SUMMARY
The mechanisms responsible for the establishment of physical domains in metazoan chromosomes are poorly understood. Here we find that physical domains in Drosophila chromosomes are demarcated at regions of active transcription and high gene density that are enriched for transcription factors and specific combinations of insulator proteins. Physical domains contain different types of chromatin defined by the presence of specific proteins and epigenetic marks, with active chromatin preferentially located at the borders and silenced chromatin in the interior. Domain boundaries participate in long-range interactions that may contribute to the clustering of regions of active or silenced chromatin in the nucleus. Analysis of transgenes suggests that chromatin is more accessible and permissive to transcription at the borders than inside domains, independent of the presence of active or silencing histone modifications. These results suggest that the higher-order physical organization of chromatin may impose an additional level of regulation over classical epigenetic marks.
Keywords: Chromatin, transcription, nucleus, epigenetics
INTRODUCTION
The issue of how the genome is organized in the three-dimensional space of the eukaryotic nucleus and how this organization affects the regulation of gene expression remains an important question (Cremer and Cremer, 2001; Lanctot et al., 2007). This organization must allow the package of the genome within the confines of the nucleus while allowing access of the transcription and replication machineries to the DNA (Henikoff, 2010; Zhao et al., 2009). The use of microscopy-based approaches has allowed us to obtain critical insights into the relationship between nuclear organization and gene expression (Bian and Belmont, 2012; Hu et al., 2009; Misteli, 2010; Schermelleh et al., 2008; Strukov et al., 2011). The recent introduction of Hi-C, an extension of the original Chromosome Conformation Capture (3C) method (Dekker et al., 2002), allows comprehensive mapping of global chromatin interactions at a resolution determined primarily by three factors - DNA fragment length, sequencing depth and genome size. (Duan et al., 2010; Lieberman-Aiden et al., 2009; Tanizawa et al., 2010). Using this approach, human chromosomes were found to be partitioned into two types of compartments correlating with active gene dense regions and repressive gene poor regions, respectively (Lieberman-Aiden et al., 2009).
Recent work in Drosophila, mouse and human systems using 5C and Hi-C has revealed that genomes are further partitioned below the megabase length scale into physical chromosome domains that correlate with active and repressive chromatin states (Dixon et al., 2012; Nora et al., 2012; Sexton et al., 2012). Due to its small genome size, Hi-C analysis of Drosophila embryonic nuclei identified physical chromosomal domains at kb level resolution. These domains are demarcated by insulator proteins and generally correlate with four distinct epigenetic chromatin states (Sexton et al., 2012). Insulators were originally characterized based on their ability to prevent enhancer-promoter interactions or to block the spreading of heterochromatin in transgene assays. More recently, insulators have been shown to tether enhancers to distant promoters, to separate different epigenetic domains, and to recruit H3K27me3 domains to Polycomb (Pc) bodies (Handoko et al., 2011; Li et al., 2011; Pirrotta and Li, 2012; Schwartz et al., 2012; Van Bortle et al., 2012). Here we describe a high resolution analysis of the arrangement of Drosophila chromosomes in Kc cells. We find that, although specific combinations of insulator proteins are enriched at domain boundaries, their role in the establishment of these domains cannot be separated from other factors such as transcription levels and gene density. Physical domains of chromosomes are distinct from epigenetic domains defined by the presence of specific histone modifications. Importantly, the higher-order compaction of the chromatin within the physical domains appears to impose an additional layer of regulation on gene expression independent of the active or silencing chromatin marks of the 10 nm chromatin fiber.
RESULTS
Partition of the Drosophila Genome into Physical Domains
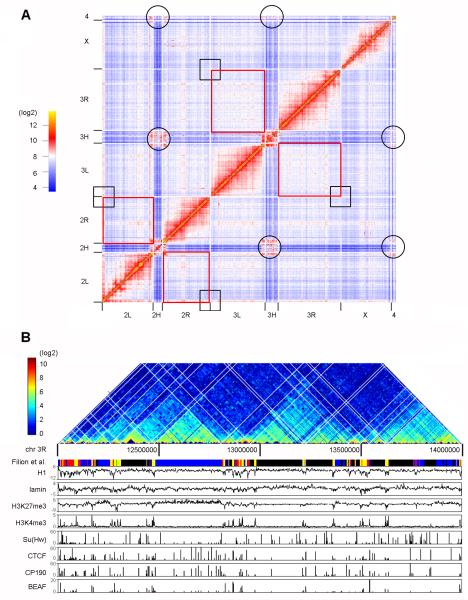
We generated Hi-C libraries using Drosophila Kc167 cells and the HindIII restriction endonuclease, which digests the fly genome into 33,004 fragments with a median size of 3.6 kb. Comparisons between technical and biological replicates show strong correlations at single fragment resolution (Pearson’s correlation r=0.991 and r=0.894, respectively) for genome wide interactions (Figure S1A). Interacting pairs were randomly chosen and confirmed by qPCR on 3C samples (Figure S1B). In total we obtained 373 million paired-end ligations (see Extended Experimental Procedures). This number of reads allows the identification of statistically significant contacts at a resolution of 4 kb within 100-140 kb regions (Figure S1C) and at 20 kb resolution within 4-9 Mb regions, depending on the chromosome (Figure S1D). The chromatin interaction heat map confirms the clustering of centromeres (circles in Figure 1A) but does not detect significant interactions between telomeres in Kc cells (black squares in Figure 1A). Contrary to observations in embryonic nuclei, intra-chromosomal inter-arm interactions (2L-2R and 3L-3R, marked by red squares in Figure 1A) show no obvious increase in fragment contact frequencies over that observed for inter-chromosomal inter-arm associations. These results are consistent with previous reports indicating that Pc domains only interact within the same arm but not with Pc domains in the other arm of the same chromosome (Tolhuis et al., 2011).
Figure 1. Partition of the Drosophila Genome into Physical Domains.
