Abstract
Tyrosine phosphorylation is a dynamic reversible post-translational modification that regulates many aspects of cell biology. To understand how this modification controls biological function, it is necessary to not only identify the specific sites of phosphorylation, but also to quantify how phosphorylation levels on these sites may be altered under specific physiological conditions. Due to its sensitivity and accuracy, mass spectrometry (MS) has widely been applied to the identification and characterization of phosphotyrosine signaling across biological systems. In this review we highlight the advances in both MS and phosphotyrosine enrichment methods that have been developed to enable the identification of low level tyrosine phosphorylation events. Computational and manual approaches to ensure confident identification of phosphopeptide sequence and determination of phosphorylation site localization are discussed along with methods that have been applied to the relative quantification of large numbers of phosphorylation sites. Finally, we provide an overview of the challenges ahead as we extend these technologies to the characterization of tyrosine phosphorylation signaling in vivo. With these latest developments in analytical and computational techniques, it is now possible to derive biological insight from quantitative MS-based analysis of signaling networks in vitro and in vivo. Application of these approaches to a wide variety of biological systems will define how signal transduction regulates cellular physiology in health and disease.
Keywords: Tyrosine phosphorylation, cellular signaling, mass spectrometry, quantification
1. The importance of tyrosine phosphorylation in cellular signaling
Cells encounter a variety of extracellular stimuli (e.g. cell-cell interactions, growth factors, nutrients, toxins) and respond by changes in cellular function such as proliferation, migration, differentiation, and invasion [1]. The principal mechanism for the transfer of information from outside the cell (typically from the local environment) to inside the cell is through phosphorylation-mediated signaling networks. Phosphorylation is one of the most abundant post-translational modifications (PTMs) in the cell, and the phosphorylation of serine, threonine, and tyrosine residues provides a dynamic mechanism which has the potential to dramatically alter protein function and activation. Phosphorylation occurs most often on serine (accounting for ~90 %), and threonine (accounting for ~10 %), with tyrosine phosphorylation accounting for only ~0.05 % in eukaryotic cells [2]. While there is a complex integration of signals between serine, theronine, and tyrosine phosphorylation, here we focus on recent advances in the identification and quantification of tyrosine phosphorylation [3].
There are currently 90 known tyrosine kinases including 58 receptor tyrosine kinases (RTKs) [4]. Dynamic signaling cascades are initiated by tyrosine kinase activation which is generally triggered by extracellular ligand binding to the RTK followed by auto-phosphorylation on conserved tyrosine residues in the intracellular portion of the receptor [5, 6]. This activation leads to the recruitment of downstream signaling molecules and potentiation of the signal throughout the cell via networks of tyrosine phosphorylation. The elucidation of signaling networks downstream of these RTKs is dependent on the identification of large numbers of phosphorylation events and their quantification upon cell perturbations with specific growth factors. Throughout these analyses it is becoming apparent that many different RTKs employ common downstream signaling molecules to facilitate functional changes [7]. For instance, tyrosine phosphorylation can significantly affect protein conformation leading to changes in protein-protein interactions and subcellular localization [8, 9]. To maintain normal cellular functions it is essential that tyrosine kinase activities are tightly regulated. Tyrosine kinase signaling activity requires attenuation and termination of signal transmission, and negative regulation occurs principally through protein tyrosine phosphatases (PTPs), receptor internalization and degradation, and negative feedback through inhibitory phosphorylation of RTKs and associated protein kinases [10–13].
Due to its critical role in cellular homeostasis, deregulation of receptor tyrosine kinase signaling by mutation and genetic alterations often leads to malignant transformation in cancer. In particular, tyrosine kinases account for 0.3 % of the genome yet contribute to a disproportionately large percentage (30 %) of the known 100 dominant oncogenes [14]. In particular, the tyrosine kinase Bcr-Abl is deregulated in leukemia, HER-2 is amplified in breast cancer, and EGFR, MET, and PDGFR are over-expression and mutated across a variety of cancers [15–17]. Thus, despite the relative low levels of tyrosine phosphorylation, it is clear that tyrosine kinases are essential in mediating cellular communication and are critically important in the regulation of cell growth and oncogenic transformation [1, 2, 4, 18]. To facilitate the molecular characterization of tyrosine phosphorylation, it is crucial to identify phosphorylation sites present within a cell system under specific conditions. In addition to site localization, relative changes in phosphorylation levels upon perturbation have an important impact on the extent of molecular change. When studying the effect of specific components in signaling pathways, it is critical to both identify and quantify phosphorylation sites to gain insight into the biological effects of tyrosine phosphorylation.
In this review we highlight the technical advances in mass spectrometry that have lead to the routine identification of tyrosine phosphorylation site localization. Furthermore, we discuss the quantitative proteomics approaches utilized for the profiling of tyrosine phosphorylation signaling across in vitro biological systems and the challenges ahead as we extend these technologies to the characterization of tyrosine phosphorylation signaling in vivo.
