Abstract
By successfully incorporating sequence diversity into proteins, combinatorial libraries have been a staple technology used in protein engineering, directed evolution, and synthetic biology for generating proteins with novel specificities and activities. However, these approaches mostly overlook the incorporations of post-translational modifications, which nature extensively uses for modulating protein activities in vivo. As an initial step of incorporating post-translational modifications into combinatorial libraries, we present a bacterial co-expression system, utilizing a recently characterized calmodulin methyltransferase (CaM KMT), to trimethylate a combinatorial library of the calmodulin central linker region. We show that this system is robust, with the successful over-expression and post-translational modification performed in E. coli. Furthermore we show that trimethylation differentially affected the conformational dynamics of the protein upon the binding of calcium, and the thermal stability of the apoprotein. Collectively, these data support that when applied to an appropriately designed protein library scaffold, CaM KMT is able to produce a post-translationally modified library of protein sequences, thus providing a powerful tool for future protein library designs and constructions.
Keywords: protein library design, high-quality libraries, synthetic biology, post-translational modification
Introduction
Synthetic biology integrates numerous disciplines towards utilizing designed proteins to produce novel phenotypic functions in a living cell; for not only addressing fundamental questions of molecular evolution, but also for downstream biotechnology applications, including: metabolic engineering for biofuel production [1]; improving/restoring contaminated environments [2]; synthesis of drugs [3] and other organic molecules [4]. As the widespread uses of synthetic biology advance, the demand for novel, designed protein systems and networks to specifically execute the desired/designed phenotypic functions will increase significantly [5–7]. While combinatorial libraries of the 20 naturally-encoded amino acids have successfully yielded peptides and proteins with novel specificities and activities, additional post-translational modifications that nature extensively utilizes to further tune protein-protein interactions in vivo are mostly overlooked in these library designs. With recent advances in the identification and understanding of the classes of enzymes responsible for catalyzing post-translational modifications, it is possible to incorporate these regulatory elements in protein library design [8, 9].
Trimethylation, an important post-translational modification that plays numerous roles in eukaryotic developmental and cellular processes, is catalyzed by the protein lysine methyltransferase (PKMT) family of enzymes, which specifically recognize and modify surface accessible lysine residues of target proteins. Well-known examples of trimethylation include histones, in which protein post-translational modification affects gene transcription to regulate numerous cellular processes [10]. Other endogenous trimethylation events, such as the trimethylation of the conserved eukaryotic calcium signaling protein calmodulin (CaM), are less understood but appear to play a significant role in cellular functioning. A limited number of studies show that trimethylated CaM, which is modified at the solvent-accessible residue Lys-115, is specific for certain developmental stages and tissue [11–13], influences activator properties of CaM with target enzymes [14] and phenotypic changes in growth and development of whole organisms [15]. Thus, incorporation of trimethylation into synthetic protein library design would provide a significant modification that nature utilizes to modulate cellular networks.
Recently, we identified an interspecies conserved class I protein lysine methyltransferase, named calmodulin-lysine N-methyltransferase (CaM KMT), responsible for the trimethylation of CaM at solvent-accessible residue Lys-115 [16]. Since bacterially expressed CaM lacks trimethylation at Lys115, we hypothesized that the utilization of this system to co-express and post-translationally modify proteins provides a unique opportunity to introduce trimethylation into a library of protein sequences.
As an initial step towards using this system as a tool to develop post-translationally modified proteins, we present the application of an E. coli co-expression system to post-translationally trimethylate a recently characterized, high-quality combinatorial library of the mammalian CaM central linker region (residues 68-92) [17]. We show that all tested library members were able to be co-expressed with CaM KMT, modified, and purified. In addition, we show that trimethylation differentially altered the conformational dynamics of the protein upon the binding of calcium, and the thermal stability of the calcium-free apoprotein. These data suggest that when applied to an appropriately designed protein library scaffold, the use of CaM KMT to produce a post-translationally modified library of sequences is possible, thus providing a powerful tool for generating post-translationally modified combinatorial libraries.
