SUMMARY
MTERF4 is the first MTERF family member shown to bind RNA, and plays an essential role as a regulator of ribosomal biogenesis in mammalian mitochondria. It forms a complex with the rRNA methyltransferase NSUN4 and recruits it to the large ribosomal subunit. In this paper, we characterize the interaction between both proteins, demonstrate that MTERF4 strongly stimulates the specificity of NSUN4 during in vitro methylation experiments and present the 2.0 Å resolution crystal structure of the MTERF4:NSUN4 protein complex, lacking 48 residues of the MTERF4 C-terminal acidic tail, bound to S-adenosyl-L-methionine, revealing the nature of the interaction between both proteins and the structural conservation of the most divergent of the human MTERF family members. Moreover, the structure suggests a model for RNA binding by the MTERF4:NSUN4 complex, providing insight into the mechanism by which an MTERF family member facilitates rRNA methylation.
INTRODUCTION
The MTERF (Mitochondrial TERmination Factor) family of proteins is conserved in the plastids and mitochondria of metazoans (Linder et al., 2005; Roberti et al., 2009). Proteins of the MTERF family, named after the human mitochondrial transcription termination factor MTERF1 (Guja and Garcia-Diaz, 2011), are believed to be nucleic acid binding proteins (Byrnes and Garcia-Diaz, 2011; Guja and Garcia-Diaz, 2011) and play various roles regulating organellar gene expression. Most species contain multiple MTERF family members (four in humans, more than thirty in some plant genomes; Babiychuk et al., 2011) and, in the cases studied, their function appears to be non-redundant. In mammalian cells, MTERF proteins have been clearly demonstrated to regulate mitochondrial gene expression (Camara et al., 2011; Park et al., 2007; Roberti et al., 2009; Wenz et al., 2009). The founding member of the family, MTERF1, mediates termination of transcription in a site-specific manner (Kruse et al., 1989). It recognizes a sequence in the mitochondrial tRNA-Leu(UUR) gene, which is downstream and adjacent to the 16S rRNA gene and promotes bidirectional transcriptional termination (Asin-Cayuela et al., 2005; Yakubovskaya et al., 2010). MTERF2 and MTERF3 have been proposed to associate with mitochondrial DNA in a non sequence-specific manner and regulate mtDNA transcription (Park et al., 2007; Roberti et al., 2009; Wenz et al., 2009).
Recently, crystal structures of MTERF1 (Jimenez-Menendez et al., 2010; Yakubovskaya et al., 2010) and MTERF3 (Spahr et al., 2010) have revealed that these proteins share a unique superhelical fold. Both proteins have a modular architecture and are composed of a series of MTERF repeats that are organized in a left-handed helical fold that conforms a large central groove capable of mediating the association with nucleic acids, suggesting that MTERF proteins belong to a larger class of tandem helical repeat nucleic acid binding proteins (Rubinson and Eichman, 2011). MTERF1 is thus far the only MTERF protein that has been crystallized in complex with nucleic acid. The crystal structure of MTERF1 in complex with the termination sequence (Yakubovskaya et al., 2010) highlights the mechanism of sequence-specific double-stranded DNA recognition by MTERF1 and reveals that binding of MTERF1 perturbs the DNA structure by bending and unwinding the duplex and everting three nucleotides. This perturbation results in a higher kinetic stability of the sequence-specific complex, and is thought to be crucial for efficient termination of transcription (Byrnes and Garcia-Diaz, 2011; Guja and Garcia-Diaz, 2011; Yakubovskaya et al., 2010). The remarkable structural similarity between MTERF1 and MTERF3 suggests that this fold, and perhaps some aspects of the nucleic acid binding mechanism, might be conserved in other MTERF family proteins.
Until recently, tandem α-helical repeat motifs, like those found in MTERF1 and MTERF3, were believed to be primarily associated with protein scaffolds or RNA recognition enzymes (Rubinson and Eichman, 2011). Thus, MTERF proteins are an example of an analogous structural solution that is dedicated to DNA binding. However, the extensive similarities with RNA binding tandem α-helical repeat proteins from the PUF family (Edwards et al., 2001; Wang et al., 2002; Wang et al., 2001), suggested the possibility that some MTERF proteins may be capable of binding RNA as well (Rubinson and Eichman, 2011; Yakubovskaya et al., 2010). Indeed, in contrast with other MTERF family members, MTERF4, the fourth human MTERF protein, was recently shown to bind RNA and be an essential regulator of translation in mammalian mitochondria by mediating a critical rRNA modification in the large mitochondrial ribosomal subunit (Camara et al., 2011). Modifications of the mitochondrial rRNAs appear to be essential for ribosome biogenesis and normal mitochondrial function. However, with the exception of the adenine dimethylation catalyzed by the TFB1M methyltransferase (Cotney et al., 2009; McCulloch et al., 2002; Metodiev et al., 2009; Raimundo et al., 2012; Seidel-Rogol et al., 2003), mitochondrial rRNA modifications are at present poorly understood. Importantly, the role of MTERF4 in this process depends on its ability to form a complex with the rRNA methyltransferase NSUN4. Unlike TFB1M, NSUN4 appears to be devoid of any ability to bind RNA in a sequence-specific manner, and therefore needs to be targeted to its methylation site (Camara et al., 2011). While the MTERF4/NSUN4 interaction with RNA has not yet been characterized and might involve interactions also with the ribosomal proteins, these observations led to a model whereby MTERF4 recruits NSUN4 to the large ribosomal subunit and mediates the interaction with the ribosomal RNA, explaining why the loss of MTERF4 leads to severe defects in ribosomal assembly and translation.
The ability of MTERF4 to bind RNA highlights the versatility of the MTERF fold and the wide spectrum of substrate specificity that it can confer, thus making MTERF4 a particularly interesting subject for more detailed structural and functional studies. In addition, the fact that a defined binding partner has been identified allows for the study of the mechanisms that facilitate this interaction. Protein-protein interactions have been postulated to be essential for the functions of MTERF proteins (Guja and Garcia-Diaz, 2011; Martin et al., 2005). However, how these interactions might be mediated by the MTERF fold, which appears to be fully engaged in nucleic acid binding, has thus far been a matter of debate.
Here, we characterize the interaction between MTERF4 and NSUN4, demonstrate that the association between these proteins leads to a strong stimulation of specificity and efficiency of in vitro mitochondrial 16S rRNA methylation, and describe the high resolution x-ray crystallographic structure of a complex between both proteins in the presence of the methyl donor S-adenosyl-L-methionine (SAM). The structure, which lacks 48 residues of the C-terminal acidic tail of MTERF4, reveals the mechanism of the interaction between the two proteins, the mechanism of SAM binding and the conservation of the MTERF fold in the most divergent of the human MTERF family members. It suggests a model that explains how MTERF4 facilitates rRNA methylation by NSUN4.