(A) Genome-wide interaction heat map at 100 kb resolution for the Drosophila genome in Kc167 cells. Black circles and squares show interactions between centromeres and telomeres, respectively. Red rectangles show interactions between chromosome arms 2L-2R and 3L-3R, respectively.
(B) Hi-C interaction frequencies displayed as a two-dimensional heat map at single fragment resolution for a 2 Mb region of chromosome 3R alongside with selected epigenetic marks and chromatin types defined by the presence of various proteins and histone modifications. The white grid on the heat map shows where the domains are partitioned.
The interaction heat map at single fragment resolution in a 2 Mb region of chromosome 3 shows distinct sub-genomic physical domains of intense local interactions (Figure 1B). To systematically map and identify these structures, we developed a Bayesian model-based probability test to optimize the domain partition of the Drosophila genome. A total of 1110 physical domains were identified covering 92% of the 130 Mb fly genome. The median domain size is 61 kb and the average size is 107 kb (Figure S2A, Table S2). We then compared the overlapping frequency of borders for the two sets of domains identified in Drosophila, here for Kc167 cells and previously for embryonic nuclei (Sexton et al., 2012). Forty two percent of domain partition sites (DPSs, sites between two adjacent physical domains) identified in Kc167 cells coincide with those mapped in embryonic nuclei, which is significantly higher than expected (Figure S2B and S2C, Fisher’s exact test, p =1.03×10−58). The observed differences between embryos and Kc cells could be due to the presence of multiple cell types in the mixed stage embryos used to map chromosome domains or could represent alterations in the physical organization of chromosomes in various cell lineages.
Physical Domain Partition Occurs Predominantly in Active Chromatin
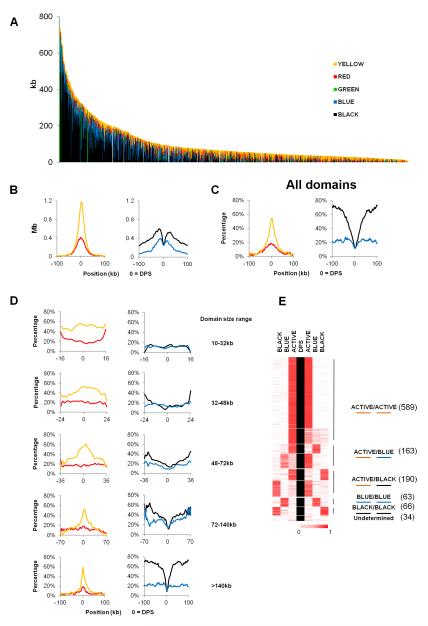
To determine whether physical chromosome domains correspond to functional domains defined by epigenetic marks we examined the composition of chromatin types within physical domains. We followed the chromatin classification established previously, in which chromatin types are defined by the presence of specific chromatin proteins and histone modifications (Filion et al., 2010). YELLOW and RED chromatin contain proteins and histone modifications characteristic of active chromatin. BLUE chromatin contains H3K27me3 and PcG proteins, GREEN chromatin contains Hp1 and Su(var)3-9, and BLACK chromatin contains Lamin (LAM) and histone H1. For domains identified in Kc167 cells the percentage of active YELLOW chromatin negatively correlates with physical domain size (Spearman correlation, r=-0.617, p<10−22) whereas the percentage of repressive BLACK and BLUE chromatin correlates positively with physical domain sizes (Spearman correlation, r=0.638 and 0.630 respectively, p<10−22) (Figure 2A); this is also the case for domains identified in embryo nuclei (Figure S2D). This observation suggests that domains rich in active chromatin tend to be smaller than those rich in silenced chromatin. We then aligned the domain boundaries and calculated the absolute size and percentage of each chromatin type in 2 kb windows flanking the DPSs up to 100 kb upstream and downstream (Figure 2B and 2C). Strikingly, YELLOW and RED chromatin are sharply enriched at boundary regions, increasing to the highest point around the DPSs (Figure 2C). In contrast, the percentage of BLACK chromatin drops sharply at boundary regions and BLUE chromatin slowly decreases to the lowest point around the DPSs (Figure 2C). The same is also true for physical domain boundaries in nuclei from Drosophila embryos (Figure S3A). GREEN chromatin shows an uneven pattern around DPSs and account for less than 5% of total chromatin at the boundaries (Figure S3B).
Figure 2. Physical Domain Boundaries Are Preferentially Formed in Regions of Active Chromatin.
(A) Size distribution of chromatin types within each physical domain. Domains are arranged in order of size, from largest (left) to smallest (right). Chromatin types (Filion et al., 2010) within domains are arranged in the order of BLACK, BLUE, GREEN, RED and YELLOW from the bottom to the top.
(B) Distribution of chromatin size for aligned domains in 2 kb windows surrounding domain partition sites (DPSs). Sizes of total chromatin (left), active chromatin types (middle) and repressive chromatin types (right) are shown.
(C) Percentage of active (RED and YELLOW) and repressive (BLUE and BLACK) chromatin surrounding DPSs for all domains. Total size of each chromatin type within each 2 kb window was divided by the total size of all chromatin in that window to obtain the percentage values.
(D) Percentage of each chromatin type surrounding DPSs for domains of different sizes (10-32, 32-48, 48-72, 72-140 and ≥140 kb) calculated as described in (C).
(E) Five groups of physical domain borders identified by clustering the percentages of ACTIVE (YELLOW & RED), BLUE and BLACK chromatin types within the first 4 kb bins flanking DPSs for each border. Borders showing similar chromatin contents are clustered and symmetrical clusters are grouped. The number of each border found in each group is listed on the right.