Materials and Methods
Materials
Unless otherwise stated, all chemicals used were reagent grade. The phenothiazine, used for the making Trifluoromethyl-10-aminopropyl phenothiazine (TAPP)-sepharose affinity resin, was synthesized at the University of Kentucky Center for Structural Biology Chemistry Core Facility. This core facility is supported in part by funds from NIH National Center for Research Resources (NCRR) grant P20 RR020171.
CaM Library Construction and Protein Purification
The design, construction, and characterization of high-quality combinatorial libraries [18] of the central linker of mammalian CaM were recently described [17]. Briefly, synthetic gene libraries encoding the native polar and nonpolar (binary patterning) periodicity of the central linker region were designed, synthesized and subcloned into the CaM expression vector, pETCaM1C. Individual protein library members lacking the introduction of lysine residues were chosen, transformed into E. coli BL21(DE3) cells, recombinantly expressed in 1L LB-kanamycin media at 37°C, and purified using a TAPP-sepharose affinity resin in a calcium-dependent manner to >95% purity [17].
CaM Methyltransferase Expression/Dual Expression
Each individual library member plasmid (pETCaM1C, [17]) was transformed into E. coli BL21(DE3)-star-RIL (Stratagene) competent cells, previously transformed with a modified pET23d expression vector, containing the SUMO-tagged human isoform of CaM KMT [16] and ampicillin resistance. Bacteria were grown in LB media and induced with 1 mM IPTG at 37 °C to simultaneously express the two proteins. Protein methylation occurs in the cell by CaM KMT, which uses bacterial AdoMet as the methyl group donor. Additional incubation of bacterial lysates at 37 °C, after cell disruption, with purified CaM KMT assures complete stoichiometric methylation.
Methylation Analysis Experiment
To analyze post-translational protein methylation, lysates from each co-expression were incubated with previously purified CaM KMT in the presence of [3H-methyl] AdoMet for 1 h, as described previously [16]. Samples were analyzed by 12.5% SDS-PAGE gel electrophoresis, and then transferred to a PVDF membrane. Samples were phosphorimaged after 24 h and 14 day exposure to measure in vitro incorporation of radiolabel.
Circular Dichroism spectroscopy
Far-UV CD spectra (250 to 190 nm) of purified proteins, in the presence (+ 1 mM CaCl2) or absence (+1 mM EGTA) of calcium, were recorded as the average of four far-UV wavelength scans with 0.5 nm steps and 8 sec averaging time using a Jasco J-810 spectrophotometer, as described previously [17]. All protein sample measurements were made in a 1 mm quartz cuvette at 25 °C. Trace Ca2+ was removed by running 5 mM HEPES buffer (pH 7.5) through a Chelex column, before addition of CaCl2 or EGTA. Additional measurements, with excess CaCl2 and EGTA, were made to guarantee complete calcium-binding saturation/elimination.
Differential scanning fluorimetry
Based on methods described previously [19–23], purified methylated and non-methylated proteins (~ 10 μM), in 20 mM HEPES buffer + 100 mM KCl, pH 7.4 + 10 mM EGTA, were incubated with SyproOrangeTM (Invitrogen) dye to give a final volume of 20 μL. Samples were placed in a 96 well plate and fluorescence was monitored (λex=465 nm; λem=580 nm) as the samples were thermally denatured between 25–95 °C at a rate of 0.5 °C/min using an Applied Biosystems RT-PCR thermocycler with Step-One software. Additional measurements in excess EGTA were made to guarantee complete removal of Ca2+ from sample. Dye and protein alone were used as baseline controls. The Tm value was calculated by fitting the experimental data to a derivation of the Boltzmann equation as described previously [22].
