SUMMARY
Centrioles are key microtubule polarity determinants. Centriole duplication is tightly controlled to prevent cells from developing multipolar spindles, a situation that promotes chromosomal instability. A conserved component in the duplication pathway is Plk4, a polo kinase family member that localizes to centrioles in M/G1. To limit centriole duplication, Plk4 levels are controlled through trans-autophosphorylation that primes ubiquitination. In contrast to Plks 1–3, Plk4 possesses a unique central region called the “cryptic polo box”. Here, we present the crystal structure of this region at 2.3 Å resolution. Surprisingly, the structure reveals two tandem, homodimerized polo boxes, PB1-PB2, that form a unique, winged architecture. The full PB1-PB2 cassette is required for binding the centriolar protein Asterless as well as robust centriole targeting. Thus, with its C-terminal polo box (PB3), Plk4 has a triple polo box architecture that facilitates oligomerization, targeting, and promotes trans-autophosphorylation, limiting centriole duplication to once per cell cycle.
INTRODUCTION
Centrioles are cylindrical, microtubule-based structures that form the core components of centrosomes and basal bodies, organelles that nucleate and spatially organize microtubules to form the mitotic spindle and cilia (Bornens 2012). Centriole number is precisely controlled with centriole duplication restricted to a single cell cycle event (Tsou and Stearns, 2006; Tsou et al., 2009; Nigg and Stearns, 2011). Centrioles exist as pairs composed of an older (mother) centriole and a daughter centriole, assembled on the mother centriole in the preceding cell cycle. G1-phase cells contain a single mother-daughter centriole pair. During S-phase, centrioles separate and duplicate, generating two mother-daughter pairs that facilitate bipolar spindle assembly. The mechanisms underlying centriole duplication define a critical step in cellular biology, as misregulation of centriole number is linked to chromosome instability and diseases including ciliopathies, male sterility, primary microcephaly, and tumorigenesis (Bettencourt-Dias et al., 2011; Rosario et al., 2010; Nigg and Raff, 2009).
Several proteins are required for centriole biogenesis. Among these are the conserved proteins Polo-like kinase 4 (Plk4/Sak), Asterless/Cep152, SAS-6, SAS-5/Ana2/STIL, Cep135, and SAS-4/CPAP (Song et al., 2008; Azimzadeh and Marshall, 2010). The order of subunit addition suggests a hierarchical centriole assembly pathway conserved across phyla. Asterless (Asl), a scaffolding protein, initially recruits Plk4 to the site of daughter centriole assembly (Hatch et al., 2010; Dzhindzhev et al., 2010; Cizmecioglu et al., 2010). Plk4 activity is upstream of the SAS proteins and primes the mother centriole for S-phase duplication (Pelletier et al., 2006; Kleylein-Sohn et al., 2007; Kitagawa et al., 2009). Daughter centriole (procentriole) assembly begins at the proximal end of the mother centriole with the formation of a nine-fold symmetric cartwheel structure composed of SAS-6 homodimers (Kitagawa et al., 2011; van Breugel et al., 2011). How Plk4 initiates centriole assembly is not well defined. In Caenorhabditis elegans, ZYG-1 (the Plk4 homolog) phosphorylates SAS-6, triggering centriole formation (Kitagawa et al., 2009), although this has not been shown in other systems. In humans, Plk4 inactivates FBXW5, a SCF (Skp, Cullin, F-box) component used to degrade SAS-6, suggesting that Plk4 initiates centriole duplication by stabilizing SAS-6 (Puklowski et al., 2011). However, Drosophila FBXW5 has no role in controlling centrosome number (Rogers et al., 2009). Thus, while species show some divergence in the duplication pathway, Plk4 has emerged as a master-regulator of centriole assembly.