The contrasting patterns of enrichment of active and repressive chromatin may be due to the fact that, as distance increases from the DPS, the number of small domains containing active chromatin decreases (Figure 2A). To address this issue, we grouped the right half (left of the DPSs) and the left half (right of the DPSs) of domains into 5 groups of increasing size, each containing the same number of 222 domains, and calculated the percentage of each chromatin type present in 2 kb windows. For small domains, since they contain mostly active chromatin, the amount of YELLOW and RED chromatin remains more or less constantly high throughout the domain; the same is true for repressive BLUE and BLACK chromatin, which it remains low throughout the domains (Figure 2D). On the other hand, for domains larger than 48 kb, YELLOW chromatin is more enriched at the highest point surrounding the DPSs (Figure 2D) whereas the fraction of BLACK chromatin increases as one moves away from the DPSs (Figure 2D). These results indicate that small domains contain mostly YELLOW chromatin. Repressive BLUE and BLACK chromatin, which constitute the majority of the genome, must then be contained within large domains. Indeed, the right half (left of the DPSs) and the left half (right of the DPSs) of large physical domains (>48 kb) show an increased enrichment of BLACK and BLUE chromatin in the internal regions, while their boundary regions are invariably enriched with active YELLOW and RED chromatin (Figure 2D).
To better categorize the domain boundary regions based on chromatin types, we calculated the percentage of each chromatin type in the first 4 kb bins flanking each DPS. Approximately 85% of boundary regions contain active YELLOW or RED chromatin at least on one side of the DPS and more than 60% have active chromatin on both sides (Figure 2F). Nevertheless, a small fraction of domain boundaries contain BLUE or BLACK chromatin on both sides. Analysis of the chromatin composition of physical domains in embryo nuclei shows similar distribution patterns of active and repressive chromatin types (Figures S3C and S3D).
Domain Boundaries are Preferentially Located at Gene-dense Regions
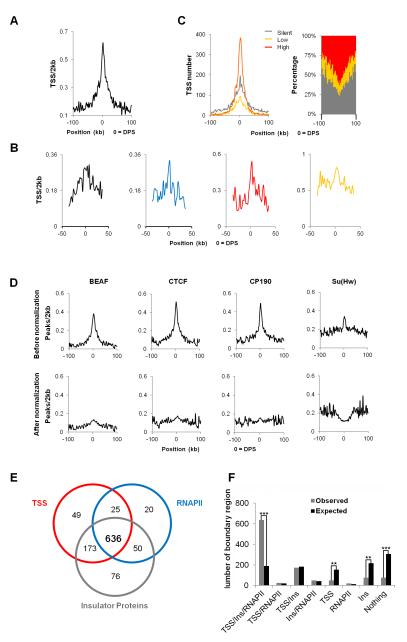
Active and repressive chromatin differ in protein binding profiles and in transcription level but, more importantly, in gene density. The preferential localization of domain boundaries at sites enriched for active chromatin suggests that the selection of domain partition sites may be rooted in the arrangement of genes in the genome. We therefore examined gene enrichment in the region surrounding DPSs and observed that gene density is highest at DPSs (Figure 3A). Analysis of gene density at domain boundaries containing different chromatin types suggests that the increase in the number of genes in inter-domain regions is true for either active or silenced chromatin (Figure 3B). Gene density also forms a sharp peak within a narrow 6 kb range at the borders of domains identified in embryo nuclei, as well as for those present in active or repressed chromatin (Figure S3E). We then examined the transcriptional status of genes located adjacent to inter-domain boundaries. Figure 3C shows that, although actively transcribed genes are enriched at the boundaries, genes that are transcribed at low levels or completely silent are also enriched in these regions. These results support the conclusion that high gene density, independent of the transcriptional state, may be one of the driving forces in the establishment of physical domain partitions in Drosophila chromosomes.
Figure 3. Domain Borders are Located in Gene-dense Regions.
(A) TSS density surrounding DPSs in Kc167 cells. Total number of TSSs in each 2 kb window flanking aligned DPSs were counted and divided by the total chromatin size in the same window.
(B) TSS density in each specific chromatin type surrounding DPSs calculated as described in (A) but only TSSs within each specific chromatin in a 2 kb window were used.
(C) Expression levels of genes surrounding DPSs. Absolute gene numbers are shown in the left panel within 2 kb windows and the percentage of genes at each expression level is shown in the right panel.
(D) Insulator protein enrichment surrounding DPSs before (upper panel) and after (lower panel) normalization against TSSs density.
(E) Venn diagram showing the number of DPSs with a given mark (TSSs, RNAPII or insulator proteins) within a +/−4 kb window surrounding DPSs. There are 76 DPSs without any of these three marks.
(F) Number of DPSs associated with a given mark (TSSs, RNAPII or insulator proteins) for observed (grey bars) and the expected (black bars) boundary regions. Statistically significant differences in the comparisons are indicated by double asterisk (**) (p < 1.00E-15) and triple asterisks (***) (p < 1.00E-80), respectively (Fisher’s exact test).
Insulator Proteins are Enriched at Domain Boundaries
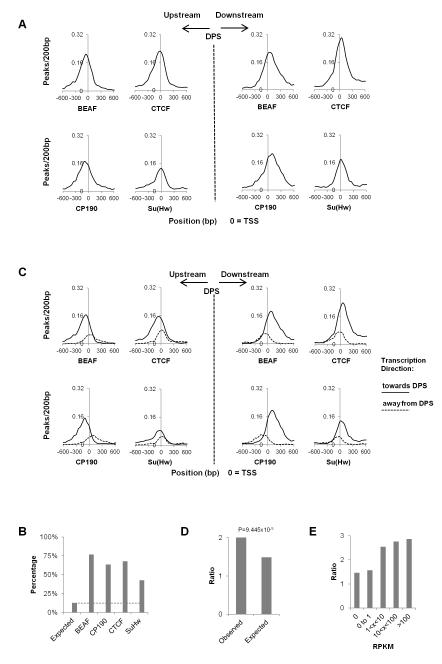
Insulator proteins BEAF, CTCF and CP190 are enriched at boundaries of physical domains in Kc cell chromosomes (Figure 3D, upper panel). This enrichment may be a consequence of their presence upstream of TSSs of active genes in the Drosophila genome (Bushey et al., 2009) and the enrichment of active genes in these regions. Indeed, normalization of the number of insulator protein binding sites relative to gene density results in a drastic reduction in their enrichment to a level only slightly higher than the genome average (Figure 3D, lower panel). Boundary regions containing TSSs associated with RNAPII and insulator proteins account for 57% (636) of all physical domain borders (Figure 3E and 3F), compared to 17% (191) of random domains (Figure 3F, Figure S4A) (Fisher’s exact test, p = 3.26×10−88). On the other hand, the number of boundary regions with either TSSs, RNAPII or insulator proteins, combination of any two, or none of them, are statistically insignificant or significantly lower than expected, which confirms that the coexistence of genes, active transcription, and insulator proteins is a signature of physical domain boundaries (Figure 3F, Figure S4A). Consistent with this idea, histone modifications characteristic of active transcription, DNaseI hypersensitive sites, and various proteins related to transcription, are also found enriched at domain boundary regions flanking DPSs (Figure S4B and S4C) more frequently than expected (Figure S4D). Surprisingly, PSC, but not other PcG members, is also found enriched at boundary regions at levels similar to those of Su(Hw) (Figure S4C and S4D). These results suggest that physical domain borders may be formed by a combination of active transcription, high gene density, and insulator proteins.