Results and Discussion
Trimethylation of Lys115 in CaM is catalyzed by an N-methyltransferase, CaM KMT, which utilizes AdoMet as a co-substrate and shows high specificity for CaM [12, 24–28]. Previous studies have shown that the CaM recognition site by CaM N-methyltransferase resides solely on the C-terminal lobe (78–148) of CaM, with residues in the loop near Lys115 being most critical for CaM N-methyltransferase recognition and function [28, 29]. While the importance of the central linker region has been relatively unexplored in these analyses, it has been demonstrated that changes to Helix 4, within the central linker region, was tolerated [29]. Based on these studies, we hypothesized that alterations to the central linker region would be tolerated by CaM KMT, thereby serving as a tool for the post-translational modification of synthetic protein library members of the CaM central linker region.
Co-expression and trimethylation of synthetic protein library members with CaM KMT
To determine if this system would be suitable for co-expression and post-translational modification with synthetic sequences of the CaM central linker region, 17 library sequences (Figure 1) were selected from high-quality combinatorial libraries of the CaM central linker region that lacked incorporation of additional lysine residues [17] and were co-transformed with CaM KMT into E. coli BL21(DE3) )-star-RIL cells (Figure 2A). SDS-PAGE analysis show all tested protein library members overexpressed with CaM KMT at levels similar to individual library sequence expression [17], and were able to be purified using TAPP-sepharose resin, demonstrating that the E. coli co-expression system is robust (Figure 2A).
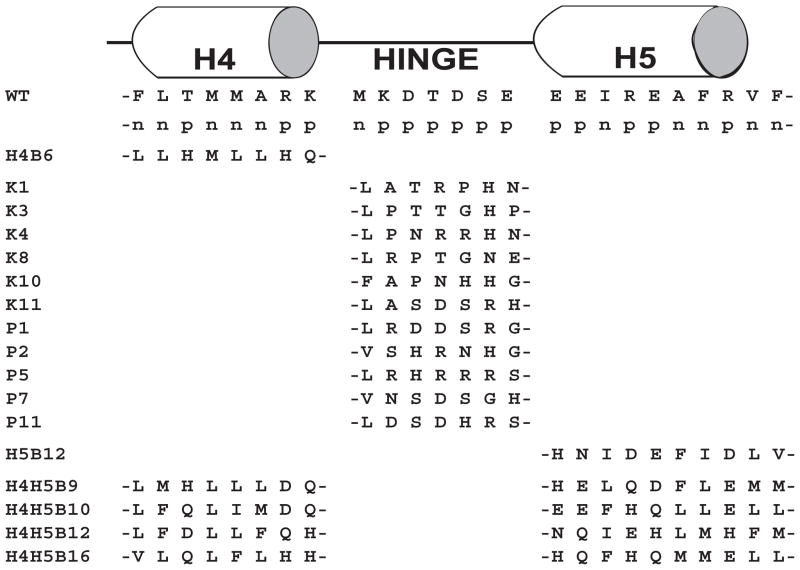
Figure 1.
Central linker library sequences co-expressed with CaM KMT. The central linker region of the mammalian CaM (positions 68–92) consist of two α-helices (Helix 4, Helix 5) separated by a seven residue hinge domain. The CaM WT sequence and the library defined nonpolar (n) and polar (p) residue positions are shown for reference [17]. Only sequences that are unique from WT are shown for clarity.