Plk4 and its binding partner Asl are required for centriole duplication (Bettencourt-Dias et al., 2005; Habedanck et al., 2005; Varmark et al., 2007) and studies in human, Drosophila and Xenopus systems show that Plk4 or Asl overexpression promotes centriole amplification as well as de novo centriole assembly (Rodrigues-Martins et al., 2007; Peel et al., 2007; Dzhindzhev et al., 2010; Eckerdt et al., 2011). Plk4 is regulated by the SCFSlimb/β-TrCP ubiquitin ligase which recognizes Plk4 after homodimer-dependent trans-autophosphorylation of the phosphodegron known as the downstream regulatory element (DRE) (Guderian et al., 2010, Cunha-Ferreira et al., 2009; Rogers et al., 2009; Brownlee et al., 2011; Holland et al., 2010). In cultured Drosophila cells, Plk4 is degraded throughout most of the cell cycle to prevent centriole amplification (Peel et al., 2007; Kleylein-Sohn et al., 2007). During M-phase however, Plk4 is dephosphorylated by Protein Phosphatase 2A, thereby stabilizing Plk4, allowing a brief mitotic debut that restricts centriole duplication to a single event per cell cycle (Brownlee et al., 2011).
Plk4 is a member of the Polo-like kinase family. Plk members 1–4 share sequence similarity to the founding member, Drosophila Polo (Plk1) (Sillibourne and Bornens, 2010). Like Polo, Plk members regulate cell-cycle events that collectively include spindle formation, the metaphase-to-anaphase transition, mitotic exit, cytokinesis, and DNA damage checkpoints. To perform these critical functions, Plk gene expression, protein expression, localization, kinase activity, and destruction are tightly regulated throughout the cell cycle (Archambault and Glover, 2009) as aberrant Plk activities contribute to chromosome instability and oncogenesis.
Plks share an amino-terminal serine/threonine kinase domain, as well as one or more ~100-residue polo box (PB) domains. Plk members 1–3 contain two carboxy-terminal PBs (Figure 1A) that interact in cis to bind phosphorylated targets, mediate localization, and activate the kinase (Lowery et al., 2005). The architecture of a PB domain consists of an anti-parallel 6-stranded β-sheet that lies across a C-terminal α-helix (Leung et al., 2002). Plk1’s tandem PBs (PB1-PB2) clamp around a phosphopeptide target with each PB contributing binding determinants (Elia et al., 2003; Cheng et al., 2003). Intriguingly, Plk4 is structurally divergent. It was annotated as containing only a single, carboxy-terminal PB, which confers homodimerization and moderate centriole localization by binding an unidentified target (Leung et al., 2002). The structure of the Plk4 PB is homodimeric and adheres to a general PB architecture, though it is formed through swapped chains of the homodimer. The homodimeric arrangement of this Plk4 PB is distinct from the tandem arrangement of the Plk1 PB1-PB2 pair, indicative that PBs adopt differential spatial arrangements.
Figure 1.
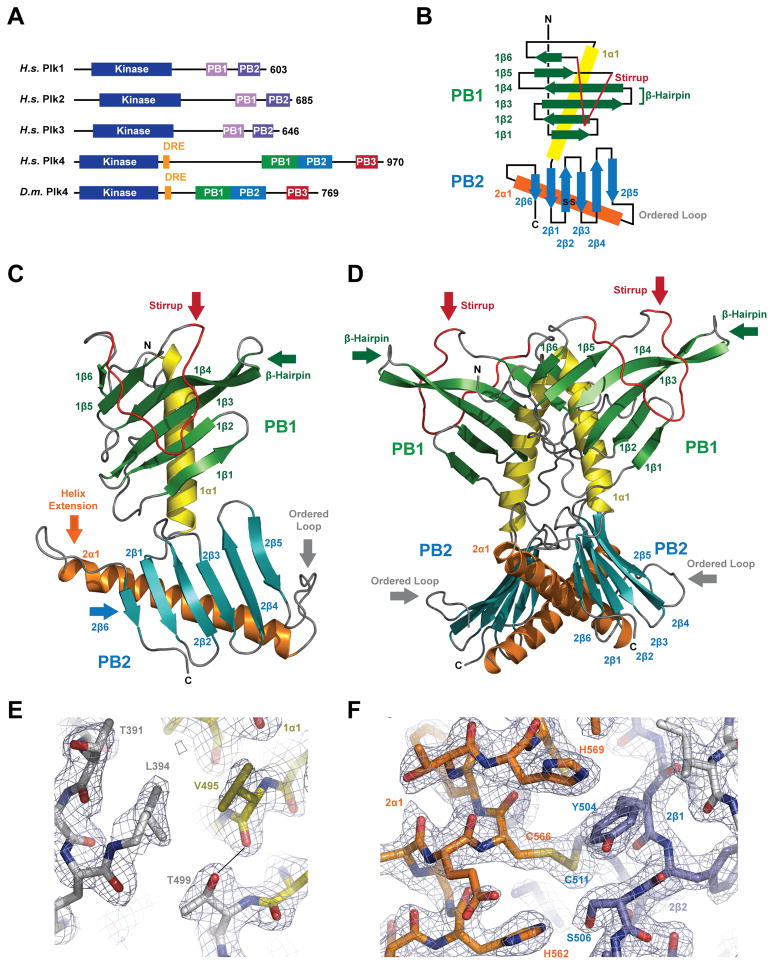