Specific Combinations of Insulator Proteins are Enriched at Physical Domain Borders
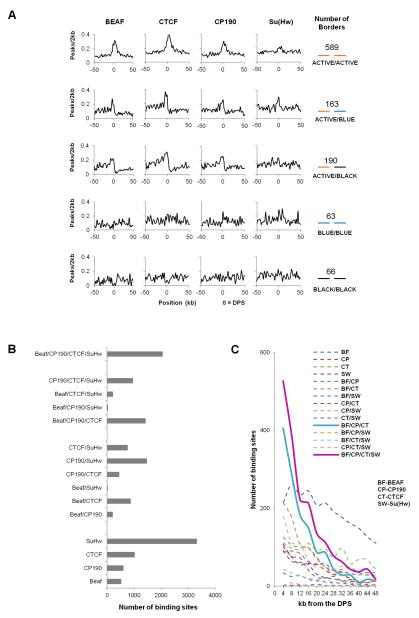
Drosophila has several insulator proteins, including BEAF, CTCF, Su(Hw) and CP190. We first examined whether the enrichment observed in Figure 3C is different at the domain borders of the various chromatin types. Figure 4A shows that BEAF and CTCF are mostly found at boundaries of active chromatin, Su(Hw) is slightly enriched at boundary regions containing BLUE chromatin, and CP190 is enriched at all boundaries except those containing BLACK chromatin (Figure 4A). We have previously shown that these four insulator proteins often co-localize at many sites through the genome (Van Bortle et al., 2012). To test whether specific combinations of insulator proteins cluster at domain boundaries, we examined the distribution of all possible combinations of these four insulator proteins. We consider that proteins co-localize if the summits of the binding peaks derived from ChIP-seq analysis are within a 300 bp window. The total number of sites for each combination of insulator proteins is highest for sites where all four insulator proteins are present together (Figure 4B). These sites may therefore represent especially strong insulators. We then plotted the distribution of single insulator protein sites as well as all possible combinations in 4kb bins with respect to the location of DPSs. Strikingly, two combinations - BEAF/CTCF/CP190 and BEAF/CTCF/Su(Hw)/CP190 - show strong enrichment at domain borders (Figure 4C).
Figure 4. Specific Combinations of Insulator Proteins are Present at Domain Boundary Regions.
(A) Single insulator protein enrichment surrounding DPSs at five groups of clustered domain borders. In each 2 kb window, total peaks for each insulator protein were counted and divided by the fraction of each chromatin type present in that window for each group of aligned borders.
(B) Number of single insulator protein sites an various protein combinations in 100 kb regions upstream and downstream of DPSs.
(C) Distribution of single insulator protein sites and various protein combinations in 4kb windows over a 48 kb region from DPSs.
Genes Adjacent to Domain Boundaries are Preferentially Transcribed Towards the Boundary
Since insulator proteins are preferentially located close to actively transcribed genes and boundary regions are enriched in transcription start sites of active genes, we examined the location of insulator proteins with respect to the DPSs and TSSs of adjacent genes. We aligned the first TSS on either side of the DPSs with the location of insulator proteins. BEAF, CTCF and CP190 are found close to TSSs but, surprisingly, are shifted distally from the TSSs with respect to the DPSs (Figure 5A). More than 60% CTCF, BEAF and CP190 sites present at boundary regions are located +/−500 bp from the TSS whereas only 42% of Su(Hw) sites are present in this regions (Figure 5B). BEAF, CTCF and CP190 are more enriched within 200 bp upstream than downstream of TSSs of active genes. The unexpected enrichment of insulator proteins more distally from the DPSs than the TSSs suggests that the adjacent genes on either side of the DPSs are more frequently transcribed towards the DPSs. To test this we divided the TSSs into two groups based on gene orientation and aligned them separately to either side of the DPSs. Two enrichment peaks were found, with the higher one corresponding to genes transcribed towards the DPSs and the lower one corresponding to genes transcribed away from the DPSs (Figure 5C). In agreement with this, the ratio between adjacent genes transcribed towards the DPSs and genes transcribed away from DPSs is significantly higher than expected (Figure 5D, Fisher exact test, p=9.445×10−5). Since the insulator proteins BEAF, CTCF and CP190 are also preferentially enriched at promoters of active genes, we wonder whether the non-random gene orientation at domain boundaries could be even higher for adjacent highly transcribed genes; this is indeed the case as shown in Figure 5E and Figure S4E. This unexpected pattern of gene orientation and insulator protein distribution may help to prevent the influence from less active internal domains on the more active boundary regions.
Figure 5. Gene Orientation Bias at Domain Boundary Regions.
(A) Enrichment of insulator protein peaks surrounding the TSSs of the genes on both sides and immediate adjacent to the DPSs. TSSs are aligned with the gene orientation unaltered.
(B) Percentage of total insulator protein peaks present in boundary regions (DPSs +/− 4kb) and located within +/−0.5 kb from TSSs.
