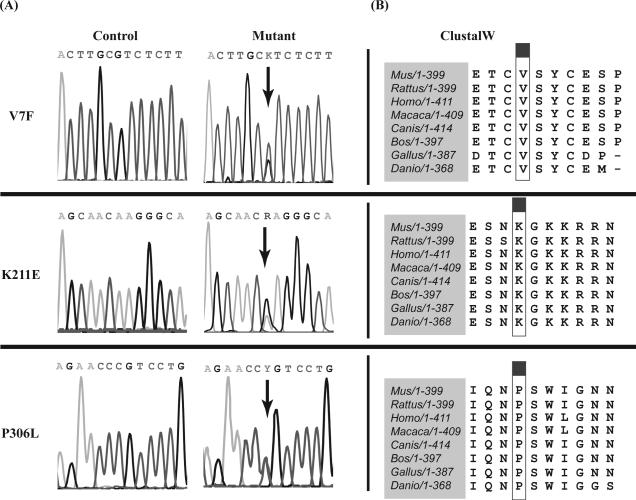
All ALX4 variants that we identified are summarized in Supp. Table S1. Previously reported SNPs were determined based on their presence in any of the following databases: HapMap, NCBI BLAST, MapBack or 1000Genomes. In order to analyze if these previously reported SNPs were overrepresented in our NSC cohort, we compared their allelic frequencies to those found in the control population obtained from either HapMap (Rel #28, Phase II + III, August 2010) or from 1000Genomes (Feb. 2009, Genome Reference Consortium). For this analysis, FINETTI software was used to test variants for deviations from Hardy-Weinberg equilibrium. We found that none of the known SNPs in our population demonstrated a significant deviation from the frequencies of either control population. Of the remaining novel SNPs, we selected those that were nonsynonymous as the best candidates for functional analysis of ALX4. Three SNPs – c.19G>T (V7F), c.631A>G (K211E) and c.917C>T (P306L) – met these criteria (Fig. 1A), although P306L was later reported as a SNP by the NHLBI Exome Sequencing Project (ss342322782). All three variants were also present in an unaffected parent. ALX4 protein sequence comparison showed that the amino acid residues at positions 7, 211 and 306, as well as the surrounding region on either side, are invariably conserved among eight vertebrae species (Fig. 1B). Additionally, we used bioinformatic tools to predict the functional consequences of the variant proteins. SIFT (Sorting Tolerant from Intolerant) predictions indicated that V7F and K211E would severely affect the function of ALX4. Conversely, P306L was not predicted to impact protein function. The PPH (PolyPhen) analysis indicates that the variants at positions 7 and 306 are potentially damaging to the protein's original structure and function, while the Grantham analyses indicates that all three polymorphic residues have moderately damaging effects compared to the wild-type residues. The combination of these different bioinformatic methods suggests that the three ALX4 variants of interest can potentially have deleterious effects on the protein function.
Figure 1.
(A) Chromatogram of each ALX4 variant, denoted by arrows. Polymorphisms were determined on the basis of double peaks and a reduction in signal intensity compared to control sequence. (B) ClustalW alignment of ALX4 protein sequence was performed in eight species -NP_031468.1 (Mus musculus), NP_001100023.1 (Rattus norvegicus), NP_068745.2.1 (Homo sapiens), XP_001113643.1 (Macaca mulatta), XP_850737.1 (Canis lupis familiaris), NP_001025475.1 (Bos taurus), NP_989493.1 (Gallus gallus), and XP_001340966.1 (Danio rerio). The boxed column denotes the site of a polymorphic variant of interest.