RESULTS & DISCUSSION
MTERF4 and NSUN4 form a tight complex
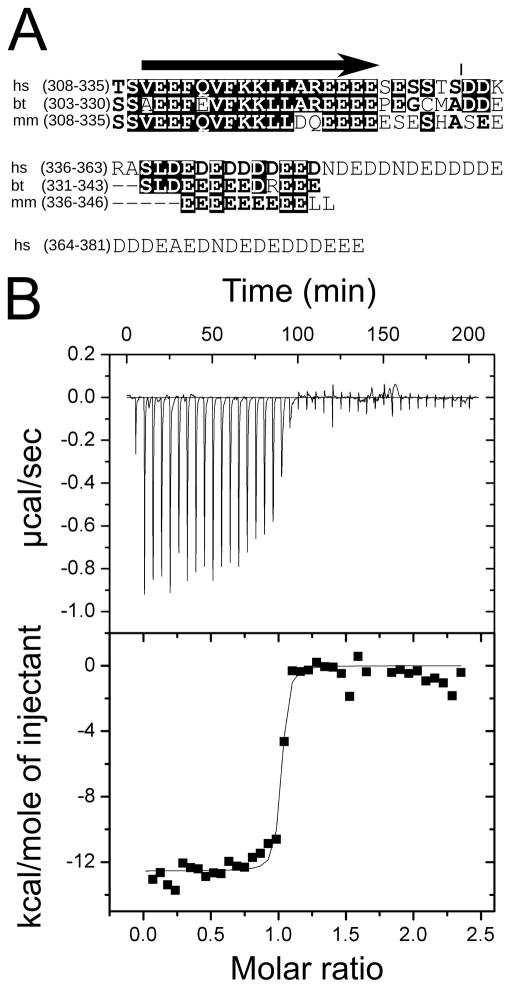
The proposed function of MTERF4 in mitochondrial ribosome biogenesis is dependent on the formation of a ternary complex with RNA and the methyltransferase NSUN4. It was previously demonstrated that MTERF4 and NSUN4 form a heterodimer in vitro (Camara et al., 2011). In order to better assess the likelihood that these proteins will form a complex in vivo, we decided to experimentally measure the stability of this complex through isothermal titration calorimetry (ITC). We expressed both human proteins in E. coli without their N-terminal targeting peptides (see Experimental Procedures) and developed a successful purification strategy for full-length NSUN4. However, we were unable to purify full-length MTERF4 in sufficient amounts due to poor protein stability. Inspection of the MTERF4 amino acid sequence reveals a 60 residue long C-terminal acidic tail of which the last 50 residues are predicted to be highly disordered (Figure 1A). Systematic truncations in this region allowed us to generate a construct missing the C-terminal 48 amino acids that resulted in the production of high yields of stable protein. We initially hypothesized that the truncated region might be involved in mediating the interaction with NSUN4. However, although it is possible that this tail might affect the interaction with NSUN4, removal of this C-terminal region did not prevent complex formation, suggesting that it might be dispensable for the interaction.
Figure 1. Interaction between MTERF4 and NSUN4.
(A) Alignment of the C-terminal regions of human (hs), mouse (mm) and bovine (bt) MTERF4. Despite differences in length, the acidic character of the C-terminus is conserved. The acidic region begins at the C-terminal end of a conserved α-helixand spans ca 30 residues, although 30 additional residues are present in the human protein (45 of the final 59 residues are acidic). The arrow represents a predicted α-helix. The vertical line represents the truncation site in the construct reported in this manuscript (B) Isothermal titration calorimetry of MTERF4 binding to NSUN4 at 25 °C. Integrated heat measurements from 3μl injections of 220 μM MTERF4 into the calorimeter cell containing NSUN4 at an initial concentration of 20μM(top panel). A standard one-site model was used for curve fitting (bottom panel). The interaction between NSUN4 and MTERF4 is characterized as an exothermic process with ΔH of −11.04 (±0.22) kcal/mol, a KD of 13.3 nM (±0.4), ΔS of −1.05 cal/mol K and a stoichiometry of N=1.08 (±0.02). See also Figure S1.
Subsequent ITC studies demonstrated that MTERF4 and NSUN4 form a stable complex with 1:1 stoichiometry (Figure 1B), and that this interaction does not depend on the presence of the SAM cofactor (Figure S1). The association of MTERF4 and NSUN4 is significantly exothermic (ΔH = −11.04 kcal/mol), whereas the entropy of association (ΔS = −1.05 cal/mol K) is very small. Low entropy of association usually indicates a smaller contribution of hydrophobic interactions to protein-protein complex formation, suggesting that the majority of the contacts between the two proteins are largely established between polar amino acids. Moreover, the affinity of MTERF4 and NSUN4 binding appears to be extremely high (KD= 13.3 nM). Such a small dissociation constant allows us to conclude that MTERF4 and NSUN4 form an obligate heterodimer in mitochondria, and that it is reasonable to consider both proteins as subunits of a stable holoenzyme as suggested previously (Camara et al., 2011). Thus, it seems unlikely that the two individual proteins have a function separate from that of the complex.