The Plk4 CPB is Composed of Tandem PB domains, PB1 and PB2. A) Polo-like kinase family domain architecture. Plk1-3 contain two PB domains, Plk4 contains three PB domains. Plk4 levels are regulated by the DRE (orange). B) Secondary structure topology diagram of Plk4’s conserved central domain: PB1 (βstrands in green, α-helix in yellow, stirrup in red, loops in black) and PB2 (β-strands in blue, α-helix in orange, loops in black). C) Tertiary structure of the Plk4 PB1-PB2 monomer colored as in B. D) Quaternary structure of homodimeric Plk4 PB1-PB2, rotated 90° relative to C. E)Stick representation of the junction between PB1 1α1 and PB2 where the T449 hydroxyl caps the 1α1 helix. F) Stick representation of the PB2 intra-domain disulfide formed between 2α1 C566 and 2β2 C511. Final 2Fo-Fc electron density contoured at 1.5σ (E,F).
Plk4 also contains a conserved central domain, hitherto called the “cryptic polo box” (CPB), which bridges the kinase domain and the carboxy-terminal PB. This region was initially identified as a centriole-targeting component, capable of binding the kinase domain in trans (Leung et al., 2002). Based on these properties, the region was named the “cryptic polo box” though it showed no apparent sequence homology to canonical PBs (Swallow et al., 2005). Recent work has identified a CPB binding partner, Asterless (Asl)/Cep152, which targets Plk4 to centrioles (Dzhindzhev et al., 2010; Hatch et al., 2010, Cizmecioglu et al., 2010). To date, the CPB has largely remained an enigma, with questions concerning its structure, function in centriole localization, and role in Plk4 activity outstanding.
Here, we present the crystal structure of the CPB, determined to a resolution of 2.3 Å. Surprisingly, this structure reveals that the CPB comprises two structurally unique PB domains, PB1 and PB2. Cellular localization and biochemical studies indicate that the entire tandem PB1-PB2 cassette is required for robust centriole localization and Asl binding. The PB1-PB2 cassette also mediates Plk4 oligomerization, and when expressed as a trans cassette, protects endogenous Plk4 from trans auto-phosphorylation and subsequent degradation. Thus, the Plk4 PB1-PB2 cassette is a unique architectural component required for Plk4 function.
RESULTS
Crystallization and Structure Determination of the Plk4 CPB
Plk4’s central region, termed the “cryptic polo box,” was analyzed using secondary structure prediction algorithms in parallel with conservation to delineate the boundary residues for structural, biochemical and cellular analysis. D. melanogaster Plk4382-602 was bacterially expressed, purified and crystallized as described in Experimental Procedures. Crystals belonged to the space group P21212. A native dataset was collected on a single crystal to a resolution of 2.3 Å. To obtain phasing information, selenomethionine (SeMet)-derivatized protein was produced but proved insoluble. We found that a SeMet- M517A mutant was marginally soluble and produced isomorphous crystals. Single wavelength peak anomalous dispersion data to 2.9 Å resolution was collected. Phases were calculated and extended to 2.3 Å. The structure was built and refined to R and Rfree factors of 18.5 and 25.5%, respectively. The final model contains two molecules of Plk4 in the asymmetric unit, comprising residues 382–596 (chain A) and 382–548, 553–597 (chain B). Crystallographic information is presented in Table 1.
Table 1.