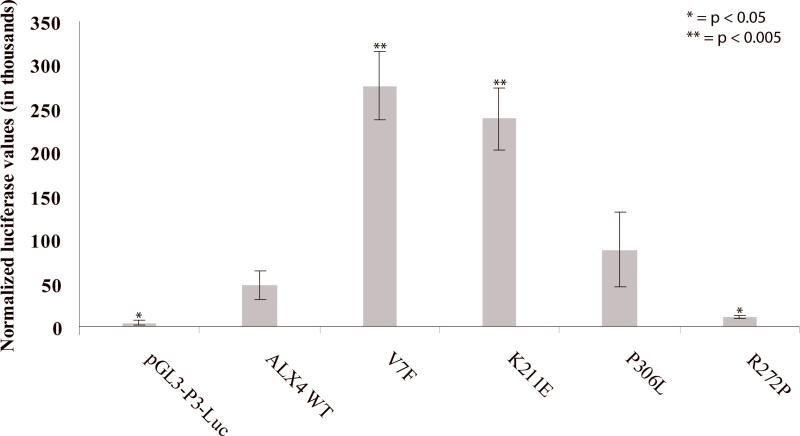
In order to test if V7F, K211E and P306L will act in the predicted gain-of-function manner, we designed a dual-luciferase assay. Full-length ALX4 cDNA was cloned into a pCMV6 expression vector (pCMV6-ALX4) and mutagenized to introduce each of the three variations (pCMV-V7F, pCMV6-K211E and pCMV6-p306L). A known loss-of-function variant, R272P, (rs104894196) that segregates with parietal foramina was also created as a control (pCMV6-R272P) (see Supp. Methods). An ALX4 binding element, P3, was cloned upstream of the minimal promoter of firefly luciferase to create a reporter vector (pGL3-P3-Luc). When transfected into control human calvarial osteoblasts, pCMV6-V7F and pCMV6-K211E resulted in a statistically significant increase in the level of luminescence – 5.8-fold and 5.0-fold, respectively, as compared to pCMV6-ALX4 (Fig. 2). The pCMV6-P306L vector also resulted in an increase (1.9-fold) in luminescence, but no statistical significance was achieved. As expected, pCMV6-R272P showed decreased levels of luminescence compared to ALX4 wild-type, consistent with a loss-of-function effect.
Figure 2.
For a dual-luciferase assay, control calvarial osteoblasts were transfected with (1) pGL3-P3-Luc reporter vector and (2) one of five ALX4 isoforms in a pCMV6 vector with minimal promoter. Transfection of pGL3-P3-Luc (P3 binding element) alone resulted in negligible luciferase production. Six independent replicates were tested for each construct and the results are expressed as the mean ± SEM. Renilla vectors were used as internal transfection controls for all experiments.
The observed increase in luciferase activity by V7F and K211E constructs suggests a GOF effect of the variants. We did not further elucidate the molecular basis of this enhancement, but several mechanisms of GOF action can be suggested. Enhanced DNA binding ability is a common result of GOF mutations, seen in Rad51 [Fortin and Symington, 2002] and Tbx5 [Postma et al., 2008], among others. This is a potential mode of action for the K211E mutation, since it is located immediately N-terminal to the amino acid residues of the homeodomain region (residues 215-273) responsible for DNA binding. Alternatively, GOF effects could be achieved though modification of secondary and tertiary structure that allows either enhanced binding of transcriptional elements and cofactors, resulting in increased levels of gene transcription or loss of interaction with known ALX4 upstream repressors, such as TWIST1 [Loebel et al., 2002].. The V7F mutation is more likely to act though one of these mechanisms, by virtue of its N-terminal location and absence of any known DNA binding domains. Aberrant interaction of ALX4 with other proteins such as LEF-1 [Boras and Hamel, 2002] and CART1 [Qu et al., 1999] may be the basis of the GOF effects. Whatever the exact means, our results indicate that ALX4 variants may contribute to NSC by altering the expression of downstream targets. Alx4 knockouts in mice have been shown to influence the transcription of Fgfr1, Fgfr2, Fgf4, Fgf8, and Spp1 (secreted phosphoprotein involved in attachment of osteoclasts to bone matrix) [Antonopoulou et al., 2004]. The impact of ALX4 on these genes supports the idea that ALX4's function is important in calvarial development and alteration of this function may negatively impact calvarial development and possibly contribute to sagittal NSC development.