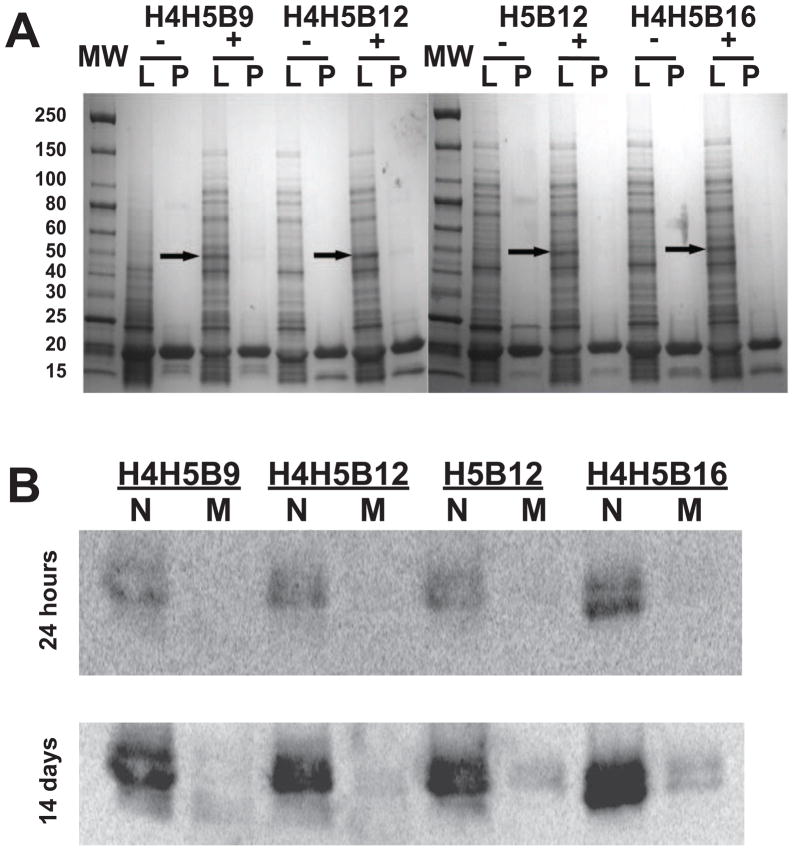
Figure 2.
E. coli co-expression and in vivo methylation analysis of representative central linker library members with CaM KMT. A) As described previously, SDS-PAGE analyses show that each central linker library sequence individually overexpressed (− lanes) in E. coli (band between 15–20 kDa; molecular weight marker, MW) and was able to be purified (P) from soluble cell lysate (L), using TAPP-sepharose calcium dependent resin. When each protein was co-expressed with CaM KMT (+ lanes; band ~ 50 kDa, arrow), the library protein still overexpressed and was able to be purified. B) To measure the success of in vivo methylation, purified library proteins from either single, non-methylated protein expression (N), and methylated CaM KMT co-expression (M) were incubated in vitro with [3H-methyl] AdoMet and purified CaM KMT for 1h. Samples were run on SDS-PAGE gels and analyzed for incorporation of radiolabel by autoradiography with 24 hours and 14 day exposure. All library protein co-expressed with CaM KMT showed trace amounts of in vitro incorporation of radiolabel following 14 day exposure, confirming nearly complete methylation of these sequences during co-expression. These results are representative of all library sequences tested.
To qualitatively test CaM KMT for the ability to trimethylate CaM central linker library members in vivo, individual library proteins co-expressed with CaM KMT were purified and assayed for incorporation of radiolabel by purified CaM KMT in vitro using [3H-methyl]-AdoMet as the methyl group donor (1). The CaM KMT was active in vivo for each library sequence examined, with in vitro incorporation of radiolabel only detectable following 14 days of exposure in purified samples (i.e. methylation was nearly complete in vivo; Figure 2B). Supportive of our in vivo methylation results, when each protein was expressed individually (i.e. no co-expression of CaM KMT); radiolabel was incorporated in all proteins (17/17) by CaM KMT in vitro (data not shown). These results support our hypothesis and earlier findings that the synthetic library diversity, introduced in the CaM central linker region, does not alter the recognition and methylation activity (at Lys-115) of CaM KMT.