MTERF4 stimulates the activity and specificity of NSUN4 on the 16S rRNA in vitro
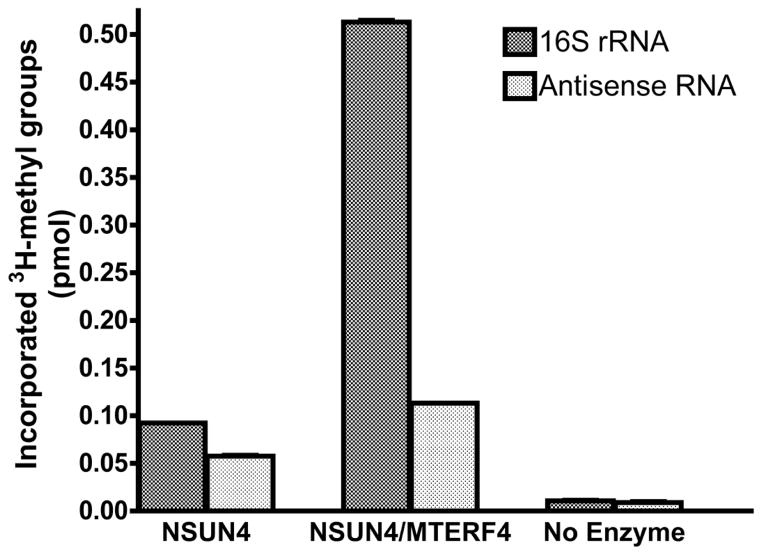
We then decided to address the functional importance of the interaction between MTERF4 and NSUN4. Since NSUN4 has been postulated to methylate the mitochondrial 16S rRNA (Camara et al., 2011), we analyzed the ability of both proteins to directly methylate in vitro transcribed 16S rRNA (see Experimental Procedures). Incubation of the purified 16S rRNA with MTERF4 and NSUN4 resulted in incorporation of 3H-methyl groups into the 16S rRNA that was significantly above background (Figure 2). This activity appeared to be mostly specific, since using an RNA antisense to the 16S rRNA as a substrate resulted in considerably lower activity, although still above that of a control with no protein, suggesting a certain degree of non-specific methylation. Interestingly, NSUN4 alone was also able to promote methylation of the 16S rRNA, but the activity observed was distinctly lower and, importantly, equivalent with both substrates, indicating that this activity was devoid of specificity. We therefore conclude that MTERF4 stimulates the activity of NSUN4 in vitro and is essential to target it to its specific methylation site, which is consistent with the proposed model of site-specific association of MTERF4 with the mitochondrial 16S rRNA. This constitutes the first formal demonstration that NSUN4 is endowed with an intrinsic RNA methyltransferase activity, and that it is capable of methylating the naked 16S rRNA.
Figure 2. MTERF4 stimulates the in vitro activity and specificity of NSUN4 on the mitochondrial 16S rRNA.
In vitro transcribed 16S mitochondrial rRNA and the corresponding antisense RNA were used for an in vitro 3H-methyl incorporation assay in order to test the activity of purified NSUN4 and the MTERF4:NSUN4 complex. The number of moles of methyl groups incorporated into the rRNA substrate was calculated from the measured disintegrations per minute (DPM). Vertical bars correspond to the mean ± SD of three independent experiments. Note that the error bars on the graph are small and may be difficult to see. The same reaction performed in the absence of enzyme was used as a control.
Crystal structure of the MTERF4:NSUN4 complex
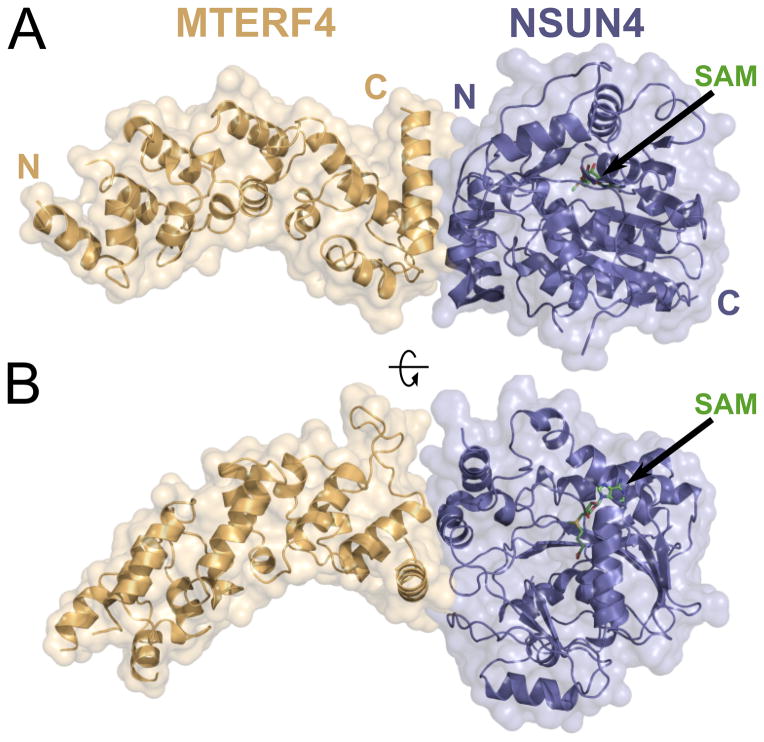
MTERF4 is the most divergent human member of the MTERF family (Roberti et al., 2009). Therefore, we initially decided to determine its structure to assess whether it indeed folds like other characterized MTERF proteins. However, all attempts to crystallize MTERF4 alone were unsuccessful. Since MTERF4 and NSUN4 interact very tightly, we then decided to attempt cocrystallization of both proteins, as such a structure could yield important functional information regarding the role of the MTERF4:NSUN4 complex. We were able to obtain native crystals of a MTERF4:NSUN4 heterodimer that, after optimization, diffracted to 2.3 Å. Attempts to solve the structure by molecular replacement using the available MTERF structures and/or methyltransferases thought to be related to NSUN4 were unsuccessful. We then prepared selenomethionione (SeMet)-derived MTERF4 protein and obtained crystals that diffracted to 2.9 Å and allowed us to perform initial phasing and build a preliminary model. Preparation of SeMet-derived NSUN4 resulted in crystals that diffracted to high resolution (2.0 Å; Table 1), contained one molecule of the complex in the asymmetric unit, and allowed us to build a complete model (Figure 3). The electron density was of sufficient quality to build a full-length model for NSUN4 (residues 38–384), with the exception of a small disordered loop (residues 111–115), and unambiguously place the bound SAM molecule. However, the lack of extensive lattice interactions in the MTERF4 region, combined with the fact that the MTERF fold likely exhibits some flexibility in the absence of its nucleic acid substrate (Jimenez-Menendez et al., 2010; Yakubovskaya et al., 2010) resulted in significant disorder in the C-terminal region of MTERF4 (distal to the interaction site). Nevertheless, we were able to assign side chains for most of the protein (residues 122–328) as well as the backbone for one additional MTERF motif. The overall structure of the complex reveals that both proteins are arranged end to end in an extended conformation and thus the interaction between them is restricted to a relatively small region. Significantly, this structure represents the first structural characterization of an MTERF protein participating in a protein-protein interaction.
Table 1.