(C) Enrichment of insulator proteins surrounding the aligned TSSs of the genes adjacent to and on either side of the DPSs.
(D) Ratios between the genes adjacent to the DPSs transcribed toward and away from the DPSs for identified and randomly created domains (p < 1.00×10−4, Fisher’s exact test).
(E) Ratios between the genes adjacent to the DPSs transcribed toward and away from the DPSs with different expression levels.
Domain Boundary Sequences are Involved in Long-range Interactions
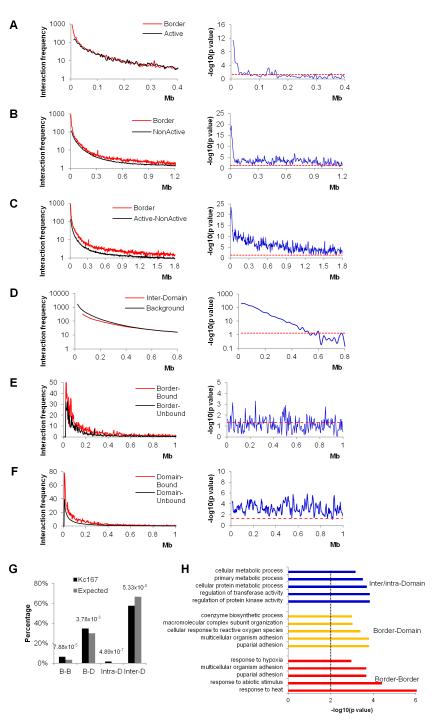
Previous work with mammalian cells has demonstrated that transcription factories can be formed by the clustering of multiple transcribed genes (Osborne et al., 2004; Schoenfelder et al., 2010). Drosophila and vertebrate insulator proteins have been shown to mediate long-range interactions (Handoko et al., 2011; Hou et al., 2010; Wood et al., 2011) and have been proposed to facilitate clustering of active genes at transcription factories and silenced genes at Polycomb (Pc) bodies (Li et al., 2011; Pirrotta and Li, 2012; Schwartz et al., 2012; Van Bortle et al., 2012). The fact that domain boundaries in Drosophila are enriched in active genes and insulator proteins suggests that domain boundaries may interact more frequently than other regions of the genome in order to cluster active or silenced genes. These interactions may be responsible for the disruption of the continuity of local chromatin condensation that results in the formation of inter-domain boundaries. To test this hypothesis, we compared interaction frequencies through the genome relative to genomic distance for four categories of 10 kb bins - interactions between bins at the boundary regions; between any two bins of active chromatin; between any two bins of inactive chromatin; and between any two bins with active chromatin in one bin and inactive chromatin in the other bin. At 10 kb resolution, interactions between bins at boundary regions are higher than interactions between bins containing active chromatin within genomic distances up to 60 kb (Figure 6A, Wilcoxon test, p<0.05). Interactions between boundary regions are also higher than interactions between bins of inactive chromatin (Figure 6B, Wilcoxon test, p<0.05) and even higher than interactions between bins containing different types of chromatin (Figure 6C, Wilcoxon test, p<0.05) within 1-2 Mb examined. Contrary to this, interaction frequencies between bins located in different domains are lower than the genome average within 500 kb (Figure 6D, Wilcoxon test, p<0.05), and are similar to the genomic background interaction frequencies at distance beyond 500 kb (Figure 6D). Comparison of interaction frequencies between borders and between paired bins of different chromatin types in embryonic nuclei shows less significant preference for border interactions, which may be due to the fact that borders for embryonic Hi-C data were called based on the average ligation frequency in a mixture of cell types (Figure S5). These results suggest that domain boundaries interact more frequently among themselves over long-distances than internal domain regions independent of the type of chromatin present at the interacting sites.
Figure 6. Long-range Interactions among Domain Boundaries.
(A) Interaction frequencies between boundary regions (Border, red line) compared to the genomic background active chromatin (Active, black line) are shown in the left panel. The right panel shows the p values for each distance examined (Wilcoxon test) with the red dashed line representing a p value of 0.05.
(B) Interaction frequencies between boundary regions (Border, red line) compared to the genomic background inactive chromatin (Non-active, black line) in the left panel. The right panel is as in (A).
(C) Interaction frequencies between boundary regions (Border, red line) compared to the genomic background active-inactive chromatin (Active-Non-active) in the left panel. The right panel is as in (A).
(D) Interaction frequencies between domain bins (Inter-Domain, red) compared to genomic background (Background, black) in the left panel. The right panel is as in (A).
(E) Interaction frequencies between fragments at boundary regions associated with BEAF, CTCF, CP190 and RNAPII simultaneously (Border-Bound, red) compared to fragments at boundary regions without the simultaneous binding of the four proteins (Border-Unbound, black). The right panel is as in (A).
(F) Interaction frequencies between fragments located inside of two different domains. Fragments associated with BEAF, CTCF, CP190 and RNAPII simultaneously (Domain-Bound, red) are compared to fragments without the simultaneous binding of the four proteins (Domain-Unbound, black). The right panel is as in (A).
(G) Percentage of interactions between boundary regions (B-B), boundary region and domain internal regions (B-D), inside domain (Intra-D) and between domains (Inter-D) identified at 20 kb resolution (black bars, Kc167). The same number interactions with the same size distribution as those identified experimentally were randomly created for each chromosome arm and reiterated 1000 times as a control (grey bars).
(H) Gene ontology analysis of genes involved in the interactions between Border-Border (red bars), Border-Domain (yellow bars) and Inter/intra-Domain (blue bars). The black dashed line indicates p=0.01.
We next examined DNA fragments simultaneously bound by BEAF, CTCF, CP190 and RNAPII (referred to as “bound”) and fragment not bound simultaneously by these proteins (referred to as “unbound”). For the more than 2,200 fragments adjacent to DPSs, interactions among bound fragments are generally more frequent than unbound fragments over nearly the whole distance range examined, but at a level not statistically significant (Figure 6E, Wilcoxon test, p>0.05). This lack of statistical significance could be due to the proximity of bound and unbound fragments at the boundaries, especially for domain borders with one side of active (YELLOW and RED) and the other side repressive (BLUE or BLACK) chromatin types. Contrary to boundary regions, bound fragments within domains show statistically significant higher inter-domain interacting frequencies than unbound fragments within about 900 kb (Figure 6F, Wilcoxon test, p<0.05).