Differential effects of trimethylation
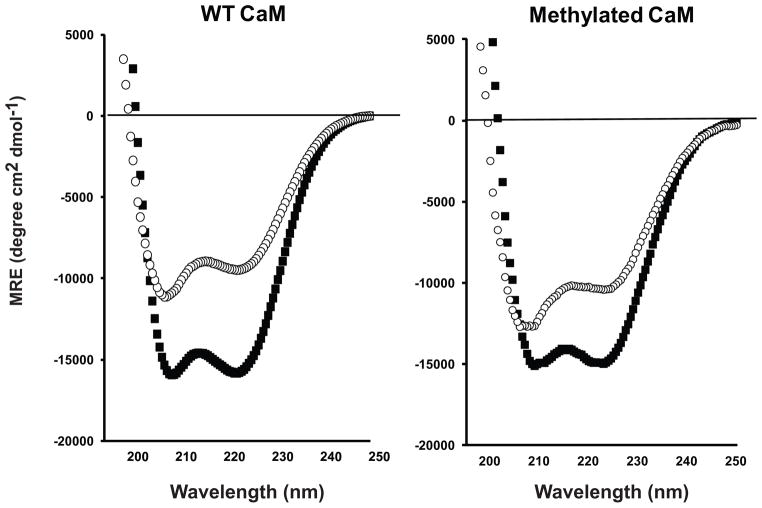
Upon the binding of Ca2+, CaM undergoes large tertiary structural changes [30–32] which are detected by far-UV circular dichroism (CD) spectroscopy [33, 34]. Using this co-expression and purification system, we have the ability to compare the effects of methylation on these allosteric changes. For bacterially expressed WT (non-methylated), characteristic changes in the CD spectra are observed upon the elimination of Ca2+ from the protein, with a 40% reduction in the mean residue ellipticity at 222 nm (Figure 3). For methylated CaM, the change in CD spectra, in the presence and absence of calcium, is less than WT, with an approximate 31% reduction at 222 nm, indicative of alterations in conformational dynamics of the protein (Figure 3). Furthermore, comparing spectra between WT and methylated-CaM shows the effect of methylation is primarily observed on the calcium-free (apoprotein) far-UV CD spectra (Figure 3). For library protein P1, the primary effect of methylation, while different than WT, is also observed on the calcium-free apoprotein spectra (Figure S1).
Figure 3.
Far-UV CD spectra of purified WT CaM and methylated CaM in the presence and absence of calcium. CD spectra for WT CaM (left) and methylated CaM (right), in 5 mM HEPES buffer pH 7.5, were recorded at 25 °C in the presence (+ 1 mM CaCl2, more negative spectra, filled squares) and absence (+ 1 mM EGTA, less negative spectra, open circles) of calcium. Additional CD measurements in excess CaCl2 and EGTA show no significant differences.
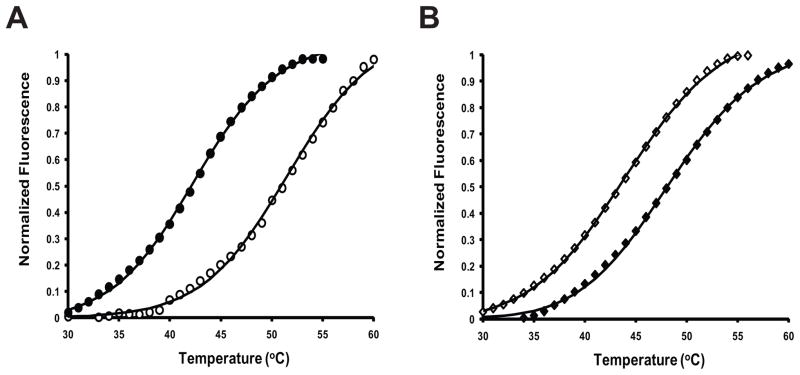
To assess changes in apoprotein structure, we measured the effects of trimethylation on the thermal stability of the WT CaM and central linker library members by utilizing differential scanning fluorimetry (DSF, Figure 4). This screening method (also referred to high throughput thermal scanning or Thermofluor) allows for the simultaneous determination of relative thermal stabilities of numerous proteins, by monitoring the binding of a hydrophobic dye SyproOrange™ as a function of the thermal unfolding of the protein in a microplate format [19–23]. For WT CaM, the non-methylated apoprotein had a Tm at approximately 51 °C, which is in relative agreement to the thermal stability of CaM determined using CD spectroscopy [35]. For the WT-Me CaM apoprotein, the Tm was determined to be 43 °C, which was 8 °C less than the non-methylated WT apoprotein Tm, as determined by DSF (Figure 4A, Table 1). When examining how methylation affects the apoprotein stabilities of randomly selected individual central linker library members, the Tm increased differentially for each sequence (Figure 4B, Table 1). This suggests that this post-translational modification, when applied to a diverse library of sequences, will have differential effects on protein structure, leading to increased diversity (beyond encoded) in function. Studies examining the relationship between methylation and alteration of function within this library of sequences are currently being pursued. In addition, these data further support earlier findings, that alterations in the central linker region do not affect CaM KMT recognition and the ability to post-translationally modify CaM [28, 29].