Crystallographic data, phasing and refinement
| Crystal | Native | SeMet M517A |
|---|---|---|
| Beamline | APS 22-ID | APS 22-BM |
| Space group | P21212 | |
| Cell: a,b,c (Å) | 86.5, 136.4, 46.5 | 87.1, 136.5, 46.6 |
| Wavelength (Å) | 1.00890 | 0.97980 |
| dmin (Å) | 2.3 (2.38-2.30) | 2.9 (3.00-2.90) |
| % Complete | 90.1 (85.2) | 100.0 (100.0) |
| <I/σI> | 21.3 (5.2) | 16.3 (3.2) |
| No. of observations | 114559 (10620) | 101205 (9570) |
| No. of independent observations | 22630 (2082) | 23604(2333) |
| Multiplicity | 5.1 (5.1) | 4.3 (4.1) |
| Rsym (%)* | 5.9 (35.9) | 8.5 (37.9) |
| Resolution (Å) at which anomalous completeness exceeds 85% for I/σI > 5, > 3, > 2 | 4.1, 3.9, 3.4 | |
| Overall log-likelihood gain★/figure of merit† | 1.4×105/0.44(0.35) | |
| Figure of merit† Centrics/Acentrics | 0.25(0.25)/0.47(0.36) | |
| Figure of merit† after density modification Centrics/Acentrics | 0.66(0.42)/0.73(0.52) | |
| Refinement (Å) | 50-2.3 (2.38-2.30) | |
| R value⋄ | 18.5 (19.8) | |
| Rfree⧧ | 25.5 (30.7) | |
| Rmsd bond lengths (Å) | 0.008 | |
| Rmsd bond angles (°) | 1.09 | |
| Mean B (min/max) (Å2) | 35.9 (9.4/141.6) | |
| No. atoms: protein/water/SO4− | 3478/184/6 | |
Values in parentheses are for the highest resolution shells unless otherwise denoted.
Rsym=ΣhΣi|Ii(h)−<I(h)>|/ΣhΣiIi(h) where Ii(h) is the integrated intensity of the ith reflection with the Miller Index h and <I(h)> is the average over Friedel and symmetry equivalents.
Log-likelihood gain value as determined by Phenix.
Figure of merit is the weighted mean of the cosine of the deviation from αbest.
R value = Σ(|Fobs|−k|Fcalc|)/Σ|Fobs|.
Rfree is calculated using a 10% subset of the data that is removed randomly from the original data and excluded from refinement.
The CPB is Composed of Two Structurally Unique Polo Box Domains
Plk4382-602forms a multi-domain structure, surprisingly composed of two tandem, structurally unique PBs (Figure 1B–D). This contrasts with the prediction that Plk4 is composed of a single C-terminal PB. In contrast to Plk1-3 that each contain two PBs, Plk4 was designated the “odd one out” (Sillibourne and Bornens, 2010), which holds true, though not because it contains one PB, but three PBs. We henceforth demarcate the Plk4382-602 PBs and assign them the sequential numbering PB1 (aa 382–499) and PB2 (aa 500–602), and propagate this scheme to the ultimate C-terminal PB, not included in our structure, assigning it PB3 (aa 660–741). We refer to the structure reported here as PB1-PB2. Both PB1 and PB2 adopt a canonical PB fold, delineated by an N-terminal anti-parallel β-sheet that packs against a C-terminal helix that runs diagonal to the β-strands. PB1 has a sequential anti-parallel β-sheet composed of the six strands 1β1-1β2-1β3-1β4-1β5-1β6 that pack against the 1α1 helix. PB1 contains two unique structural attributes. First, the 1β3-1β4 strands form an extended hairpin off the β-sheet giving PB1 a winged structure (Figure 1C–D, green arrows). Second, a sixteen-residue segment that we term the “stirrup” bridges 1β5 and 1β6 and blankets one face of the β-sheet, effectively sandwiching the PB1 β-sheet between 1α1 and the stirrup (Figure 1C–D, red arrows). The 1β4 portion of the winged β-hairpin buttresses the N-terminal flank of the stirrup, implicating a structural co-dependence between these unique PB elements. The 1α1 helix completes PB1, followed by a two amino acid linker that bridges PB1 and PB2: threonine 499 and proline 500. T499 is conserved across species and caps the 1α1 helix by forming a hydrogen bond between the T499 γ-hydroxyl and the K496 backbone carbonyl (Figure 1E). The invariant proline P500 makes a jog in the backbone, offsetting the first strand of the PB2 sheet, 2β1, from the 1α1 axis.