To further understand the role of long range interactions in chromosome organization, we examine all interactions at 20 kb resolution and identified 1,703 statistically significant contacts (Table S3). Associations among boundary regions are significantly higher than expected, further confirming that boundary regions preferentially interact among themselves (Figure 6G, Fisher’s exact test, p<1.00×10−4). At the same time, the frequency of interactions between domains is lower (Figure 6G, Fisher’s exact test, p<1.00×10−7). Taken together, these results show preferential contacts among boundary regions that may disrupt the continuity of local chromatin interaction and create a “weak” point in the genome identified as a physical domain partition in the Hi-C analysis. For small domains, primarily composed of active chromatin, this analysis suggests their preferential clustering may be due to the enrichment of active gene transcription and insulator proteins.
We then carried out gene ontology (GO) analysis for genes involved in different groups of interactions (Figure 6H, 5 categories with lowest p values are shown). Interestingly, genes with border-border interactions are mostly enriched in processes responsive to environmental or physiological stress, while genes with inter/intra-domain interactions are primarily enriched in metabolic processes. Consistently, genes with border-domain interactions are enriched in processes similar either to genes with border-border or to inter/intra-domain interactions (Figure 6H). It is possible that the presence of stress-inducible at domain borders allows them to be rapidly induced and co-regulated in response to environmental or physiological stimulation.
Domain Borders are More Accessible and Permissive to Transcription than Internal Regions
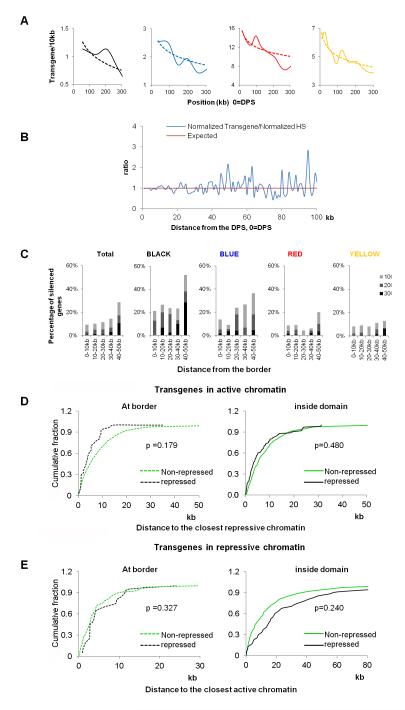
Physical domains in chromosomes arise from a high number of interactions between sequences confined to a specific region of the chromosome. It is possible that these interactions result in a higher degree of compaction of the DNA inside the domains with respect to the border regions. To test this possibility, we examined a data set of 2,852 random P element insertions that carry a white reporter gene expressed in the eye pigment cells. The expression level and insertion site for each transgene has been reported previously(Babenko et al., 2010). Several additional datasets of 29,419 P element insertion sites were also included in the analysis (Bellen et al., 2011; Spradling et al., 2011; Venken et al., 2011). Most transgenes are inserted into YELLOW and RED chromatin but a smaller number of them are also inserted into repressive BLACK and BLUE chromatin (Figure S6A), suggesting that regions of the chromosome with histone modifications characteristic of active chromatin are more accessible than those containing silencing marks. When we examined the distribution of transgene insertion sites with respect to the location of physical domains, we found that most transgenes map close to DPSs (Figure S6A), and insertion rates decrease for most chromatin types as the distance from the DPSs increases, which correlates with enrichment in DNase I hypersensitive sites (Figure 7A and 7B, Figure S6B and S6D). This suggests that, independent of the chromatin type, the DNA in the physical domain boundary regions is more accessible than that in the domain internal regions. We then examined the expression levels of the transgenes in relation to their location with respect to domain boundaries. The results indicate that transgene expression is higher for those inserted in boundary regions, and decreases as the insertion site moves towards the interior of physical domains. This is true independent of the chromatin type (Figure 7C, Figure S6C). However, since inactive chromatin is more enriched away from domain boundaries, it is possible that the increased repression of transgenes in active chromatin is due to their presence close to repressive chromatin inside domains. Similarly, for transgenes inserted in repressive chromatin, the increased repression may be due to their distance far away from active chromatin. To test if this is the case, we divided transgenes into four groups (within DPS+/−10kb, or beyond this range, and at the border, or inside domains) and examined their distance distribution relative to the closest repressive or active chromatin type. The results show that there is no statistically significant difference (all p values > 0.15, KS test) in their relative distances to the closest repressive or active chromatin types, suggesting that the increased repression observed for transgenes present inside domains is not due to their proximity to repressive or active chromatin (Figure 7D and 7E). These results suggest that domain boundaries represent more accessible regions of the genome. Importantly, in addition to the type of chromatin defined by classical epigenetic marks, the location of the DNA within a physical domain may then serve as an additional “structural epigenetic mark” for genome function.
Figure 7. Analysis of Transgene Insertion and Expression in Physical Domains.
(A) Transgene insertion rates in each chromatin type relative to DPSs. The insertion rates were calculated by dividing the total transgene number in each chromatin type present in bins containing the same size of each specific chromatin.
(B) Two step normalization of insertion rate for transgenes against DNaseI hypersensitive sites (HS) density relative to the DPS.
(C) Percentage of transgenes repressed at different levels in 10 kb bins from DPSs in all domains (Total) and in domains containing various chromatin types.
(D) Cumulative percentage of transgene distribution relative to the closest repressive chromatin type. Transgenes repressed and non-repressed at borders are shown in the left panel and those repressed and non-repressed inside domains are shown on the right. Statistical significance was calculated using the Kolmogorov-Smirnov test (KS-test).