Figure 4.
Thermal denaturation curves of non-methylated (open symbols) and methylated (filled symbols) WT CaM (A; circles) and library protein K4 (B; diamonds) in the absence of calcium (+1 mM EGTA), as measured by DSF. Tm values (Table 1) show that methylation has differential effects on the thermal stability of the apo (calcium free) form of WT and central linker library CaM proteins.
Table 1.
The thermal denaturation of methylated and non-methylated WT and Hinge CaM protein library members, as determined by DSF. All proteins were measured in triplicate, in 20 mM HEPES buffer + 100 mM KCl, pH 7.4, with 2 mM EGTA. The Tm was calculated as described previously [19].
| Protein | Non-methylated Tm (°C) | Methylated Tm (°C) | ΔTm (°C) |
|---|---|---|---|
| WT | 51 | 43 | −8 |
| P1 | 38 | 39 | +1 |
| P2 | 47 | 48 | +1 |
| P5 | 52 | 54 | +2 |
| K4 | 44 | 48 | +4 |
Conclusions
We have presented a novel dual-expression system, utilizing a recently characterized CaM KMT, to enable the post-translational modification of a library of CaM sequences during bacterial expression. All tested library proteins were able to be over-expressed with CaM KMT and modified. Furthermore, trimethylation had differential effects on the allosteric changes associated with the binding of calcium, and the thermal stability of the apoprotein. These findings support that this recently identified CaM KMT, is able to incorporate modifications extensively used in nature for regulating protein activity, and provides a novel and valuable tool for CaM combinatorial protein library design.
Supplementary Material
Highlights.
E. coli co-expression system for post-translationally modifying protein libraries
Library proteins were over-expressed with CaM KMT and post-translationally modified
Trimethlyation differentially affected protein stability and allosteric changes
Acknowledgments
BC conducted research as part of his Georgetown College Honors Thesis Project and was supported by a summer undergrad research fellowship from the Howard Hughes Medical Institute program (GCPALS). MLB conducted research as part of his University of Kentucky Honors Thesis project. MLB and ED were supported in part by summer fellowships from the University of Kentucky Office of Undergraduate Research. Other support was obtained partially from a pilot project grant from the NIH National Center for Research Resources (NCRR) grant P20 RR020171 (LHB), the Kentucky Science and Engineering Foundation (Grant Agreement # KSEF-148-502-207-201 with the Kentucky Science and Technology Corporation) (LHB), and the University of Kentucky College of Medicine Startup funds (LHB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee SK, Chou H, Ham TS, Lee TS, Keasling JD. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Current Opinion in Biotechnology. 2008;19:556–563. doi: 10.1016/j.copbio.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 2.Lovley DR. Cleaning up with genomics: Applying molecular biology to bioremediation. Nat Rev Microbiol. 2003;1:35–44. doi: 10.1038/nrmicro731. [DOI] [PubMed] [Google Scholar]
- 3.Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MCY, Withers ST, Shiba Y, Sarpong R, Keasling JD. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 4.Marshall AL, Alaimo PJ. Useful Products from Complex Starting Materials: Common Chemicals from Biomass Feedstocks. Chem-Eur J. 2010;16:4970–4980. doi: 10.1002/chem.200903028. [DOI] [PubMed] [Google Scholar]