Given the fact that all variants are familial and the parents are unaffected, it is likely the observed ALX4 variants are low penetrance mutations that predispose, but do not cause NSC by themselves. Similar low penetrance mutations have been observed in other candidate genes for NSC by other investigators [Johnson et al., 2000; Seto et al., 2007]. The same type of incomplete penetrance and variable expressivity has also been seen in syndromic CS disorders such as Meunke syndrome [Lajeunie et al., 1999].. This further supports the idea that the genetic etiology of all forms of CS is complex and other genetic or environmental factors are likely to contribute to its etiology in concert with the predisposing genetic variants. For example, the proband with the K211E polymorphism in ALX4 was born prematurely at 26 weeks of gestation and had metopic and right squamosal suture fusion along with sagittal NSC. It has been suggested that prematurity is a risk factor for NSC [Sanchez-Lara et al., 2010]. Both parents were evaluated by a clinical geneticist and found to be unaffected. Similarly, the proband carrying the P306L variation who had single suture sagittal NSC was born from pregnancy with reduced amniotic fluid levels and it has been well documented that increased intrauterine pressure is a contributing factor for CS [Sanchez-Lara et al., 2010]. This proband's family history is significant for small uterus size of the mother as well as micrognathia, corrected surgically at age 20. However, neither parent has CS by clinical assessment. No risk factors were identified for the proband with the V7F variant, who also had single suture sagittal NSC. The family history was reported as unremarkable, and although the parents were not available for a clinical assessment, neither was reported to have any form of CS. All three probands were evaluated post-surgically and were confirmed not to have associated anomalies or dysmorphisms and had appropriate-for-age development.
It has been reported that osteoblast or osteoclast gene families such as bone morphogenic proteins [Hogan, 1996] and Wnt-family proteins [Kuhl and Wedlich, 1997] interact with ALX4. Variants in these genes may contribute to accelerated osteoblast differentiation along with mutations in ALX4. Indeed, Wnt signaling induction of TWIST1 and BMP2 induction of RUNX2 tightly control the transition of mesenchymal cells into osteoblast precursors [Marie et al., 2002]. It is also known that MSX1 and MSX2 control the size of the osteoprogenitor cell population [Kim et al., 1998]. As ALX4 and MSX2 share many characteristics and both cause the phenotype of parietal foramina, one can postulate that ALX4 could also be involved in controlling the balance between osteoprogenitor cells’ proliferation and differentiation.
Our work indicates that a small fraction of probands with NSC may be impacted by GOF variants in the coding region of ALX4. However, variations in the non-coding regulatory regions of ALX4 may also result in altered expression of the gene, thus causing accelerated differentiation of calvarial suture osteoblasts. We have previously performed candidate gene association studies using a cohort of 89 case-parent trios with isolated sagittal NSC and observed an association to ALX4 (rs1828656, p = 0.0091; rs1869480, p = 0.0091) [Boyadjiev et al., 2007]. Future quantitative analysis of ALX4 expression in osteoblasts from NSC patients may demonstrate a broader impact of ALX4 in the etiology of craniosynostosis.
Supplementary Material
Acknowledgments
The authors are highly indebted to all families who contributed to this study. S.A. Boyadjiev is partially funded through a Children's Miracle Network Endowed Chair and through grants K23 DE00462, R03 DE016342, and R01 DE016886 from NIDCR/NIH and M01-RR00052 from NCRR/NIH. Grant obtained by M. L. Cunningham (R01 DE018227) also contributed to this project. We are indebted to Bridget Wilson, Nisha Issac and Elijah Cherkez for patient recruitment, Jia Liu for bench work and John Graham, Jeffrey L. Marsh, Andrew O. M. Wilkie, Jayesh Panchal, James Boggan, Travis Tollefson, Craig Senders, Craig A. Vander Kolk, George Jallo and Maria Passos-Bueno for contributing clinical information and biospecimens for this project. All authors have confirmed that they have no conflicts of interest with regards to this manuscript.