(E) Cumulative percentage of transgenes in repressive chromatin types relative to the closest active chromatin type. Transgenes distribution and statistical significance are plotted and calculated as in (D).
DISCUSSION
The use of Hi-C to map intra- and inter-chromosomal interactions in metazoan genomes has given important insights into the organization of the chromatin fiber in eukaryotic nuclei (Dixon et al., 2012; Kalhor et al., 2012; Lieberman-Aiden et al., 2009; Nora et al., 2012; Sexton et al., 2012). One important conclusion from these studies is that eukaryotic chromosomes are organized into a series of chromatin domains, perhaps formed by a series of local interactions among various regulatory sequences and the genes they control. Long-range interactions between chromatin domains may result in additional levels of folding to create larger domains (Bau et al., 2011; Lieberman-Aiden et al., 2009; Mirny, 2011). These results complement and converge with evidence suggesting that specific sequences come together in the nucleus in the process of, or with the purpose of, carrying out various nuclear processes. For example, actively transcribed genes and their regulatory sequences have been shown to colocalize at transcription factories (Cook, 2010; Osborne et al., 2004; Schoenfelder et al., 2010; Tolhuis et al., 2002), whereas genes silenced by PcG proteins converge at repressive factories termed Pc bodies (Bantignies et al., 2011). It is unclear whether these associations are a consequence of self-organizing principles with no functional outcomes i.e. they result from interactions among multiprotein complexes present at active or silenced genes, or they play a functional role in gene expression and are mediated by structural proteins specifically involved in mediating inter- and intra-chromosomal interactions (Misteli, 2007).
A critical roadblock in understanding the principles governing the folding of metazoan genomes is the identification of proteins or forces responsible for the formation of chromosome domains and the boundaries that separate these structures. Results from the analysis of mixed cell populations in Drosophila embryos indicate a correlation between the formation of domain boundaries and the presence of insulator proteins and the transcription factor Chromator (Sexton et al., 2012). Similar results in mouse and human cells find a high degree of correlation between the presence of CTCF and housekeeping genes and the formation of domain boundaries (Dixon et al., 2012; Nora et al., 2012)}.
To further explore the mechanisms of physical domain partition in metazoans, we carried out a Hi-C analysis using Drosophila Kc167 cells. We find that physical domains do not exactly correlate with functional domains defined by epigenetic marks. Furthermore, domain boundaries usually form at regions enriched for active histone modifications such as H3K4me3 but also form in regions enriched for silencing marks such as H3K27me3 and LAM. The common theme among domain boundaries, even those present in regions enriched for H3K27me3 and LAM, is a high density of actively transcribed genes. The likely causal role of transcription in the establishment of domains boundaries is underscored by the formation of multiple small physical domains in regions of the genome enriched for active genes. Regions of the genome enriched for silenced chromatin form large domains, with boundaries between these domains often forming when closely spaced and transcribed genes are present at the domain borders. The high correlation between gene density, transcription and the formation of domain boundaries help explain why these domains are conserved across different cell types of the same or different species (Dixon et al., 2012).
In agreement with these observations, RNAPII, transcription factors and insulator proteins are also found enriched at the borders of domains. Drosophila insulator proteins, with the exception of Su(Hw), are preferentially located adjacent to promoter regions of actively transcribed genes (Bushey et al., 2009). It is then possible that insulators play an active role in the formation of domain boundaries and that the observed increase in actively transcribed genes in these regions is a consequence of their close association with insulator proteins. Alternatively, active transcription in regions of high gene density may be the driving force behind the formation of physical domains and the enrichment of insulator proteins at the boundaries may be a result of their presence adjacent to these genes. Given the demonstrated role of insulators in mediating interactions between different sequences in the genome it is possible that a combination of these two possibilities is actually responsible for domain formation. An interesting observation that may offer additional clues as to the role of insulators in the formation of physical domains is the specific enrichment of clusters of insulator proteins at the boundaries. Drosophila insulator proteins Su(Hw), BEAF and CTCF bind specific DNA sequences and recruit CP190 and Mod(mdg4); these two proteins then interact with each other and/or themselves to bridge contacts between distant sites (Yang and Corces, 2012). The presence of multiple insulator DNA binding proteins would, presumably, make for a stronger insulator, able to mediate more frequent long-distance interactions. This hypothesis is supported by the observation that long-distance interactions involving domain boundaries are significantly higher than expected. These interactions can bring together highly transcribed regions, offering a mechanism to explain the formation of transcription factories (Schoenfelder et al., 2010).
An important question is whether this differential compaction of the chromatin between the inside and the borders of physical domains has an effect on gene expression. We have addressed this issue by examining the insertion frequency and the expression levels of a large collection of P element transgenes. The frequency of transgene insertion is much higher at the borders of the domains than in the interior, independent of the type of chromatin, suggesting that the DNA inside physical domains is more compacted than at the borders. Furthermore, independent of the epigenetic marks present in the chromatin, transgenes inserted in the region surrounding the domain boundaries are less repressed than those inserted in the domain interior. Therefore, the physical compaction of DNA arising from the higher-order organization of the chromatin may add a different layer of regulatory information superimposed on that resulting from classical epigenetic marks.
EXPERIMENTAL PROCEDURES
Hi-C and data analysis
Hi-C experiments were carried out as described (Lieberman-Aiden et al., 2009) with modifications. Control 3C experiments were carried out to validate that Hi-C libraries (Figure S1 and Table S1). Paired reads were aligned to the Drosophila reference genome (Dm3) using Bowtie 0.12.7 (Langmead et al., 2009). GC content and fragment length effects were normalized as described (Yaffe and Tanay, 2011) (See Extended Experimental Procedures).