- 5.Baker D, Church G, Collins J, Endy D, Jacobson J, Keasling J, Modrich P, Smolke C, Weiss R. Engineering life: building a fab for biology. Sci Am. 2006;294:44–51. doi: 10.1038/scientificamerican0606-44. [DOI] [PubMed] [Google Scholar]
- 6.Purnick PEM, Weiss R. The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol. 2009;10:410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 7.Wingler LM, Cornish VW. Reiterative Recombination for the in vivo assembly of libraries of multigene pathways. Proceedings of the National Academy of Sciences. 2011;108:15135–15140. doi: 10.1073/pnas.1100507108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angelini A, Heinis C. Post-translational modification of genetically encoded polypeptide libraries. Curr Opin Chem Biol. 2011;15:355–361. doi: 10.1016/j.cbpa.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Davis BG. Mimicking Posttranslational Modifications of Proteins. Science. 2004;303:480–482. doi: 10.1126/science.1093449. [DOI] [PubMed] [Google Scholar]
- 10.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nature Reviews Genetics. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh SH, Roberts DM. Analysis of the state of posttranslational calmodulin methylation in developing pea-plants. Plant Physiol. 1990;93:880–887. doi: 10.1104/pp.93.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rowe PM, Wright LS, Siegel FL. Calmodulin n-methyltransferase - partial-purification and characterization. Journal of Biological Chemistry. 1986;261:7060–7069. [PubMed] [Google Scholar]
- 13.Takemori N, Komori N, Thompson JN, Yamamoto MO, Matsumoto H. Novel eye-specific calmodulin methylation characterized by protein mapping in Drosophila melanogaster. Proteomics. 2007;7:2651–2658. doi: 10.1002/pmic.200700343. [DOI] [PubMed] [Google Scholar]
- 14.Roberts DM, Burgess WH, Watterson DM. Comparison of the nad kinase and myosin light chain kinase activator properties of vertebrate, higher-plant, and algal calmodulins. Plant Physiol. 1984;75:796–798. doi: 10.1104/pp.75.3.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts DM, Besl L, Oh SH, Masterson RV, Schell J, Stacey G. Expression of a calmodulin methylation mutant affects the growth and development of transgenic tobacco plants. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:8394–8398. doi: 10.1073/pnas.89.17.8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magnani R, Dirk LMA, Trievel RC, Houtz RL. Calmodulin methyltransferase is an evolutionarily conserved enzyme that trimethylates Lys-115 in calmodulin. Nat Commun. 2010;1:43. doi: 10.1038/ncomms1044. [DOI] [PubMed] [Google Scholar]
- 17.Bradley LH, Bricken ML, Randle C. Expression, purification, and characterization of proteins from high-quality combinatorial libraries of the mammalian calmodulin central linker. Protein Expression and Purification. 2011;75:186–191. doi: 10.1016/j.pep.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bradley LH, Kleiner RE, Wang AF, Hecht MH, Wood DW. An intein-based genetic selection allows the construction of a high-quality library of binary patterned de novo protein sequences. Protein Eng Des Sel. 2005;18:201–207. doi: 10.1093/protein/gzi020. [DOI] [PubMed] [Google Scholar]
- 19.Lavinder JJ, Hari SB, Sullivan BJ, Magliery TJ. High-Throughput Thermal Scanning: A General, Rapid Dye-Binding Thermal Shift Screen for Protein Engineering. Journal of the American Chemical Society. 2009;131:3794–3795. doi: 10.1021/ja8049063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Layton CJ, Hellinga HW. Thermodynamic Analysis of Ligand-Induced Changes in Protein Thermal Unfolding Applied to High-Throughput Determination of Ligand Affinities with Extrinsic Fluorescent Dyes. Biochemistry. 2010;49:10831–10841. doi: 10.1021/bi101414z. [DOI] [PubMed] [Google Scholar]