Data Collection and Refinement Statistics
| Native-MTERF4:SeMet-NSUN4 | |||
|---|---|---|---|
| Data collection
| |||
| Space group | C 1 2 1 | ||
| Cell dimensions | |||
| a, b, c (Å) | 298.68, 53.25, 53.04 | ||
| αβγ (°) | 90.00, 98.76, 90.00 | ||
| Peak | Inflection | Remote | |
|
|
|||
| Wavelength (Å) | 0.9790 | 0.9795 | 0.9500 |
| Resolution (Å) | 46.82–2.00 (2.06–2.00) | 49.21–2.09 (2.10–2.09) | 46.84–2.18 (2.19–2.18) |
| Rmerge | 0.058 (0.623) | 0.041 (0.548) | 0.049 (0.546) |
| I/σI | 23.7 (2.4) | 26.3 (3.0) | 25.6 (3.0) |
| Completeness (%) | 99.9 (99.6) | 99.8 (99.8) | 99.9 (99.3) |
| Redundancy | 6.6 (6.1) | 6.7 (6.8) | 6.7 (6.8) |
|
| |||
| Refinement
| |||
| Resolution (Å) | 46.82–2.00 | ||
| Unique reflections | 55,979 | ||
| Rwork/Rfree | 0.1688/0.1960 | ||
| No. atoms | |||
| Total | 4633 | ||
| Protein | 4195 | ||
| Water | 301 | ||
| SAM | 26 | ||
| B-factors | |||
| Protein | 54.8 | ||
| SAM | 20.0 | ||
| Water | 48.4 | ||
| RMS deviations | |||
| Bond lengths (Å) | 0.010 | ||
| Bond angles (°) | 1.29 | ||
| Ramachandran | |||
| Favored (%) | 95.0 | ||
| Outliers (%) | 0 | ||
|
| |||
| PDB ID | 4FZV | ||
Values in parentheses are for highest-resolution shell.
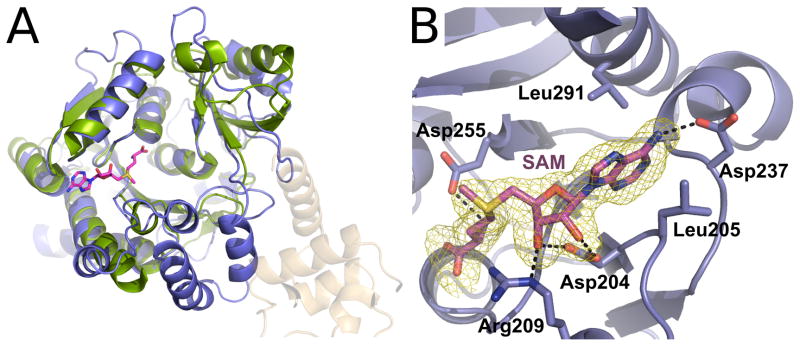
Figure 3. Crystal structure of the MTERF4:NSUN4 complex.
(A) Global view of the MTERF4:NSUN4 complex. MTERF4 is rendered in orange. NSUN4 is blue. The molecular surface is rendered transparent. N, N-terminal end; C, C-terminal end. The black arrow points to the position of the SAM residue. (B) A 90° rotation of the complex aroundthe long axis.
MTERF4 adopts a canonical MTERF fold
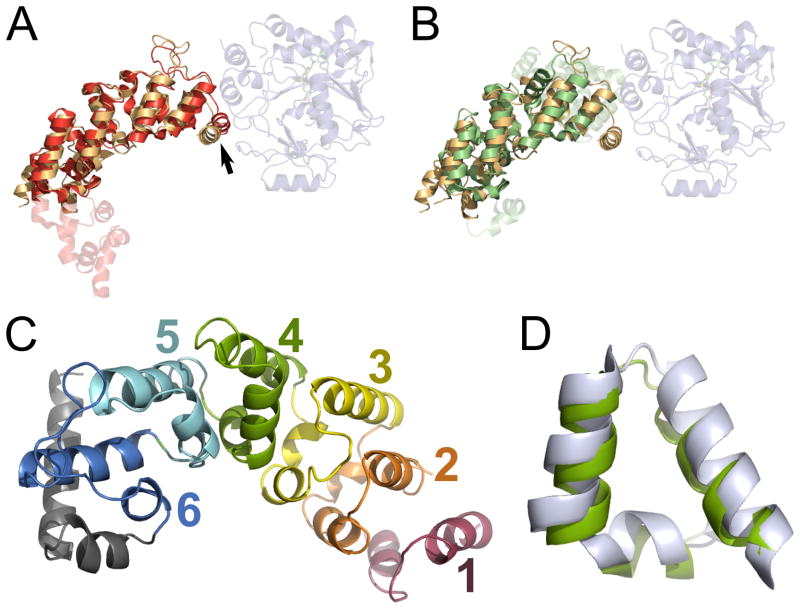
MTERF1 and MTERF3 have been shown to adopt very similar modular superhelical folds composed of repeating MTERF motifs, despite the fact that MTERF3 was crystallized in the absence of its nucleic acid substrate. This fold has been postulated to be a common feature of MTERF proteins and the basis for their ability to bind nucleic acids. Our current structure reveals that the MTERF fold is clearly conserved in MTERF4 and is not perturbed by the interaction with NSUN4 (Figure 3). Overlays with the MTERF3 (Figure 4A) and MTERF1 (Figure 4B) structures reveal extensive structural similarity between MTERF4 and the other MTERF proteins. However, MTERF4 displays much higher structural homology to MTERF3 (rmsd of 2.37 Å for 165 C-α atoms) than MTERF1 (rmsd of 4.33 Å for 165 C-α atoms; Figure 4), although this might simply be a consequence of the fact that both MTERF3 and MTERF4 were crystallized in the absence of nucleic acid. Six MTERF motifs can be recognized in MTERF4 (Figures 4C, S2), and these motifs are identical to those found in MTERF1 or MTERF3 (Figure 4D). However, the C-terminal MTERF motif in MTERF4 (6 in Figure 4C) is distorted, as the second α-helix in the motif is replaced by a large loop, as seen in MTERF3 (Figure 4A). Moreover, two additional α-helices are present C-terminal to the last MTERF motif. These two helices are present in both MTERF4 and the C-terminus of MTERF3. Interestingly, these two additional helices are involved in the interaction between MTERF4 and NSUN4 (see below).
Figure 4. MTERF4 adopts the canonical MTERF fold.
(A) Overlay of MTERF4 (orange) and MTERF3 (red). The rmsd was 2.37 Å for 165 C-α atoms. The arrow points to the α-helix involved in heterodimer formation. (B) Overlay of MTERF4 (orange) and MTERF1 (green). The rmsd was 4.33 Å for 165 C-α atoms. (C) Five canonical MTERF motifs (1–5) could be resolved in MTERF4. Only the backbone could be assigned for motif 1 (see text). The final C-terminal MTERF motif (6) is distorted and followed by two α-helices(gray). See also Figure S2. (D) Structural conservation of the MTERF motif. Overlay of MTERF motifs from MTERF4 (green) and MTERF1 (light gray).