2. Phosphotyrosine enrichment
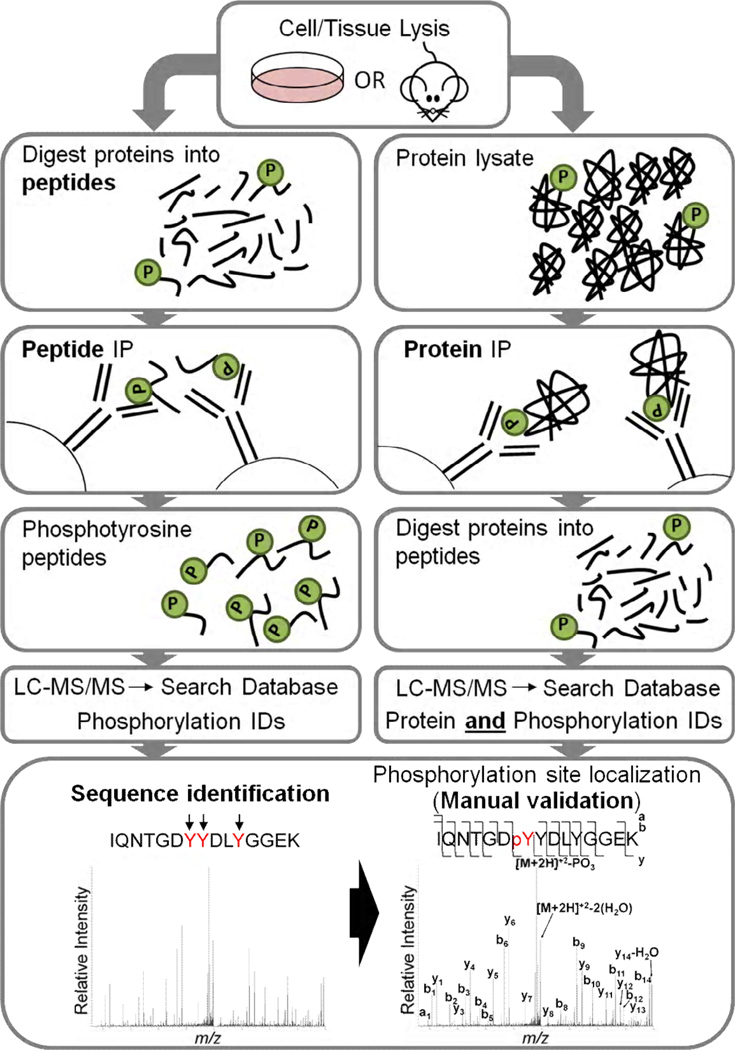
Mass spectrometry (MS) is a sensitive and specific analytical tool that has widely been applied to the identification and characterization of PTMs across biological systems. MS analyses typically require the separation of proteins and/or their constituent peptides, often using chromatographic separation, prior to mass spectrometric analysis. Mass analysis of a single purified protein can permit the assessment of the type of PTM present, such as the first analysis of tyrosine phosphorylation by mass spectrometry over two decades ago using an in vitro recombinant protein [19]. However, more detailed tandem MS analysis, usually at the peptide level, is required to enable site localization of the modification [20]. Due to the complexity of biological systems and the distinctly low levels of tyrosine phosphorylation, it is essential to specifically enrich for tyrosine phosphorylated proteins or peptides, thereby drastically reducing sample complexity prior to mass spectrometric analyses [21]. It is likely that a combination of the larger size of phosphotyrosine (compared to phosphoserine and phosphothreonine) and the relatively low abundance has led to higher affinity pan-specific antibodies for phosphotyrosine compared to phosphoserine or phosphothreonine. These pan-specific high affinity anti-phosphotyrosine monoclonal antibodies have proved to be a critical tool for the enrichment of tyrosine phosphorylation [22]. Typically, antiphosphotyrosine antibodies are bound to phosphotyrosine proteins or peptides and immobilized to protein agarose material prior to washing of the agarose to remove nonspecifically bound proteins or peptides. Phosphotyrosine proteins or peptides are then eluted from the specific antibody in a low pH solution (Fig. 1). When this strategy is applied to enrich phosphotyrosine containing proteins, elution from the beads is typically followed by proteolysis prior to LC-MS/MS analysis of the peptides. Unfortunately, proteolysis of the enriched proteins generates many peptides, the vast majority of which are non-phosphorylated. As a result, ionization of the phosphorylated peptides can be suppressed by the non-phosphorylated peptides, and identification of the phosphorylated peptides can be obscured, as the bulk of the instrument time is consumed by identifying nonphosphorylated peptides. (Fig. 1) [21, 23, 24]. To obtain site specific information, antiphosphotyrosine antibodies have more recently been used to enrich for phosphotyrosine peptides prior to MS analysis (Fig. 1) [25, 26]. In this approach, proteins are subjected to proteolysis using a suitable enzyme (typically trypsin) to generate peptides prior to phosphotyrosine enrichment and liquid chromatography tandem mass spectrometry (LC-MS/MS). Hundreds of tyrosine phosphorylation sites can be identified from complex biological mixtures following tryptic digestion [25, 26]. Yet, it is obvious that substantial portions of the phosphorylation sites present in the cell are missed due to tryptic fragments that are not in the m/z range for efficient ionization and/or fragmentation by MS. This has been identified extensively across ‘bottom-up’ proteomics and more recently numerous proteases, such as Lys-C, Glu-C, and Asp-N, have been used to increase the overall coverage of proteins and potential sites of modification [25]. One can envisage that the use of these alternative proteases in the identification of phosphotyrosine sites will yield additional and complementary information. Phosphotyrosine affinity methods are often coupled to immobilized-metal affinity chromatography (IMAC) which allows the capture of phosphate groups based on their interaction with the immobilized metal ion and their negative charge [24, 26–28]. Although ferric ions immobilized on chelating resin are the most common metal ions used for the selective enrichment of phosphorylated peptides, multiple other metal ions have been employed, with varying degrees of specificity. IMAC can be used as a single stage enrichment step prior to LC-MS/MS analysis to identify phosphorylation sites in biological systems. However, due to the much greater abundance of phosphoserine and phosphothreonine in the cell relative to phosphotyrosine, this strategy does not typically result in the identification of large numbers of phosphotyrosine sites.
Fig. 1.
Tyrosine phosphorylation enrichment prior to MS analysis. The most common methods of tyrosine phosphorylation enrichment are peptide enrichment (left) and protein enrichment (right). Protein enrichment consists of cell or tissue lysis followed by protein IP using anti-phosphotyrosine antibodies. Proteins are then digested into peptides using trypsin (or other suitable proteases) and analyzed by LC-MS/MS. Using this approach; proteins are identified, along with some phosphorylation sites. Peptide enrichment consists of cell or tissue lysis followed by digestion with trypsin (or other suitable proteases) and peptide IP using anti-phosphotyrosine antibodies. Tyrosine phosphorylated peptides are subsequently analyzed by LC-MS/MS. Peptides from both approaches are searched against a relevant sequence database using a search algorithm. The bottom panel shows the tyrosine phosphorylated peptide IQNTGDYYDLYGGEK which has 3 tyrosine residues that could be phosphorylated (indicated by arrows). Manual validation leads to the unequivocal identification of Y7 as the phosphorylation residue by the assignment of a, b and y series fragment ions.
3. Tyrosine phosphorylation site localization by MS/MS
3.1 Fragmentation methods
Following enrichment for phosphotyrosine peptides, samples are analyzed by MS; site specific information is provided by fragment ion spectra generated through tandem mass spectrometry (MS/MS). For many years collision induced dissociation (CID) has been the predominant method for MS/MS of peptides and proteins with and without PTMs. When undergoing CID, peptides or proteins undergo energetic collisions with inert gas molecules, depositing a large amount of energy into the peptide. This energy is randomized over many vibrational degrees of freedom before dissociation takes place at the most labile bonds, typically resulting in fragmentation at the amide bonds along the peptide backbone. The resulting fragment ions are predominantly b- and y-type ions containing the N- or C-terminus of the peptide, respectively, and the series of fragment ions from a given peptide permits the deduction of the precursor peptide sequence.
When analyzing phosphorylated peptides, identification of serine and threonine phosphorylation sites is impeded during CID type fragmentation due to the facile cleavage of the phosphate group, typically resulting in neutral loss of 98 Da (H3PO4) from the precursor ion and often from the fragment ions [29, 30]. To improve fragmentation efficiency, a neutral loss-triggered MS3 or multistage activation experiment can be performed whereby the precursor ion mass and the neutral-loss mass (e.g. the m/z ratio of the precursor minus the neutral loss of H3PO4) undergo fragmentation either consecutively or simultaneously [31]. While these approaches can improve the sensitivity and potentially the dynamic range of the MS/MS spectrum (through fragmenting the dominant neutral-loss species), the MS/MS spectra of peptides containing serine- and threonine-phosphorylation are typically more complex and more challenging to correctly assign as compared to MS/MS spectra of non-modified peptides.