Physical Domain Partition
We developed a probability model-based method assuming that the number of paired- end tags linking two loci follows a Poisson distribution with different intensity rates for intra-domain loci pairs and inter-domain loci pairs:
Here xij represents the number of tags linking loci i and j; dij represents the distance in terms of genomic coordinates between the two loci; i ~ j indicates that loci i and j are located within the same domain. The parameter β0 represents the background intensity rate for the paired-end tags; β1 > β0 represents the elevated intensity rate; γ0 and γ1 represent the decay rate of tag counts that is assumed to be linear with the genomic distance between the loci. The overall likelihood of observing all the intra-chromosomal paired-end tags can be written as follows:
. To estimate B, which represents the location of the boundary points, we used a Markov chain Monte Carlo (MCMC) strategy. Detailed description of the method can be found in the Extended Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Physical chromosome domains are generally flanked by active chromatin
The borders and interior of domains are gene-rich and gene-poor, respectively
Insulators and transcription factors are enriched at domain boundaries
Domain borders are more permissive to transcription independent of epigenetic marks
ACKNOWLEGMENTS
We would like to thank Drs. Hugo Bellen, Roger Hoskins and Robert Levis for help in collecting the data sets of P element insertions. We also thank Dr. Hao Wu for help with computational analyses and Dr. Chintong Ong for critical reading and suggestions on the manuscript. Research reported in this publication was supported by the National Institutes of Health under award numbers R01GM035463 to VC and R01HG005119 to ZQ. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACESSION NUMBERS Sequence data have been deposited in NCBI’s Gene Expression Omnibus under accession number GSE38468.
SUPPLEMENTAL INFORMATION Supplemental information, included Extended Experimental Procedures, six figures and four tables, can be found with this article.
REFERENCES
- Babenko VN, Makunin IV, Brusentsova IV, Belyaeva ES, Maksimov DA, Belyakin SN, Maroy P, Vasil’eva LA, Zhimulev IF. Paucity and preferential suppression of transgenes in late replication domains of the D. melanogaster genome. BMC Genomics. 2010;11:318. doi: 10.1186/1471-2164-11-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–226. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- Bau D, Sanyal A, Lajoie BR, Capriotti E, Byron M, Lawrence JB, Dekker J, Marti-Renom MA. The three-dimensional folding of the alpha-globin gene domain reveals formation of chromatin globules. Nat Struct Mol Biol. 2011;18:107–114. doi: 10.1038/nsmb.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, He Y, Carlson JW, Evans-Holm M, Bae E, Kim J, Metaxakis A, Savakis C, Schulze KL, et al. The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics. 2011;188:731–743. doi: 10.1534/genetics.111.126995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Q, Belmont AS. Revisiting higher-order and large-scale chromatin organization. Curr Opin Cell Biol. 2012 doi: 10.1016/j.ceb.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009;23:1338–1350. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PR. A model for all genomes: the role of transcription factories. J Mol Biol. 2010;395:1–10. doi: 10.1016/j.jmb.2009.10.031. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Summary: The nucleus--a close-knit community of dynamic structures. Cold Spring Harb Symp Quant Biol. 2010;75:607–615. doi: 10.1101/sqb.2010.75.051. [DOI] [PubMed] [Google Scholar]
- Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci U S A. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Kireev I, Plutz M, Ashourian N, Belmont AS. Large-scale chromatin structure of inducible genes: transcription on a condensed, linear template. J Cell Biol. 2009;185:87–100. doi: 10.1083/jcb.200809196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2012;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HB, Muller M, Bahechar IA, Kyrchanova O, Ohno K, Georgiev P, Pirrotta V. Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Mol Cell Biol. 2011;31:616–625. doi: 10.1128/MCB.00849-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny LA. The fractal globule as a model of chromatin architecture in the cell. Chromosome Res. 2011;19:37–51. doi: 10.1007/s10577-010-9177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Misteli T. Higher-order genome organization in human disease. Cold Spring Harb Perspect Biol. 2010;2:a000794. doi: 10.1101/cshperspect.a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Pirrotta V, Li HB. A view of nuclear Polycomb bodies. Current opinion in genetics & development. 2012;22:101–109. doi: 10.1016/j.gde.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Linder-Basso D, Kharchenko PV, Tolstorukov MY, Kim M, Li HB, Gorchakov AA, Minoda A, Shanower G, Alekseyenko AA, et al. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 2012 doi: 10.1101/gr.138156.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Bellen HJ, Hoskins RA. Drosophila P elements preferentially transpose to replication origins. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15948–15953. doi: 10.1073/pnas.1112960108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strukov YG, Sural TH, Kuroda MI, Sedat JW. Evidence of activity-specific, radial organization of mitotic chromosomes in Drosophila. PLoS Biol. 2011;9:e1000574. doi: 10.1371/journal.pbio.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa H, Iwasaki O, Tanaka A, Capizzi JR, Wickramasinghe P, Lee M, Fu Z, Noma K. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic Acids Res. 2010;38:8164–8177. doi: 10.1093/nar/gkq955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Blom M, Kerkhoven RM, Pagie L, Teunissen H, Nieuwland M, Simonis M, de Laat W, van Lohuizen M, van Steensel B. Interactions among Polycomb domains are guided by chromosome architecture. PLoS Genet. 2011;7:e1001343. doi: 10.1371/journal.pgen.1001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Van Bortle K, Ramos E, Takenaka N, Yang J, Wahi J, Corces V. Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains. Genome Res. 2012 doi: 10.1101/gr.136788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW, Levis RW, Spradling AC, Hoskins RA, et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods. 2011;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AM, Van Bortle K, Ramos E, Takenaka N, Rohrbaugh M, Jones BC, Jones KC, Corces VG. Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Mol Cell. 2011;44:29–38. doi: 10.1016/j.molcel.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe E, Tanay A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat Genet. 2011;43:1059–1065. doi: 10.1038/ng.947. [DOI] [PubMed] [Google Scholar]
- Yang J, Corces VG. Insulators, long-range interactions, and genome function. Curr Opin Genet Dev. 2012;22:86–92. doi: 10.1016/j.gde.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Bodnar MS, Spector DL. Nuclear neighborhoods and gene expression. Curr Opin Genet Dev. 2009;19:172–179. doi: 10.1016/j.gde.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.