NSUN4 is structurally related to m5C methyltransferases and binds SAM
NSUN4 exhibits a classical Rossman-like fold that is characteristic of SAM-dependent methyltransferases, containing a core seven-stranded β-sheet (Figure S3; Martin and McMillan, 2002). Structural homology searches with the DALI server (Holm and Rosenstrom, 2010) indicated that, as suggested previously on the basis of sequence homology (Camara et al., 2011), NSUN4 shares extensive structural homology with m5C methyltransferases. The lowest rmsd (2.3 Å for 235 C-α atoms) was obtained with the tRNA methyltransferase Trm4 from M. jannaschi (PDB 3A4T; Figure 5A), although similar overlays can be obtained with other available m5C methyltransferase structures (PDB IDs 2FRX, 3M6V, 2YXL, 1IXK, etc). Lower structural homology was found to other classes of methyltransferases. Despite the fact that the structural overlay is extensive between Trm4 and NSUN4, NSUN4 contains a number of extensions that are not conserved in other m5C methyltransferases. Interestingly, the majority of these insertions are directly involved in the interaction with MTERF4 (see below).
Figure 5. NSUN4 is related to m5C methyltransferases.
(A)Overlay of NSUN4 (blue) with the Trm4 m5C methyltransferase from M. jannaschii (green; PDB 3A4T). The rmsd was 0.78 Å for 116 C-α atoms. The SAM molecule corresponding to the MTERF4:NSUN4 structure is shown (magenta). See also Figure S3. (B) View of the SAM binding pocket in NSUN4. SAM (magenta) is bound in a positively charged binding pocket and establishes several interactions with NSUN4 side chains. A simulated annealing Fo-Fc omit electron density map is shown (yellow) contoured at 4σ. Hydrogen bonds are shown as dashed lines.
The active site of NSUN4 displayed clear density for the bound SAM molecule (Figure 5B). As could be expected, SAM is located in a position analogous to the one observed in the Trm4 structure. NSUN4 has a relatively open SAM binding site that is similar to that of other methyltransferases. The SAM molecule is bound in a negatively charged pocket and makes extensive contacts with protein side chains (Figure 5B), with the S-methyl group oriented towards a large groove in the surface of the protein. The bound SAM is stabilized by a variety of hydrogen-bonding and van der Waals interactions. The N6 amino group of the adenosine is stabilized by a hydrogen bond with Asp237, while the adenine ring is sandwiched between two leucine residues. The ribose is bound through a bidentate interaction between the O2′ and O3′ groups and Asp204 and an additional hydrogen bond between the O3′ group and Arg209. The methionine moiety is stabilized by interactions with the main chain as well as a hydrogen bond with Asp255.
A conserved interaction motif adjacent to the MTERF fold
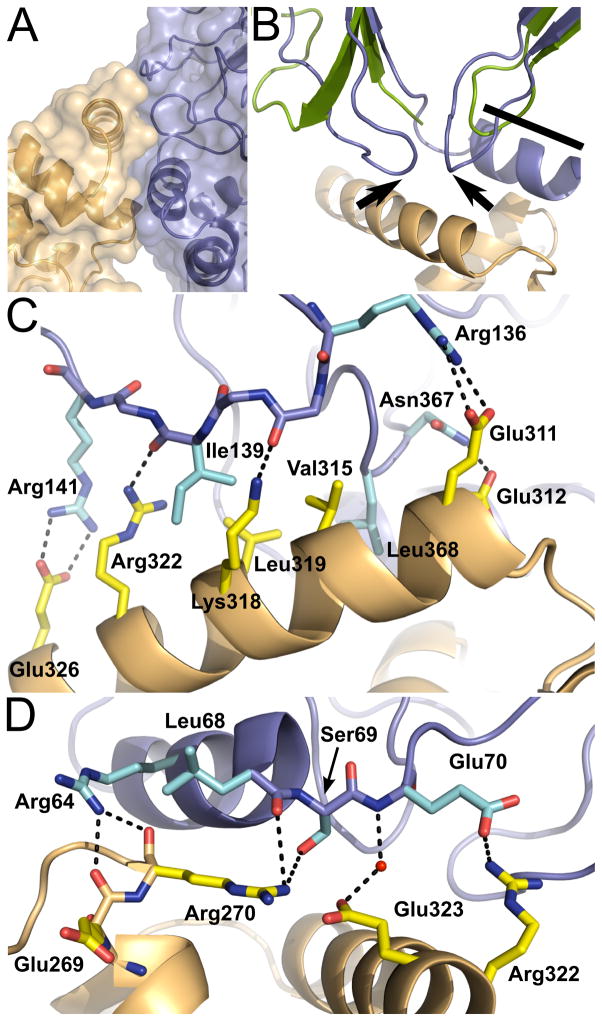
All the regions involved in the interaction between MTERF4 and NSUN4 are well ordered, clearly revealing the molecular details of the interaction between both proteins. This is the first structure depicting a protein-protein interaction involving an MTERF family member, and it is particularly interesting that the interaction involves only regions of MTERF4 that are C-terminal to the conserved MTERF motifs. As a consequence, most of the fold remains available to participate in RNA binding. This observation is consistent with the idea that the MTERF fold itself is dedicated to nucleic acid binding. MTERF4 interacts with NSUN4 utilizing the last distorted MTERF motif in the C-terminus of the protein (6 in Figure 4C) as well as the two α-helices C-terminal to this motif (Figure 6A). In NSUN4, the interaction involves an α-helix (residues 58–69; Figures 6B, 6D) and two small non-conserved loops (residues 132–142 and 364–375; Figures 6B, 6C). It is interesting that all of the regions of NSUN4 that are involved in the interactions are not conserved in Trm4. The aforementioned α-helix is part of an N-terminal extension (although this extension is present in some m5C methyltransferases other than Trm4), while the two loops are present in Trm4 but are longer in NSUN4.
Figure 6. The interaction with NSUN4 involves the region C-terminal of the MTERF fold.