Tyrosine phosphorylation is largely exempt from these issues and the phosphate group typically remains attached to b- and y-series fragment ions. In fact, in most cases, CID-based MS/MS spectra of peptides containing tyrosine phosphorylation closely resemble the MS/MS spectra of their non-phosphorylated analogues, with the addition of 80 Da to the precursor and appropriate fragment ions. For this reason, CID-fragmentation is typically used to identify the sequence and site of phosphorylation for tyrosine phosphorylated peptides. However, some tyrosine phosphorylated peptides lose 80 Da (HPO3) from the precursor ion, and infrequently from fragment ions, during CID, which can complicate the MS/MS spectrum and hamper the amount of sequence information that is detected for these peptides.
Tyrosine phosphorylated peptides can be selectively detected by MS using the specific phosphotyrosine immonium ion at m/z 216.043 [32]. Precursor ion scanning with a quadrupole ToF instrument was initially used to detect peptide precursor masses that produced the m/z 216.043 immonium ion following CID; these peptide precursor masses were then subjected to CID fragmentation to identify the sequence and site of phosphorylation [32]. This approach works well for analysis of semi-complex mixtures; for instance, to map sites of tyrosine phosphorylation in individual proteins. In complex mixtures such as cell lysates, the number of tyrosine phosphorylated peptides can begin to overwhelm this approach, and alternate strategies, including immunoaffinity enrichment of tyrosine phosphorylated peptides, are recommended.
For data dependent MS/MS acquisition, the m/z 216.043 immonium ion can be a useful indicator of phosphotyrosine peptides, depending on the instrument type and scan mode. If the peptide is fragmented by CID in an ion trap (either quadrupole or linear quadrupole ion trap), the immonium ion is often produced at fairly low levels due to the higher energy fragmentation pathway and/or multiple fragmentation events needed to generate this ion. Under these conditions, the immonium ion can also be lost due to the decreased stability of ions below 30% of the selected precursor ion in ion trap instruments (note that the ‘low-mass cutoff’ depends on the q-value; more details are available elsewhere [33]). Use of the 216.043 immonium ion to determine the presence of tyrosine phosphorylated peptides is most common on either quadrupole TOF instruments or Orbitrap instruments, where fragmentation is performed by higher-energy C-trap dissociation (HCD), a version of CID, and detection of the fragment ions occurs in the high resolution Orbitrap [29, 34]. In both of these instruments, peptide fragmentation occurs by accelerating the precursor ion into a collision cell filled with inert gas. The precursor ion typically undergoes a small number of higher-energy collisions, and the resulting fragment ions are then transferred to the mass detector, either the TOF or the Orbitrap, to generate the MS/MS spectrum. For both instruments, while the 216.043 immonium ion indicates the presence of a tyrosine phosphorylated peptide, not all tyrosine phosphorylated peptides produce 216.043, so the absence of this immonium ion is not diagnostic.
Tyrosine phosphorylated peptides can also be identified by other fragmentation methods, including electron transfer dissociation (ETD). ETD fragmentation involves the reaction of multiply protonated precursor peptides with singly charged anions, resulting in fragmentation at N-Cα bonds and generating c- and z- type ions [35]. This fragmentation method has generally been applied to analysis of large peptides or intact proteins, and has the added benefit that labile post-translational modifications are retained on the peptide, facilitating the sequence and site identification of these PTMs. At this point in time, ETD fragmentation is not as efficient as CID/HCD and therefore has not typically been applied to the analysis of low-abundance tyrosine phosphorylation.
3.2 Database searching
To identify peptide sequences, MS/MS spectra are searched against a database containing the protein sequences from the species of interest. A variety of database searching algorithms are currently available and applicable to the identification of tyrosine phosphorylated peptides, with the most common of these being MASCOT and SEQUEST [36–38]. Virtually all search algorithms are based on a comparison of a theoretical (in silico) fragment ion spectrum to the experimental MS/MS spectrum, using different comparison and scoring metrics. For instance, SEQUEST performs a cross-correlation of theoretical and experimental spectra, while MASCOT uses probability based scoring to determine the likelihood of a given match. Given the comparison between experimental and theoretical fragment ion spectra, the mass accuracy for the precursor ion and for the fragment ions is an important factor. The precursor ion mass accuracy determines the width of the search window; as the accuracy improves, the number of possible peptides matching the correct mass decreases, thereby improving the likelihood of matching the correct spectra and eliminating many potential false positives. At the limit, in the absence of PTM’s and amino acid mutations, the mass accuracy could theoretically be sufficient to identify a peptide sequence simply through very high mass accuracy precursor ion measurements. However, in reality, one cannot rule out either amino acid mutations or protein PTMs, both of which will radically expand the size of the computational protein database, leading to potentially orders of magnitude increase in the number of possible matches within a particular search window defined by the mass accuracy. The best solution to overcome uncertainty in the peptide sequence assignment is to have a high quality MS/MS spectrum to accompany high mass accuracy precursor ion measurements.
In the case of phosphotyrosine identification, enrichment is typically carried out prior to MS analysis, thereby significantly increasing the likelihood that an identified peptide will contain a phosphorylated tyrosine residue. However, since the analysis will generally contain some amount of non-phosphorylated peptides and since peptides will have both phosphorylated and non-phosphorylated tyrosines, it is necessary to search for ‘variable’ modifications, greatly increasing the size of the database, as each tyrosine has to be considered in the phosphorylated and non-phosphorylated form when generating the theoretical MS/MS spectra. Additionally, since some peptides are phosphorylated on nearby serine and threonine residues (e.g. in the activation loop of many kinases), it is suggested that variable phosphorylation of serine and threonine is also included in the database search, thereby increasing the search space even further. As the search space increases, the demand on high quality MS/MS spectra also increases, due to the increased number of potential matches to any given precursor ion match.
To increase the confidence in the identifications, it is important to understand the factors that go into the scoring algorithm for a given database and the potential error associated with the identifications made using a given algorithm and set of heuristics. For instance, in the comparison of the theoretical spectrum and the experimental spectrum, as precursor ion mass increases, the length of the peptide increases, and the number of potential fragment ions increases. In general, the score for a given peptide match increases with the length of the peptide, due to additional potential fragment ion matches. Thus, the threshold for ‘good’ sequence identifications should be tied to the precursor ion mass, with lower thresholds applied to smaller peptides. To gain statistical insight into the number of potential false positives, many groups have turned to false discovery rates (FDR), which can be calculated for each of these algorithms. FDR calculations enable the determination of the percentage of the total identifications that are likely false positives, where a mass spectrum is incorrectly matched to a peptide sequence in the database. The most common method of assessing FDR is to search the data of interest against the proper database and an alternative database (referred to as a decoy) where the amino acid sequences are either scrambled or reversed. The numbers of identifications that occur upon searching the decoy database are assumed to be due to random chance as these amino acid sequences do not occur naturally in the biological system. The ‘identifications’ resulting from the decoy database search are then imposed onto the search results from the database containing the correct protein sequences, and the number of false positives, and therefore the FDR rate, can be calculated [39]. There are several assumptions and flaws in this approach, and FDR calculations based on commonly used decoy databases dramatically under represent the true percentage of incorrect identifications [40]. Even if the FDR were accurate, this approach only provides an estimate of the number of potential false positives, and does not identify the spectra that may be incorrectly assigned.