References
- Antonopoulou I, Mavrogiannis LA, Wilkie AO, Morriss-Kay GM. Alx4 and Msx2 play phenotypically similar and additive roles in skull vault differentiation. J Anat. 2004;204(6):487–99. doi: 10.1111/j.0021-8782.2004.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boras K, Hamel PA. Alx4 binding to LEF-1 regulates N-CAM promoter activity. J Biol Chem. 2002;277(2):1120–7. doi: 10.1074/jbc.M109912200. [DOI] [PubMed] [Google Scholar]
- Boyadjiev SA, Kimonis V, Cunningham M, ICC Genetic analysis of non-syndromic craniosynostosis. Orthod Craniofac Res. 2007;10(3):129–37. doi: 10.1111/j.1601-6343.2007.00393.x. [DOI] [PubMed] [Google Scholar]
- Desai J, Shannon ME, Johnson MD, Ruff DW, Hughes LA, Kerley MK, Carpenter DA, Johnson DK, Rinchik EM, Culiat CT. Nell1-deficient mice have reduced expression of extracellular matrix proteins causing cranial and vertebral defects. Hum Mol Genet. 2006;15(8):1329–41. doi: 10.1093/hmg/ddl053. [DOI] [PubMed] [Google Scholar]
- Fortin GS, Symington LS. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes. EMBO J. 2002;21(12):3160–70. doi: 10.1093/emboj/cdf293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan BL. Bone morphogenetic proteins in development. Curr Opin Genet Dev. 1996;6(4):432–8. doi: 10.1016/s0959-437x(96)80064-5. [DOI] [PubMed] [Google Scholar]
- Howard T, Paznekas W, Green E, Chiang L, Ma N, Luna R.O.d., Delgado CG, Gonzalez-Ramos M, Kline A, Jabs E. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat Genet. 1997;15(1):36–41. doi: 10.1038/ng0197-36. [DOI] [PubMed] [Google Scholar]
- Jabs E, Muller U, Li X, Ma L, Luo W, Haworth I, Klisak I, Sparkes R, Warman M, Mulliken J, Snead M, Maxson R. A mutation in the homeodomain of the human MSX2 gene in a family affected with autosomal dominant craniosynostosis. Cell. 1993;75:443–450. doi: 10.1016/0092-8674(93)90379-5. [DOI] [PubMed] [Google Scholar]
- Johnson D, Wall S, Mann S, Wilkie A. A novel mutation, Ala315Ser, in FGFR2: a gene-environment interaction leading to craniosynostosis? Eur J Hum Genet. 2000;8(8):571–577. doi: 10.1038/sj.ejhg.5200499. [DOI] [PubMed] [Google Scholar]
- Kim H, Rice D, Kettunen P, Thesleff I. FGF-, BMP-, and Shh-mediated signalling pathways in the regulation of cranial suture morphogenesis and calvarial bone development. Development. 1998;125:1241–1251. doi: 10.1242/dev.125.7.1241. [DOI] [PubMed] [Google Scholar]
- Kimonis V, Gold JA, Hoffman TL, Panchal J, Boyadjiev SA. Genetics of craniosynostosis. Semin Pediatr Neurol. 2007;14(3):150–61. doi: 10.1016/j.spen.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Kuhl M, Wedlich D. Wnt signalling goes nuclear. Bioessays. 1997;19(2):101–4. doi: 10.1002/bies.950190204. [DOI] [PubMed] [Google Scholar]
- Lajeunie E, Crimmins DW, Arnaud E, Renier D. Genetic considerations in nonsyndromic midline craniosynostoses: a study of twins and their families. J Neurosurg. 2005;103(4 Suppl):353–6. doi: 10.3171/ped.2005.103.4.0353. [DOI] [PubMed] [Google Scholar]
- Lajeunie E, El Ghouzzi V, Le Merrer M, Munnich A, Bonaventure J, Renier D. Sex related expressivity of the phenotype in coronal craniosynostosis caused by the recurrent P250R FGFR3 mutation. J Med Genet. 1999;36(1):9–13. [PMC free article] [PubMed] [Google Scholar]
- Loebel DA, O'Rourke MP, Steiner KA, Banyer J, Tam PP. Isolation of differentially expressed genes from wild-type and Twist mutant mouse limb buds. Genesis. 2002;33(3):103–13. doi: 10.1002/gene.10091. [DOI] [PubMed] [Google Scholar]
- Marie PJ, Debiais F, Hay E. Regulation of human cranial osteoblast phenotype by FGF-2, FGFR-2 and BMP-2 signaling. Histol Histopathol. 2002;17(3):877–85. doi: 10.14670/HH-17.877. [DOI] [PubMed] [Google Scholar]