(A)Region of interaction between MTERF4 and NSUN4. The C-terminal distorted MTERF motif (6 in Figure 4C) of MTERF4 is involved in the interaction. Contacts are mainly established with one α-helix and two loops in NSUN4. The molecular surface is rendered transparent. (B) The two loops in NSUN4 involved in the interaction with MTERF4 are not conserved in other m5C methyltransferases. Overlay of NSUN4 with the methyltransferase Trm4 from M. jannaschii (green), showing the two non-conserved loops (black arrows) and the N-terminal helix involved in the interaction (black bar). (C) Interactions between MTERF4 and NSUN4. Most interactions are established between a C-terminal α-helix in MTERF4 (orange) and one of the non-conserved loops in NSUN4 (blue; see text). NSUN4 side chains are cyan. MTERF4 side chains are yellow. Hydrogen bonds are shown as dashed lines. (D) Additional region of interaction between MTERF4 and NSUN4. Contacts also involve an α-helix in NSUN4 (in addition to the two loops in NSUN4 shown in panel C). NSUN4 side chains are cyan. MTERF4 side chains are yellow. Hydrogen bonds are shown as dashed lines.
In accordance with our thermodynamic data, most of the contacts established between MTERF4 and NSUN4 are electrostatic, although they surround a patch of hydrophobic residues that forms the center of the interaction surface (Figure 6C). The contact surface between both proteins has an area of 732.5 Å2, and involves 73 atoms in 18 residues.
Residues in the C-terminal α-helix of MTERF4 are responsible for establishing most of the contacts with NSUN4 and are thus likely crucial for the interaction (Figure 6C). Importantly, as mentioned above, this helix, and the region surrounding it, seems to be conserved in MTERF3 (arrow in Figure 4A). This suggests that this region might be a conserved motif to mediate protein-protein interactions in MTERF3 and MTERF4. Moreover, a similar C-terminal extension is likely present in MTERF2 (Yakubovskaya et al., 2010) and thus could similarly constitute a potential interaction site. Interestingly, the only human MTERF protein in which this motif is definitely absent is the transcriptional terminator MTERF1, perhaps highlighting the fact that this protein is able to mediate transcriptional termination in the absence of a protein partner.
A model for RNA binding
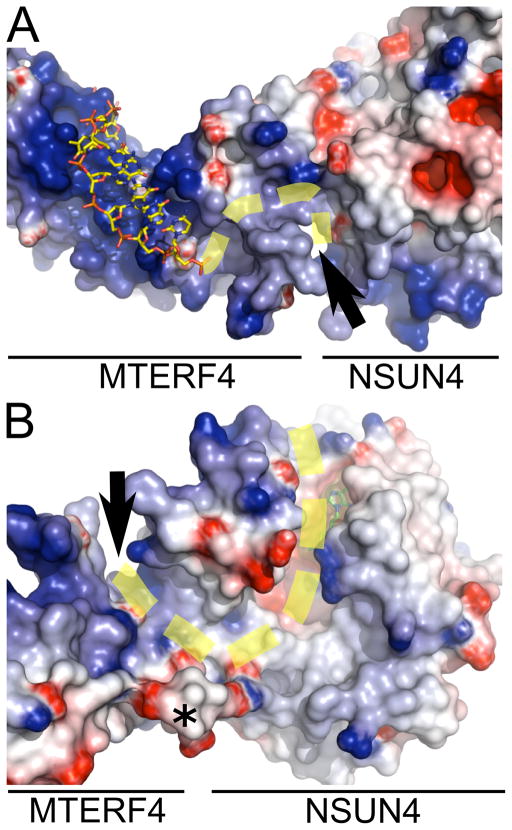
MTERF1 binds double-stranded DNA through its central groove. However, the vast majority of the contacts are established with only one of the two DNA strands, suggesting that a similar mechanism could be employed by MTERF4 to associate with a single strand of RNA. Calculation of an electrostatic surface potential map for MTERF4 revealed a large basic patch in the expected interaction region based on the MTERF1 structure. Accordingly, an overlay of MTERF1 and MTERF4 suggests a binding mode that is consistent with the electrostatic potential map (Figure 7A).
Figure 7. A proposed model for RNA binding.
(A) MTERF4 has a basic surface in the central groove postulated to mediate nucleic acid binding. Part of one of the DNA chains in the MTERF1 structure (3MVA) is shown (yellow). The RNA could bind in a similar mode and track through a groove (black arrows) present in the complex that leads to the SAM binding site. The proposed path of the RNA is shown as a dashed yellow line. A different orientation is shown in (B), highlighting the groove in NSUN4. SAM is shown in green. The last residue modeled in the C-terminus of MTERF is shown with an asterisk. The electrostatic surface potential map was generated with Delphi (Honig and Nicholls, 1995) and is colored from −7 kTe−1 (blue) to +7 kTe−1 (red).
Significantly, a large prominent groove can be clearly seen that is formed first by the MTERF4:NSUN4 interaction region and then by NSUN4 alone and that leads into the SAM binding pocket. This readily suggests a potential path for the RNA (yellow dashed line in Figure 7) and a model for how MTERF4 can direct binding of the RNA substrate by NSUN4. This model is consistent with the orientation of the SAM cofactor in the groove. Moreover, the model suggests that, in addition to the extensive structural conservation between MTERF proteins, the mechanism of nucleic acid binding might also be conserved, at least in its general features. Given the strongly structured nature of rRNAs, it is possible that the RNA molecule bound by MTERF4 might exhibit some double-stranded character, increasing the similarities with the MTERF1 binding mechanism. The binding groove in MTERF4 is sufficiently wide to accommodate a second RNA strand (Figure 7A).
It is interesting to note that the long acidic MTERF4 C-terminal tail that is absent in our structure would be expected to be located near the putative RNA binding groove (asterisk in Figure 7B). It is tempting to speculate that a function of this tail might be to increase the binding specificity by competing for the same binding surface with the RNA, as has been previously shown for other proteins (Marintcheva et al., 2008). Alternatively, it is also possible that this acidic tail serves to mediate additional protein-protein interactions that might help coordinate the MTERF4:NSUN4 complex with the rest of the mitochondrial rRNA modification machinery.