3.3 Phosphorylation site determination
Once the peptide sequence has been accurately determined, the next challenge is to localize the site of phosphorylation. While some peptides have only a single modifiable residue, the majority of peptides have multiple potential sites of modification (serines, threonines, and tyrosines). In these cases, search algorithms are often not able to accurately determine the site of modification, and in fact, accurate site localization by MS can be non–trivial, as confident localization of phosphorylation requires the presence of specific fragment ions flanking the modified amino acid.
Accurate site localization is essential for biological insight, as the sequence motif surrounding the phosphorylation site can be used to suggest the identity of the kinase that may have phosphorylated the site. Perhaps more importantly, functional validation of the phosphorylation site is often performed through mutation of the modified site to either a non-phosphorylatable analogue (e.g. phosphotyrosine to phenylalanine) or a phosphomimetic residue (e.g. phosphotyrosine to glutamine); in these cases knowing the exact site of modification is absolutely critical, especially given the effort necessary to perform each mutation and test the resulting phenotypic effect. At a larger scale, there is a significant proportion of phosphorylation site data that is catalogued in online databases such as phosphosite (www.phosphosite.org/) and phosida (www.phosida.de/). In many cases the error associated with modification site localization determinations are not typically taken into consideration on curation of these large-scale databases [41]. Site localization errors then can be propagated when these large scale databases are queried for phosphorylation motifs, for structural insights, and for evolutionary conservation of phosphorylation. Therefore, the error associated with incorrect site localization has the potential to affect not only single experiments, but also large-scale bioinformatic studies.
The need for more accurate site identification has led to the development of software methods aimed at calculating the confidence in phosphorylation site assignments of each serine, threonine, and tyrosine residue present within a peptide sequence. As one example, the post translational modification (PTM) scoring tool in the MSQuant software calculates theoretical spectra for all possible phosphorylated versions of each peptide. The algorithm then assigns the most intense ions per 100 m/z in each fragmentation spectrum and calculates the best match [42]. As an alternative, the Ascore algorithm (ascore.med.harvard.edu/) calculates probability scores for each phosphorylation site localization, with the most likely site determined based on the presence and intensity of site-specific fragment ions [43]. Finally, the MASCOT delta score is a commonly used and easily applied approach [44]. When routinely searching phosphorylation data a score is calculated for all different possible phosphorylation sites on a phosphopeptide. As the phosphorylation site with the highest score is the most likely to be correct, this method calculates the difference between the top scoring phosphorylation site and those alternate sites with lower scores. Confidence in site assignment therefore increases with increasing differential between the top site and the next highest score [44].
3.4 Manual Validation
Despite the latest developments, all phosphorylation site localization and peptide identification algorithms have an associated error that cannot be accurately calculated [40]. This error is further compounded by the fact that almost all phosphorylation site identifications are ’one-hit wonders’, where the identification of the site is based on a single (or multiple replicate) MS/MS spectrum. Since sequence and site identification is critical for biological insight, manual validation of tyrosine phosphorylation sites is strongly recommended [45]. The aim of manual sequence validation is to assign fragment ions that are indicative of the full peptide sequence, and if possible, to identify all of the fragment ions that are present in the MS/MS spectra [45]. It is worth noting that peptides may fragment in an unpredictable way, making it difficult to identify all of the fragment ions that are present within a MS/MS spectrum. In this case, to definitively identify peptides and phosphorylation sites a long standing option has been the generation of chemically synthesized phosphorylated peptides matching the identified sequence from the biological sample. The MS/MS spectrum of the synthetic phosphopeptide can then be compared to the original MS/MS spectrum to confirm the putative identification. In selected cases, coelution or MS3 experiments can also be performed to further strengthen the confidence in the identification [46]. Manual validation is manageable for data sets comprised of hundreds of phosphorylation sites, but as the scan speed and sensitivity of the instrumentation improves, the number of phosphorylation sites that are routinely identified continues to increase, making manual validation more challenging. To address this challenge, we have developed a computational approach, based on manually validated spectra, that automatically removes many of the false-positive identifications from the searching algorithm [47]. Moving forward it is vital to pursue a more rigorous approach that combines software and manual validation to understand the biological importance of identified phosphorylation sites.
4. Quantitative analyses
Protein tyrosine kinases and phosphatases regulate physiological events at a molecular scale by altering the level of phosphorylation at selected sites on given proteins, thereby affecting protein function, protein-protein interaction, and potentially protein stability. To properly decipher these events, in addition to sequence and site identification, quantitative information, either relative or absolute, regarding the level of phosphorylation under different conditions is required. Relative quantification of phosphorylation can be carried out as an extension of the enrichment of phosphotyrosine peptides (Fig.1).