- 21.Arai R, Kobayashi N, Kimura A, Sato T, Matsuo K, Wang AF, Platt JM, Bradley LH, Hecht MH. Domain-Swapped Dimeric Structure of a Stable and Functional De Novo Four-Helix Bundle Protein, WA20. The Journal of Physical Chemistry B. 2012;116:6789–6797. doi: 10.1021/jp212438h. [DOI] [PubMed] [Google Scholar]
- 22.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protocols. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 23.Cummings MD, Farnum MA, Nelen MI. Universal Screening Methods and Applications of ThermoFluor®. Journal of Biomolecular Screening. 2006;11:854–863. doi: 10.1177/1087057106292746. [DOI] [PubMed] [Google Scholar]
- 24.Morino H, Kawamoto T, Miyake M, Kakimoto Y. Purification and properties of calmodulin-lysine n-methyltransferase from rat-brain cytosol. J Neurochem. 1987;48:1201–1208. doi: 10.1111/j.1471-4159.1987.tb05647.x. [DOI] [PubMed] [Google Scholar]
- 25.Han CH, Richardson J, Oh SH, Roberts DM. Isolation and kinetic characterization of the calmodulin methyltransferase from sheep brain. Biochemistry. 1993;32:13974–13980. doi: 10.1021/bi00213a030. [DOI] [PubMed] [Google Scholar]
- 26.Pech LL, Nelson DL. PURIFICATION AND CHARACTERIZATION OF CALMODULIN (LYSINE-115) N-METHYLTRANSFERASE FROM PARAMECIUM-TETRAURELIA. Biochim Biophys Acta-Gen Subj. 1994;1199:183–194. doi: 10.1016/0304-4165(94)90114-7. [DOI] [PubMed] [Google Scholar]
- 27.Wright LS, Bertics PJ, Siegel FL. Calmodulin N-methyltransferase - Kinetics, mechanism, and inhibitors. Journal of Biological Chemistry. 1996;271:12737–12743. doi: 10.1074/jbc.271.22.12737. [DOI] [PubMed] [Google Scholar]
- 28.Cobb JA, Han CH, Wills DM, Roberts DM. Structural elements within the methylation loop (residues 112-117) and EF hands III and IV of calmodulin are required for Lys(115) trimethylation (vol 340, pg 417, 1999) Biochemical Journal. 1999;341:863–863. [PMC free article] [PubMed] [Google Scholar]
- 29.Cobb JA, Roberts DM. Structural requirements for N-trimethylation of lysine 115 of calmodulin. Journal of Biological Chemistry. 2000;275:18969–18975. doi: 10.1074/jbc.M002332200. [DOI] [PubMed] [Google Scholar]
- 30.Babu YS, Sack JS, Greenhough TJ, Bugg CE, Means AR, Cook WJ. Three-dimensional structure of calmodulin. Nature. 1985;315:37–40. doi: 10.1038/315037a0. [DOI] [PubMed] [Google Scholar]
- 31.Kretsinger RH, Rudnick SE, Weissman LJ. Crystal structure of calmodulin. J Inorg Biochem. 1986;28:289–302. doi: 10.1016/0162-0134(86)80093-9. [DOI] [PubMed] [Google Scholar]
- 32.Zhang M, Tanaka T, Ikura M. Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat Struct Biol. 1995;2:758–767. doi: 10.1038/nsb0995-758. [DOI] [PubMed] [Google Scholar]
- 33.Craig TA, Watterson DM, Prendergast FG, Haiech J, Roberts DM. Site-specific mutagenesis of the alpha-helices of calmodulin. Effects of altering a charge cluster in the helix that links the two halves of calmodulin. J Biol Chem. 1987;262:3278–3284. [PubMed] [Google Scholar]
- 34.Putkey JA, Ono T, VanBerkum MF, Means AR. Functional significance of the central helix in calmodulin. J Biol Chem. 1988;263:11242–11249. [PubMed] [Google Scholar]
- 35.Sorensen BR, Shea MA. Interactions between Domains of Apo Calmodulin Alter Calcium Binding and Stability. Biochemistry. 1998;37:4244–4253. doi: 10.1021/bi9718200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.