CONCLUSIONS
In this paper, we have described the structure of a MTERF4:NSUN4 complex that is essential to direct the activity of the NSUN4 methyltransferase to its methylation site in the mitochondrial 16S rRNA. This novel structure is the first example of a protein-protein interaction that involves an MTERF protein. MTERF family members have been previously postulated to interact with other proteins to help control their regulatory activities (Martin et al., 2005) or explain the strong polarity observed in transcription termination by MTERF1 (Byrnes and Garcia-Diaz, 2011; Guja and Garcia-Diaz, 2011). Our structure reveals that the interaction does not involve the MTERF fold, which is likely exclusively devoted to nucleic acid binding. Rather, the interaction mostly involves a region C-terminal to the MTERF fold that appears to be conserved in other MTERF proteins, suggesting that they could utilize an analogous binding mode to mediate protein-protein interactions. Moreover, the structure suggests a model for how the MTERF4:NSUN4 complex utilizes the MTERF fold to associate with RNA and direct methylation by NSUN4. This highlights how an MTERF protein can cooperate with a separate enzymatic activity by endowing it with substrate and/or sequence specificity. In this respect, it is important to note that NSUN4 appears devoid of any intrinsic sequence specificity, therefore suggesting that this specificity is conferred by the MTERF fold. Future experiments are needed to more fully understand how these proteins, despite sharing a very similar fold, can nevertheless have very different substrate specificities, including affinity for a different nucleic acid.
EXPERIMENTAL PROCEDURES
Protein Cloning and Purification
Wild-type MTERF4 (residues 66–332) and NSUN4 (residues 26 –384) were cloned into pTEV-HMBP3, allowing expression of a fusion with his-tagged maltose binding protein (MBP) that is cleavable with TEV protease. Proteins were overexpressed in Arctic Xpress (DE3) E. coli cells (Stratagene) at 16°C for 20 hr. MTERF4 and NSUN4 were purified using ProBond Resin (Invitrogen), followed by overnight TEV protease cleavage and Heparin chromatography. Subsequently, ProBond Resin was used to remove TEV. The MTERF4-NSUN4 complex was created by mixing the two proteins in equimolar concentration after the Heparin column. All proteins were subjected to gel filtration on a Superdex 200 16/60 column (GE Healthcare) in a buffer containing 20 mM Hepes, pH 8.0, 2.5% glycerol, 0.5 M KCl, 1mM DTT and 1 mM EDTA. The gel filtration profile shows that MTERF4 and NSUN4 form a complex of 70–80kDa.
SeMet-substituted protein was expressed in the Arctic Xpress (DE3) E. coli, which is not auxotrophic for methionine. Methionine biosynthesis was inhibited by growth conditions as described previously (Van Duyne et al., 1993). The protein was subsequently purified as described above. Proteins were concentrated using a 10,000 MWCO Amicon Ultra-15 device up to 20mg/ml. Concentrated proteins were stored in 20 mM HEPES (pH 8.0), 300 mM KCl, 2.5% glycerol, and 1mM DTT.
Crystallization and Structure Determination
MTERF4:NSUN4 crystals were grown by hanging drop vapor diffusion at room temperature in 4 μL drops of a 1:1 ratio of protein and crystallization solution (0.1–0.2M Mg Formate and 50 mM Bis-Tris, pH 5.0–6.0) in the presence of SAM. Crystals were cryoprotected with 35% ethylene glycol and flashed cooled in liquid nitrogen. Diffraction data were collected on beamlines X25 and X29 of the National Synchrotron Light Source (Upton, NY) at the peak, inflection point and high-energy remote wavelengths of the K edge of selenium. Datasets were processed using XDS (Kabsch, 2010) and Scala as implemented in the autoPROC pipeline (Vonrhein et al., 2011). All seven selenium sites in NSUN4 were identified with SHELX (Sheldrick, 2008) and phases were determined via Multiwavelegth Anomalous Dispersion (Hendrickson, 1997). Phases were calculated to 2.0 Å using SHARP (Vonrhein et al., 2007) and automated modeling building with ARP-wARP (Langer et al., 2008) produced a starting model consisting of 75% of NSUN4 and 30% of MTERF4. Manual model building was carried out in COOT (Emsley and Cowtan, 2004) using the experimental map and the complete final model was refined with PHENIX (Terwilliger, 2002). Model quality was assessed using MOLPROBITY (Davis et al., 2007).
Binding Measurements
ITC experiments were performed with a VP-ITC calorimeter (Microcal). NSUN4 (20–25 μM) was titrated with 10 μl injections of 200–250 μM MTERF4. Samples were prepared by dialyzing all interacting components against a buffer containing 20 mM HEPES, pH 8.0, 200 mM KCl, 2.5% glycerol, 1 mM EDTA. All runs were made at a constant stirring speed of 310 rpm, and all experiments were performed at 25 °C. Data were analyzed using the ORIGIN software.
In Vitro Methylation Assay
A 1.5kb fragment of the human 16S mitochondrial DNA was cloned through PCR of HeLa mitochondrial DNA in the pBlueScriptII SK+ vector between the XhoI and EcoRI sites. 16S ribosomal RNA was transcribed using T7 RNA polymerase using DNA linearized with EcoRI as a template. The corresponding antisense RNA was transcribed using T3 RNA polymerase using DNA linearized with XhoI. The resulting products were purified using the RNeasy Mini Kit (Qiagen). In vitro methylation assays were carried out according to a published procedure (Kuratani et al., 2010)with some modifications. 3H-SAM (MP Biomedical) was used as the methyl donor in the reaction. The methylation reaction was carried out in 100μl and contained 25mM Hepes pH 8.0, 50mM KCl, 0.1mM EDTA, 5mM MgCl2, 1mM DTT, 2.5% glycerol, 5 pmol RNA and 70 pmol SAM (78Ci/mmol). The reaction was initiated by adding the enzyme (0.5–5 pmol). A time course with different enzyme concentrations was carried out to identify linear conditions. Final experiments were carried out using 2 pmol of enzyme (NSUN4, MTERF4 or MTERF4:NSUN4 holoenzyme). Reactions with no enzyme were performed as a control. After 30 min, samples were transferred to 5ml of 10% trichloroacetic acid (TCA). The acid-insoluble fraction was then transferred to filter paper (Whatman), washed 3 times with 5ml of ice-cold 10% TCA and with 100% ethanol. The radioactivity on each filter disk was measured on a scintillation counter (Beckman Coulter). The amount of methyl groups incorporated into the RNA was calculated from the measured decays per minute (DPM). Experiments were independently repeated three times.
Supplementary Material
HIGHLIGHTS.
MTERF4 enhances the specificity of in vitro methylation by NSUN4
We present the first structure of the human MTERF4:NSUN4:SAM complex at 2.0 Å.
MTERF4 binds NSUN4 via its C-terminal region adjacent to the conserved MTERF fold
The structure explains how MTERF4 helps direct NSUN4 to its methylation site
Acknowledgments
The authors wish to thank Drs. Clemens Vonrhein and Annie Héroux for insightful discussions on X-ray data collection and processing as well as the NSLS Protein Crystallography group for beamline support. NSLS beamlines ×25 and ×29 are mainly supported by the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and the National Center for Research Resources of the National Institutes of Health. This work was supported by NIH R00-ES015421, R01-GM100021, and a UMDF grant to M.G.-D.