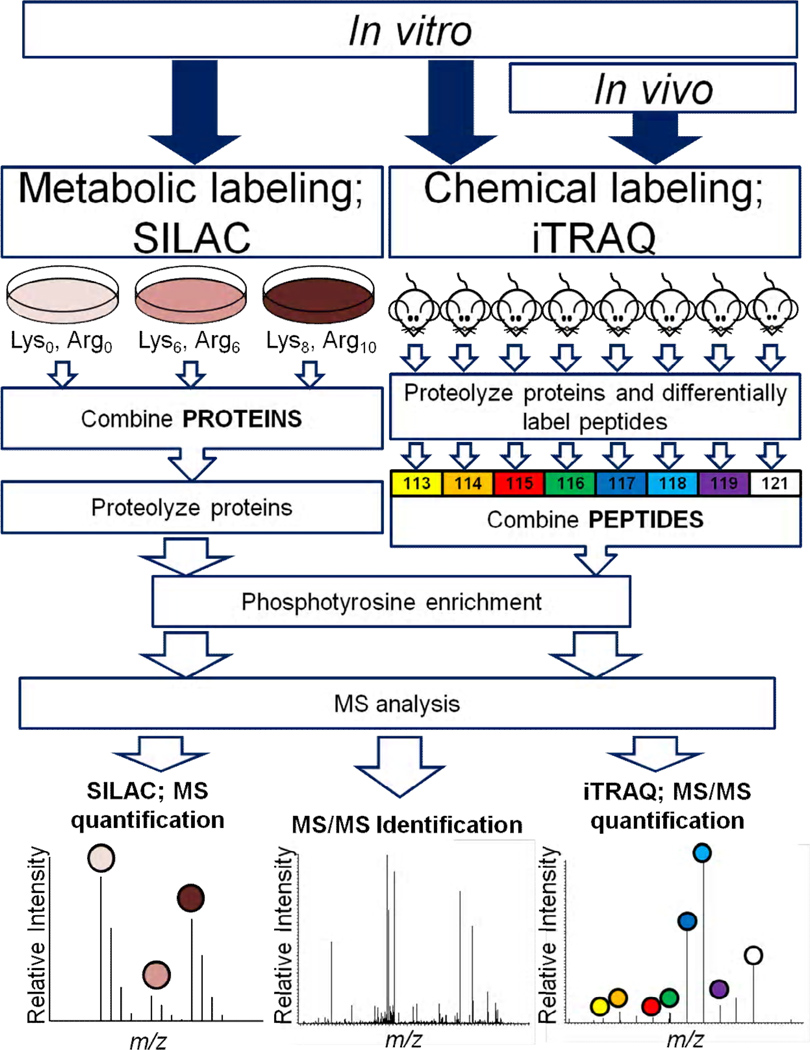
Although there are variations in the labeling agents used and the stage in the analytical workflow at which the analyte is labeled, quantification of phosphotyrosine generally consists of two or more samples being differentially labeled and combined prior to mass spectrometric analysis. Numerous methods are available for stable isotope labeling for quantitative analysis using mass spectrometry. These methods fall broadly into two categories: metabolic labeling, where proteins are labeled with stable isotopes during cell culture, and chemical labeling, where proteins or peptides derived from complex biological samples are differentially labeled with stable isotopes or mass tags using chemical reactions, typically after cell lysis (Fig. 2). The most widely applied variation of metabolic labeling is stable isotope labeling in cell culture (SILAC) [23]. During SILAC, cells are grown in the presence of isotopically labeled amino acids. 13C6-arginine and 13C6-lysine are commonly used to introduce a consistent mass difference in all tryptic peptides, with addition of 15N to these amino acids enabling comparison of three samples. In this case, the ‘light’ samples are unlabeled, ‘medium’ peptides are labeled with 13C6-arginine and 13C6-lysine, and ‘heavy’ peptides are labeled with 13C615N4-arginine and 13C615N2-lysine. MS analysis of ‘medium’ or ‘heavy’ labeled samples combined with samples from unlabeled cells provides accurate quantification based on relative intensities of precursor peptide ions derived from each sample, with each peptide mass differing by the number or arginines or lysines in the peptide sequence (Fig. 2). The clear benefit of this methodology is that it allows the labeling of large amounts of protein, thus allowing the use of a large amount of starting material to circumvent sensitivity limitations. Furthermore, as the samples can be combined much earlier in the sample preparation workflow, (i.e. cell lysates can be combined directly with each other) there is minimal variation associated with slight differences in processing of the various samples. Although initially limited to cell culture, SILAC labeling of whole organisms has been developed through application of an exclusive diet of 13C6-substituted versions of amino acids over multiple generations [48, 49]. Full incorporation of stable isotopically labeled amino acids is achievable, but the prohibitively high cost of the diet coupled to the extensive time required for complete labeling limits the application of in vivo SILAC.
Fig. 2.
Relative quantification of tyrosine phosphorylation by mass spectrometry. Metabolic labeling and chemical labeling are routinely applied to in vitro quantitative analyses while chemical labeling is the most common quantitative strategy for in vivo quantification (as indicated by the arrows). During SILAC, cells from up to three different conditions are cultured in different SILAC-labeled media (either unlabeled; Lys0, Arg0, labeled with 13C6-arginine and 13C6-lysine; Lys6, Arg6 or labeled with 13C615N4-arginine and 13C615N2-lysine; Lys8, Arg10). Cell lysates are combined, proteolyzed to peptides and subjected to a phosphotyrosine IP to enrich for phosphotyrosine peptides prior to analysis by LC–MS/MS. SILAC quantification data is dependent on the integration of MS peak intensities or chromatographic peak area for differentially labeled peptides. For iTRAQ labeling, up to eight biological samples are lysed in detergent or chaotropic agents; proteins are proteolyzed to peptides and tryptic peptides from individual samples are differentially labeled with isobaric mass tags. Labeled samples are then combined and analyzed by LC–MS/MS. Peptide quantification is determined by comparison of iTRAQ reporter ion intensities (m/z 113–121). Peptide identification is determined by the MS/MS spectrum in both approaches.
Chemical tagging strategies provide a useful and accurate alternative to metabolic labeling. There are multiple available strategies for the chemical labeling of proteins and peptides, ranging from simple acetylation or methylation reagents to more complex, multiplexed options such as isobaric tags for relative and absolute quantification (iTRAQ) and tandem mass tags (TMT). Currently, iTRAQ quantification is the most common method applied to the quantitative analysis of phosphotyrosine; using this approach it is now possible to quantify up to 8 independent samples within a single experiment. iTRAQ labeling consists of differential labeling of peptides through the reaction of an isobaric mass tag with all free ε-amine groups present in the sample, namely the N-terminus of a peptide chain and lysine side chains. The iTRAQ labels are isobaric and co-elute chromatographically, thus peptides labeled with the different iTRAQ isoforms elute as a single peak (with associated isotope envelope) in the MS spectrum. However, upon fragmentation (CID, HCD) the iTRAQ moiety dissociates, producing characteristic fragment ions of m/z 114–117 for 4plex iTRAQ and m/z 113–121 for 8 plex iTRAQ (discounting 120 due to the presence of the phenylalanine immonium ion) in the MS/MS spectrum. The relative intensities of each of these reporter ions enable relative determination of peptide or phosphorylation levels in the respective biological samples (Fig. 2). This quantification strategy has been used to quantify tyrosine phosphorylation signaling networks with site-specific resolution in a variety of different biological applications. For instance, we have used this approach to generate temporal dynamic profiles of tyrosine phosphorylation sites from cells stimulated with EGF [26], insulin [50], or anti-CD3 to stimulate the T-cell receptor [51]. We have also used iTRAQ to characterize signaling networks and study the effects of point mutations in EGFRvIII expressing glioma cell lines [52, 53]. Since iTRAQ labeling occurs post-lysis, this technique is generally applicable to all biological samples, including direct comparisons of human tissues. However, the total amount of biological material that can be labeled and analyzed in a single experiment is limited by the reagent cost, and thus the MS sensitivity required to comprehensively characterize and quantify tyrosine phosphorylation signaling is significantly increased.