- Mavrogiannis LA, Taylor IB, Davies SJ, Ramos FJ, Olivares JL, Wilkie AO. Enlarged parietal foramina caused by mutations in the homeobox genes ALX4 and MSX2: from genotype to phenotype. Eur J Hum Genet. 2006;14(2):151–8. doi: 10.1038/sj.ejhg.5201526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill AE, Bochukova EG, Brugger SM, Ishii M, Pilz DT, Wall SA, Lyons KM, Wilkie AO, Maxson RE., Jr. Cell mixing at a neural crest-mesoderm boundary and deficient ephrin-Eph signaling in the pathogenesis of craniosynostosis. Hum Mol Genet. 2006;15(8):1319–28. doi: 10.1093/hmg/ddl052. [DOI] [PubMed] [Google Scholar]
- Mundlos S, Otto F, Mundlos C, Mulliken J, Aylsworth A, Albright S, Lindhout D, Cole W, Henn W, Knoll J, Owen M, Mertelsmann R, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- Passos-Bueno MR, Serti Eacute AE, Jehee FS, Fanganiello R, Yeh E. Genetics of craniosynostosis: genes, syndromes, mutations and genotype-phenotype correlations. Front Oral Biol. 2008;12:107–43. doi: 10.1159/000115035. [DOI] [PubMed] [Google Scholar]
- Postma AV, van de Meerakker JB, Mathijssen IB, Barnett P, Christoffels VM, Ilgun A, Lam J, Wilde AA, Lekanne Deprez RH, Moorman AF. A gain-of-function TBX5 mutation is associated with atypical Holt-Oram syndrome and paroxysmal atrial fibrillation. Circ Res. 2008;102(11):1433–42. doi: 10.1161/CIRCRESAHA.107.168294. [DOI] [PubMed] [Google Scholar]
- Qu S, Tucker SC, Zhao Q, deCrombrugghe B, Wisdom R. Physical and genetic interactions between Alx4 and Cart1. Development. 1999;126(2):359–69. doi: 10.1242/dev.126.2.359. [DOI] [PubMed] [Google Scholar]
- Rice D, Aberg T, Chan Y, Tang Z, Kettunen P, Pakarinen L, Maxson R, Thesleff I. Integration of FGF and TWIST in calvarial bone and suture development. Development. 2000;127:1845–1855. doi: 10.1242/dev.127.9.1845. [DOI] [PubMed] [Google Scholar]
- Sanchez-Lara PA, Carmichael SL, Graham JM, Jr., Lammer EJ, Shaw GM, Ma C, Rasmussen SA. Fetal constraint as a potential risk factor for craniosynostosis. Am J Med Genet A. 2010;152A(2):394–400. doi: 10.1002/ajmg.a.33246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto ML, Hing AV, Chang J, Hu M, Kapp-Simon KA, Patel PK, Burton BK, Kane AA, Smyth MD, Hopper R, Ellenbogen RG, Stevenson K, et al. Isolated sagittal and coronal craniosynostosis associated with TWIST box mutations. Am J Med Genet A. 2007;143(7):678–86. doi: 10.1002/ajmg.a.31630. [DOI] [PubMed] [Google Scholar]
- Shevde NK, Bendixen AC, Maruyama M, Li BL, Billmire DA. Enhanced activity of osteoblast differentiation factor (PEBP2alphaA2/CBFa1) in affected sutural osteoblasts from patients with nonsyndromic craniosynostosis. Cleft Palate Craniofac J. 2001;38(6):606–14. doi: 10.1597/1545-1569_2001_038_0606_eaoodf_2.0.co_2. [DOI] [PubMed] [Google Scholar]
- Spruijt L, Verdyck P, Van Hul W, Wuyts W, de Die-Smulders C. A novel mutation in the MSX2 gene in a family with foramina parietalia permagna (FPP). Am J Med Genet A. 2005;139(1):45–7. doi: 10.1002/ajmg.a.30923. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Thiele H, Baldermann C, Kruger A, Giannakudis I, Dorr S, Werner N, Kunz J, Rappold GA, Hansmann I. Phenotypic findings due to trisomy 7p15.3-pter including the TWIST locus. Am J Med Genet. 2001;103(1):56–62. doi: 10.1002/ajmg.1512. [DOI] [PubMed] [Google Scholar]
- Zhang X, Carpenter D, Bokui N, Soo C, Miao S, Truong T, Wu B, Chen I, Vastardis H, Tanizawa K, Kuroda S, Ting K. Overexpression of Nell-1, a craniosynostosis-associated gene, induces apoptosis in osteoblasts during craniofacial development. J Bone Miner Res. 2003;18(12):2126–34. doi: 10.1359/jbmr.2003.18.12.2126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.