Footnotes
ACCESSION NUMBERS
Coordinates and structure factor files have been deposited in the RCSB Protein Data Bank with accession code 4FZV.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asin-Cayuela J, Schwend T, Farge G, Gustafsson CM. The human mitochondrial transcription termination factor (mTERF) is fully active in vitro in the non-phosphorylated form. J Biol Chem. 2005;280:25499–25505. doi: 10.1074/jbc.M501145200. [DOI] [PubMed] [Google Scholar]
- Babiychuk E, Vandepoele K, Wissing J, Garcia-Diaz M, De Rycke R, Akbari H, Joubes J, Beeckman T, Jansch L, Frentzen M, et al. Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc Natl Acad Sci U S A. 2011;108:6674–6679. doi: 10.1073/pnas.1103442108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes J, Garcia-Diaz M. Mitochondrial transcription: How does it end? Transcription. 2011;2:32–36. doi: 10.4161/trns.2.1.14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camara Y, Asin-Cayuela J, Park CB, Metodiev MD, Shi Y, Ruzzenente B, Kukat C, Habermann B, Wibom R, Hultenby K, et al. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011;13:527–539. doi: 10.1016/j.cmet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Cotney J, McKay SE, Shadel GS. Elucidation of separate, but collaborative functions of the rRNA methyltransferase-related human mitochondrial transcription factors B1 and B2 in mitochondrial biogenesis reveals new insight into maternally inherited deafness. Hum Mol Genet. 2009;18:2670–2682. doi: 10.1093/hmg/ddp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–289. doi: 10.1016/s0092-8674(01)00318-x. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Guja KE, Garcia-Diaz M. Hitting the brakes: Termination of mitochondrial transcription. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagrm.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson WmOC. Phase determination from multiwavelength anomalous diffraction measurements. Meth Enzymol. 1997:494–523. doi: 10.1016/S0076-6879(97)76074-9. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- Jimenez-Menendez N, Fernandez-Millan P, Rubio-Cosials A, Arnan C, Montoya J, Jacobs HT, Bernado P, Coll M, Uson I, Sola M. Human mitochondrial mTERF wraps around DNA through a left-handed superhelical tandem repeat. Nat Struct Mol Biol. 2010;17:891–893. doi: 10.1038/nsmb.1859. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse B, Narasimhan N, Attardi G. Termination of transcription in human mitochondria: identification and purification of a DNA binding protein factor that promotes termination. Cell. 1989;58:391–397. doi: 10.1016/0092-8674(89)90853-2. [DOI] [PubMed] [Google Scholar]
- Kuratani M, Hirano M, Goto-Ito S, Itoh Y, Hikida Y, Nishimoto M, Sekine S, Bessho Y, Ito T, Grosjean H, Yokoyama S. Crystal structure of Methanocaldococcus jannaschii Trm4 complexed with sinefungin. J Mol Biol. 2010;401:323–333. doi: 10.1016/j.jmb.2010.06.046. [DOI] [PubMed] [Google Scholar]
- Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder T, Park CB, Asin-Cayuela J, Pellegrini M, Larsson NG, Falkenberg M, Samuelsson T, Gustafsson CM. A family of putative transcription termination factors shared amongst metazoans and plants. Curr Genet. 2005;48:265–269. doi: 10.1007/s00294-005-0022-5. [DOI] [PubMed] [Google Scholar]
- Marintcheva B, Marintchev A, Wagner G, Richardson CC. Acidic C-terminal tail of the ssDNA-binding protein of bacteriophage T7 and ssDNA compete for the same binding surface. Proc Natl Acad Sci U S A. 2008;105:1855–1860. doi: 10.1073/pnas.0711919105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JL, McMillan FM. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr Opin Struct Biol. 2002;12:783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- Martin M, Cho J, Cesare AJ, Griffith JD, Attardi G. Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell. 2005;123:1227–1240. doi: 10.1016/j.cell.2005.09.040. [DOI] [PubMed] [Google Scholar]
- McCulloch V, Seidel-Rogol BL, Shadel GS. A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol Cell Biol. 2002;22:1116–1125. doi: 10.1128/MCB.22.4.1116-1125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metodiev MD, Lesko N, Park CB, Camara Y, Shi Y, Wibom R, Hultenby K, Gustafsson CM, Larsson NG. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Park CB, Asin-Cayuela J, Camara Y, Shi Y, Pellegrini M, Gaspari M, Wibom R, Hultenby K, Erdjument-Bromage H, Tempst P, et al. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130:273–285. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- Raimundo N, Song L, Shutt TE, McKay SE, Cotney J, Guan MX, Gilliland TC, Hohuan D, Santos-Sacchi J, Shadel GS. Mitochondrial stress engages E2F1 apoptotic signaling to cause deafness. Cell. 2012;148:716–726. doi: 10.1016/j.cell.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberti M, Polosa PL, Bruni F, Manzari C, Deceglie S, Gadaleta MN, Cantatore P. The MTERF family proteins: mitochondrial transcription regulators and beyond. Biochim Biophys Acta. 2009;1787:303–311. doi: 10.1016/j.bbabio.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Rubinson EH, Eichman BF. Nucleic acid recognition by tandem helical repeats. Curr Opin Struct Biol. 2011 doi: 10.1016/j.sbi.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel-Rogol BL, McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat Genet. 2003;33:23–24. doi: 10.1038/ng1064. [DOI] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Spahr H, Samuelsson T, Hallberg BM, Gustafsson CM. Structure of mitochondrial transcription termination factor 3 reveals a novel nucleic acid-binding domain. Biochem Biophys Res Commun. 2010;397:386–390. doi: 10.1016/j.bbrc.2010.04.130. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC. Automated structure solution, density modification and model building. Acta Crystallogr D Biol Crystallogr. 2002;58:1937–1940. doi: 10.1107/s0907444902016438. [DOI] [PubMed] [Google Scholar]
- Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, Bricogne G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Mol Cell. 2001;7:855–865. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- Wenz T, Luca C, Torraco A, Moraes CT. mTERF2 regulates oxidative phosphorylation by modulating mtDNA transcription. Cell Metab. 2009;9:499–511. doi: 10.1016/j.cmet.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yakubovskaya E, Mejia E, Byrnes J, Hambardjieva E, Garcia-Diaz M. Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription. Cell. 2010;141:982–993. doi: 10.1016/j.cell.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.