5. Reproducibility: ‘Discovery’ and ‘Targeted’ approaches
Many of the tyrosine phosphorylation site identifications are carried out using data dependent acquisition (DDA) experiments, in which the top n-most (typically n = 5–20) abundant ions from the full scan mass spectrum (MS1 scan) are isolated and subjected to MS/MS. Unfortunately, due to variability in the timing of selection, isolation, and fragmentation between separate analyses, the reproducibility of peptide and phosphorylation site identification between analyses tends to be poor. We have previously shown that the experimental reproducibility of DDA experiments is 34% across four replicate analyses for iTRAQ-based quantification of temporal tyrosine dynamics following EGFR stimulation [54]. While iTRAQ multiplexing improves the reproducibility (e.g. through identification and quantification of 4 or 8 samples in a single analysis), comparison across multiple iTRAQ-based MS analyses still faces these same issues. With this poor reproducibility, it becomes challenging to get quantification for every phosphorylation site across multiple biological replicates. To address this issue, we have previously applied multiple reaction monitoring (MRM) to quantify tyrosine phosphorylation using a triple quadrupole instrument. In MRM, the first quadrupole isolates a precursor ion with a particular m/z which is then fragmented in the collision cell (second quadrupole), while the third quadrupole is set to pass only selected fragment ions. In this way, only peptide precursor and fragment ions of the correct mass (m/z ratio) are detected, and the instrument can spend additional time targeting these selected transitions. Due to the increased sensitivity of identification and quantification associated with increased detection time for the selected transitions, MRM is often used for quantification of low-level species in biological matrices. Since this approach is targeted to quantify particular peptides and fragment ions, a priori knowledge such as chromatographic elution time, m/z ratios of peptide precursor ions and m/z ratios of characteristic fragments generated during MS/MS are required. In general, one or more ‘discovery-mode’ DDA analyses are performed to generate the list which is then imported into the MRM method for targeted quantification across multiple replicates or different conditions. In one application, phosphorylated peptides identified from two iTRAQ DDA experiments were collated to construct a tailormade MRM method to quantify over 200 tyrosine phosphorylation sites [54]. Using this approach the inter-experimental reproducibility across four replicate analyses increased to 88%, thereby enabling the multiplexed analysis of two iTRAQ experiments reflecting seven different time points in an EGF time course [54]. With the increased speed of the latest generation of mass spectrometers, it should now be possible to generate full-scan MS/MS spectra for a large list of precursor peptide targets, improving the MRM approach through the acquisition of high-resolution fragment ion spectra and thereby decreasing false-positive or contaminated MRM data.
6. Biological Insight from Informatics
Once tyrosine phosphorylation sites have been identified and quantified, the next challenge is to gain biological insight from these complex quantitative data sets. To this end, multiple bioinformatics approaches have been developed to extract information from well-characterized sites with the goal of inferring putative function for poorly characterized proteins and phosphorylation sites. In two different studies, we have applied clustering methods to distinguish subgroups of phosphorylation sites based on quantitative phosphorylation profiles to generate hypotheses for further functional validation [26, 52]. To further interrogate phosphorylation data, we have developed PTMscout (ptmscout.mit.edu/) as an on-line repository enabling the comparison of data to previously identified sites [55]. PTMscout enables the generation of PTM motifs that may be present within the dataset of interest, generates gene ontology annotations, and applies Scansite predictions (scansite.mit.edu/) [56] to identify kinases that may be responsible for the phosphorylation of each identified phosphorylation site. To gain additional insight into the connection between signaling and cell physiology bioinformatics methods can be used correlate quantitative cell based phenotypic measurements (i.e. cell migration and proliferation) with phosphorylation data gathered under the same conditions [54]. For example, partial least squares regression (PLSR) modeling enabled the identification of nine tyrosine phosphorylation sites on six proteins whose temporal dynamic measurements provided sufficient information to predict cell invasion and migration in HER2 over-expressing human mammary epithelial cells (HMECs) [57, 58]. PLSR also enables the assignment of putative functions to novel phosphorylation sites, and provides quantitative predictions for the effect of deleting or inhibiting selected phosphorylation sites. Although PLSR has predominantly been applied to analysis of signaling and phenotype of cells in culture, it is also possible to apply this technique to cells and tissues in vivo, providing that quantitative phenotypic data is available for these tissues.
7. Characterization of tyrosine phosphorylation signaling events in vivo
The majority of phosphotyrosine analyses have been carried out using in vitro model systems, most likely due to the ease of generating the large sample amounts associated with isolation and characterization of low-abundance tyrosine phosphorylation sites. Despite many insights gained from studying these in vitro model systems, the role of tyrosine phosphorylation in the pathogenesis of multiple diseases, including many cancers, has highlighted the need for comprehensive analyses of tyrosine phosphorylation across disease states in vivo to enable the relevant identification of new drug targets. Tyrosine kinases and phosphatases have been identified to be deregulated across many cancer types through genetic profiling, leading to the development of single agent kinase inhibitors. Although tumors initially respond to treatment with these single agent inhibitors, these treatments are often not curative and these tumors eventually become resistant to these kinase specific drugs. It is thought that resistance may be due to the presence of tumor heterogeneity and tumor-stroma interactions in the tumor microenvironment, both of which are virtually impossible to recapitulate in an in vitro cell system. To fully understand the relevance and regulation of tyrosine phosphorylation in the context of human cancer pathogenesis, it is essential to move towards the analysis of patient tumors.
As one transitions from in vitro to in vivo analysis of tyrosine phosphorylation, sample procurement and processing becomes a critical factor. Due to the rapid signaling response to hypoxia and stress, it is essential to freeze the tissue within seconds to minutes of resection when characterizing physiological phosphorylation. However, samples that are routinely stored in tissue repositories are often not frozen within this time-frame; in many cases the delay between resection and freezing is not documented. Regardless, there have recently been a few attempts at semi-quantitative profiling of tyrosine phosphorylation sites across in vivo model systems and patient tumor tissues [59, 60]. The largest of these studies was a phosphotyrosine profiling screen which was carried out across 41 non-small cell lung cancer (NSCLC) cell lines and 150 NSCLC tumors [59]. In this study a total of 4500 phosphotyrosine sites were identified on more than 2700 proteins, a truly astounding number [59]. Semi-quantitative label-free analysis was used to quantify tyrosine phosphorylated peptides and the resulting tyrosine kinase profiles were then clustered to reveal distinct groups of tumors [59, 60]. While these results were intriguing, it is worth noting that label-free quantification requires reproducible sample processing and chromatographic separation of peptides prior to MS analysis. Although this technique has been applied to the quantification of proteins from complex samples with modest success [61], it cannot be applied accurately to the quantification of PTMs due to the enrichment methods that are typically required prior to MS analysis. Enrichment steps introduce an additional level of variation based on a complex number of variables, including sample concentrations, incubation times, antibody amounts, batch-to-batch variation in antibody quality, and irreproducibility in non-comprehensive immunoprecipitations. Furthermore, tyrosine phosphorylation occurs at low abundance, and therefore there are typically only a few MS/MS events per peptide, representing a regime in which spectral counting approaches, in which the quantification is based on the number of MS/MS spectra, are notoriously poor.
Moving forward, it is essential that rigorous methods of quantification are applied to the characterization of tissue samples. Coupling isotopic labeling strategies to targeted MS based approaches will help increase sample to sample reproducibility when attempting to profile large numbers of in vivo tissue samples. In addition to improving quantification and reproducibility, we must also increase our confidence in phosphorylation site identification through extensive manual and computational validation of MS data. Finally, improved bioinformatic and computational modeling tools are needed to provide the framework to understand the vastly different functional roles of tyrosine phosphorylation. In the near future, the rigorous application of these tools and techniques to quantify signaling networks in biological samples will reveal critical insights into the regulation and deregulation of in vivo signaling networks in health and disease.
Highlights.
In this review, we highlight the critical issues that need to be addressed as mass spectrometry based analysis of tyrosine phosphorylation signaling moves from in vitro samples to the in vivo setting.
We provide background information regarding the various technologies underlying quantitative analysis of tyrosine phosphorylation by mass spectrometry
We discuss the application of isotopic labeling strategies for quantitative mass spectrometry of in vitro and in vivo samples
The importance of site specific identification and the challenges underlying automated site identification are highlighted, along with the need for manual validation of at least the most biologically relevant phosphorylation sites
Acknowledgements
The authors would like to thank Dr Amanda Del Rosario for critical reading of the manuscript.
Abbreviations
- CID
collision induced dissociation
- DDA
Data dependent acquisition
- ETD
Electron transfer dissociation
- FDR
False discovery rate
- HMEC
Human mammary epithelial cells
- iTRAQ
isobaric tags for relative and absolute quantification
- MRM
Multiple reaction monitoring
- MS
Mass spectrometry
- MS/MS
tandem mass spectrometry
- PLSR
Partial least square regression
- PTM
post-translational modification
- PTP
Protein tyrosine phosphatase
- RTK
receptor tyrosine kinase
- SILAC
stable isotope labeling in cell culture
- TMT
Tandem mass tags
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase-activity. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T, Sefton BM. Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc Natl Acad Sci. 1980;77:1311–1315. doi: 10.1073/pnas.77.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Posada J, Cooper JA. Molecular signal integration. Interplay between serine, threonine, and tyrosine phosphorylation. Mol Biol Cell. 1992;3:583–592. doi: 10.1091/mbc.3.6.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 5.Yarden Y, Schlessinger J. Self-phosphorylation of epidermal growth factor receptor: evidence for a model of intermolecular allosteric activation. Biochemistry. 1987;26:1434–1442. doi: 10.1021/bi00379a034. [DOI] [PubMed] [Google Scholar]
- 6.Schlessinger J. Cell Signaling by Receptor Tyrosine Kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 7.Kratchmarova I, Blagoev B, Haack-Sorensen M, Kassem M, Mann M. Mechanism of Divergent Growth Factor Effects in Mesenchymal Stem Cell Differentiation. Science. 2005;308:1472–1477. doi: 10.1126/science.1107627. [DOI] [PubMed] [Google Scholar]
- 8.Yaffe MB. Phosphotyrosine-binding domains in signal transduction. Nat Rev Mol Cell Biol. 2002;3:177–186. doi: 10.1038/nrm759. [DOI] [PubMed] [Google Scholar]
- 9.Pawson T, Nash P. Protein–protein interactions define specificity in signal transduction. Genes Dev. 2000;14:1027–1047. [PubMed] [Google Scholar]
- 10.Hunter T. Protein kinases and phosphatases: The Yin and Yang of protein phosphorylation and signaling. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 11.Hunter T, Ling N, Cooper JA. Protein kinase C phosphorylation of the EGF receptor at a threonine residue close to the cytoplasmic face of the plasma membrane. Nature. 1984;311:480–483. doi: 10.1038/311480a0. [DOI] [PubMed] [Google Scholar]
- 12.Bowen S, Stanley K, Selva E, Davis RJ. Constitutive phosphorylation of the epidermal growth factor receptor blocks mitogenic signal transduction. J Biol Chem. 1991;266:1162–1169. [PubMed] [Google Scholar]
- 13.Dikic I, Giordano S. Negative receptor signalling. Curr Opin Cell Biol. 2003;15:128–135. doi: 10.1016/s0955-0674(03)00004-8. [DOI] [PubMed] [Google Scholar]
- 14.Futreal PA, Kasprzyk A, Birney E, Mullikin JC, Wooster R, Stratton MR. Cancer and genomics. Nature. 2001;409:850–852. doi: 10.1038/35057046. [DOI] [PubMed] [Google Scholar]
- 15.Rubbi L, Titz B, Brown L, Galvan E, Komisopoulou E, Chen SS, et al. Global Phosphoproteomics Reveals Crosstalk Between Bcr-Abl and Negative Feedback Mechanisms Controlling Src Signaling. Sci Signal. 2011;4:ra18. doi: 10.1126/scisignal.2001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slamon D, Clark G, Wong S, Levin W, Ullrich A, McGuire W. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 17.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 18.Hunter T. The Croonian lecture 1997. The phosphorylation of proteins on tyrosine: its role in cell growth and disease. Proceedings B. 1998;353:583–605. doi: 10.1098/rstb.1998.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schroeder W, Covey T, Hucho F. Identification of phosphopeptides by mass spectrometry. FEBS Lett. 1990;273:31–35. doi: 10.1016/0014-5793(90)81044-o. [DOI] [PubMed] [Google Scholar]
- 20.Rossomando AJ, Wu J, Michel H, Shabanowitz J, Hunt DF, Weber MJ, et al. Identification of Tyr-185 as the site of tyrosine autophosphorylation of recombinant mitogen-activated protein kinase p42mapk. Proc Natl Acad Sci. 1992;89:5779–5783. doi: 10.1073/pnas.89.13.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandey A, Podtelejnikov AV, Blagoev B, Bustelo XR, Mann M, Lodish HF. Analysis of receptor signaling pathways by mass spectrometry: Identification of Vav-2 as a substrate of the epidermal and platelet-derived growth factor receptors. Proc Natl Acad Sci. 2000;97:179–184. doi: 10.1073/pnas.97.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross AH, Baltimore D, Eisen HN. Phosphotyrosine-containing proteins isolated by affinity chromatography with antibodies to a synthetic hapten. Nature. 1981;294:654–656. doi: 10.1038/294654a0. [DOI] [PubMed] [Google Scholar]
- 23.Blagoev B, Ong S-E, Kratchmarova I, Mann M. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat Biotech. 2004;22:1139–1145. doi: 10.1038/nbt1005. [DOI] [PubMed] [Google Scholar]
- 24.Salomon AR, Ficarro SB, Brill LM, Brinker A, Phung QT, Ericson C, et al. Profiling of tyrosine phosphorylation pathways in human cells using mass spectrometry. Proc Natl Acad Sci. 2003;100:443–448. doi: 10.1073/pnas.2436191100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotech. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, et al. Time-resolved Mass Spectrometry of Tyrosine Phosphorylation Sites in the Epidermal Growth Factor Receptor Signaling Network Reveals Dynamic Modules. Mol Cell Proteomics. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Andersson L, Porath J. Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal Biochem. 1986;154:250–254. doi: 10.1016/0003-2697(86)90523-3. [DOI] [PubMed] [Google Scholar]
- 28.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotech. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 29.Schreiber TB, Mäusbacher N, Breitkopf SB, Grundner-Culemann K, Daub H. Quantitative phosphoproteomics – an emerging key technology in signal-transduction research. Proteomics. 2008;8:4416–4432. doi: 10.1002/pmic.200800132. [DOI] [PubMed] [Google Scholar]
- 30.Palumbo AM, Tepe JJ, Reid GE. Mechanistic Insights into the Multistage Gas-Phase Fragmentation Behavior of Phosphoserine- and Phosphothreonine-Containing Peptides. J Proteome Res. 2008;7:771–779. doi: 10.1021/pr0705136. [DOI] [PubMed] [Google Scholar]
- 31.Schroeder MJ, Shabanowitz J, Schwartz JC, Hunt DF, Coon JJ. A Neutral Loss Activation Method for Improved Phosphopeptide Sequence Analysis by Quadrupole Ion Trap Mass Spectrometry. Anal Chem. 2004;76:3590–3598. doi: 10.1021/ac0497104. [DOI] [PubMed] [Google Scholar]
- 32.Steen H, Küster B, Fernandez M, Pandey A, Mann M. Detection of Tyrosine Phosphorylated Peptides by Precursor Ion Scanning Quadrupole TOF Mass Spectrometry in Positive Ion Mode. Anal Chem. 2001;73:1440–1448. doi: 10.1021/ac001318c. [DOI] [PubMed] [Google Scholar]
- 33.Jonscher KR, Yates JR., III The Quadrupole Ion Trap Mass Spectrometer—A Small Solution to a Big Challenge. Anal Biochem. 1997;244:1–15. doi: 10.1006/abio.1996.9877. [DOI] [PubMed] [Google Scholar]
- 34.Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Higher-energy C-trap dissociation for peptide modification analysis. Nat Meth. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 35.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 37.Yates JR, Eng JK, McCormack AL, Schieltz D. Method to Correlate Tandem Mass Spectra of Modified Peptides to Amino Acid Sequences in the Protein Database. Anal Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 38.Ducret A, Oostveen IV, Eng JK, Yates JR, Aebersold R. High throughput protein characterization by automated reverse-phase chromatography/electrospray tandem mass spectrometry. Protein Sci. 1998;7:706–719. doi: 10.1002/pro.5560070320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Meth. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 40.Chen Y, Zhang J, Xing G, Zhao Y. Mascot-Derived False Positive Peptide Identifications Revealed by Manual Analysis of Tandem Mass Spectra. J Proteome Res. 2009;8:3141–3147. doi: 10.1021/pr900172v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gnad F, Gunawardena J, Mann M. PHOSIDA 2011: the posttranslational modification database. Nucleic Acid Res. 2011;39:D253–D260. doi: 10.1093/nar/gkq1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, et al. Global, In Vivo, and Site-Specific Phosphorylation Dynamics in Signaling Networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 43.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotech. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 44.Savitski MM, Lemeer S, Boesche M, Lang M, Mathieson T, Bantscheff M, et al. Confident Phosphorylation Site Localization Using the Mascot Delta Score. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.003830. M110.003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nichols AM, White FM. Manual Validation of Peptide Sequence and Sites of Tyrosine Phosphorylation from MS/MS Spectra. 2009;492:143–160. doi: 10.1007/978-1-59745-493-3_8. [DOI] [PubMed] [Google Scholar]
- 46.Lee J, Xu Y, Chen Y, Sprung R, Kim SC, Xie S, et al. Mitochondrial Phosphoproteome Revealed by an Improved IMAC Method and MS/MS/MS. Mol Cell Proteomics. 2007;6:669–676. doi: 10.1074/mcp.M600218-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lahesmaa-Korpinen A-M, Carlson SM, White FM, Hautaniemi S. Integrated data management and validation platform for phosphorylated tandem mass spectrometry data. Proteomics. 2010;10:3515–3524. doi: 10.1002/pmic.200900727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krüger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, et al. SILAC Mouse for Quantitative Proteomics Uncovers Kindlin-3 as an Essential Factor for Red Blood Cell Function. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 49.Westman-Brinkmalm A, Abramsson A, Pannee J, Gang C, Gustavsson MK, von Otter M, et al. SILAC zebrafish for quantitative analysis of protein turnover and tissue regeneration. J Proteomics. 2011;75:425–434. doi: 10.1016/j.jprot.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 50.Schmelzle K, Kane S, Gridley S, Lienhard GE, White FM. Temporal Dynamics of Tyrosine Phosphorylation in Insulin Signaling. Diabetes. 2006;55:2171–2179. doi: 10.2337/db06-0148. [DOI] [PubMed] [Google Scholar]
- 51.Iwai LK, Benoist C, Mathis D, White FM. Quantitative Phosphoproteomic Analysis of T Cell Receptor Signaling in Diabetes Prone and Resistant Mice. J Proteome Res. 2010;9:3135–3145. doi: 10.1021/pr100035b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, et al. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci. 2007;104:12867–12872. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang PH, Miraldi ER, Xu AM, Kundukulam VA, Del Rosario AM, Flynn RA, et al. Phosphotyrosine signaling analysis of site-specific mutations on EGFRvIII identifies determinants governing glioblastoma cell growth. Mol BioSystems. 2010;6:1227–1237. doi: 10.1039/c001196g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc Natl Acad Sci. 2007;104:5860–5865. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naegle KM, Gymrek M, Joughin BA, Wagner JP, Welsch RE, Yaffe MB, et al. PTMScout: A web resource for analysis of high-throughput post-translational proteomic studies. Mol Cell Proteomics. 2010;9:2558–2570. doi: 10.1074/mcp.M110.001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acid Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolf-Yadlin A, Kumar N, Zhang Y, Hautaniemi S, Zaman M, Kim H-D, et al. Effects of HER2 overexpression on cell signaling networks governing proliferation and migration. Mol Syst Biol. 2006;2:54. doi: 10.1038/msb4100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar N, Wolf-Yadlin A, White FM, Lauffenburger DA. Modeling HER2 Effects on Cell Behavior from Mass Spectrometry Phosphotyrosine Data. PLoS Comput Biol. 2007;3:e4. doi: 10.1371/journal.pcbi.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, et al. Global Survey of Phosphotyrosine Signaling Identifies Oncogenic Kinases in Lung Cancer. Cell. 2007;131:1190–11903. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 60.Drake JM, Graham NA, Stoyanova T, Sedghi A, Goldstein AS, Cai H, et al. Oncogene-specific activation of tyrosine kinase networks during prostate cancer progression. Proc Natl Acad Sci. 2012;109:1643–1648. doi: 10.1073/pnas.1120985109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, et al. Comparison of Label-free Methods for Quantifying Human Proteins by Shotgun Proteomics. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]