Plk4 PB2 also contains unique features that deviate from the canonical PB fold, including secondary structure elements that flank the β-sheet as well as an extended 2α1 helix. The PB2 β-sheet proceeds in an anti-parallel fashion, consecutively snaking through strands 2β1-2β2-2β3-2β4-2β5. Instead of forming a sixth consecutive β-strand, as is the case with PB1, PB2 2β5 is followed by an ordered loop that connects 2β5 with the 2α1 helix (Figure 1C–D, grey arrows). The 2β5-2α1 loop is stabilized by a hydrogen bond between the conserved D542 δO and the M546 backbone amide as well as van der Waals interactions between hydrophobic residues in the loop, the 2β5 strand, and the 2α1 helix. The C-terminal region of this loop is variable across species and contains a seven-residue insert in higher eukaryotes. The 2α1 helix runs diagonal to the β-sheet, spanning 40 Å and extending 13 Å past the 2β1 strand (Figure 1C, orange arrow). The helix-sheet interaction is stabilized both by a hydrophobic core as well as a disulfide bond between C511 in 2β2 and C566 in 2α1 (Figure 1F). The 2α1 helix leads into a loop that curls back towards 2β1, effectively stabilizing the extended 2α1 helix by packing F588 against the β-sheet-2α1 hydrophobic core. The loop terminates at an invariant proline, P589, that facilitates a bend in the backbone, leading into PB2’s final β-strand, 2β6, which runs parallel to 2β1 and completes the PB2 β-sheet and the PB1-PB2 cassette (Figure 1C, blue arrow). The location and polarity of the Plk4 2β6 strand is unique, normally running anti-parallel to β5 in other PB structures.
Collectively, PB1 and PB2 form a composite structure, with a conserved core interface that buries 329 Å2 and limits molecular flexibility to peripheral loop regions. Relative to PB1, PB2 is rotated approximately 120°. The protomers align well with 1.2 Å rmsd across 214 Cα atoms. PB1 and PB2 individually align to their dimeric mate with a Cα rmsd equal to 1.1 and 1.2 Å respectively. Structural differences between the protomers, indicative of molecular flexibility, localize to loop regions, specifically the stirrup’s C-terminal region, the 2β4-2β5 loop, and the 2β5-2α1 loop (Figure 2A, red arrows). As a second metric for structural flexibility, we mapped B factors on the PB1-PB2 structure. The structure’s Cα B-factors range from 14–105 Å2, with increased levels in the stirrup, the 2β4-2β5 loop, the 2β5-2α1 loop, and the 2α1-2β6 loop, correlating with structural differences noted in the alignment of the protomers (Figure 2B, arrows). The core regions of PB1 and PB2 as well as the T499-P500 bridge exhibit low temperature factors and little structural divergence when protomers are compared, indicative that the relative arrangement of PB1 and PB2 is static.
Figure 2.
Plk4 PB1-PB2 is an Asymmetric Homodimer with Plastic Stirrups and Loops. A) Superposition of Plk4 PB1-PB2 protomers A and B showing plasticity in the stirrup as well as the 2β4-2β5 and 2β5-2α1 loops. Plk4 colored as in Figure 1B, shown in cartoon format. B) Plk4 PB1-PB2 protomer A backbone colored and scaled according to Cα B-factor values ranging from 14 (dark blue) to 105 (red) Å2. High B-factors correlate with the structurally plastic loop segments between protomers A and B (shown in A). The region bridging PB1 and PB2 shows little structural divergence and is dominated by low B-factor values. Plk4 PB1-PB2 protomer A (C), and protomer B (D) shown in sphere format; oriented as shown in the inset. Residues involved in homodimerization from both protomers, unique to protomer A, and unique to protomer B, are colored dark grey, purple, and raspberry, respectively. E) Size exclusion chromatography – multi-angle light scattering analysis of H6-Plk4 PB1-PB2 injected at 19 μM (red trace) and 28 μM (green trace)(100 μl). Y-axis at left displays molecular weight (kDa), Y-axis at right displays normalized differential refractive index, X-axis displays time component of the run. (See Figure S1).
Plk4 PB1-PB2 Forms a Pseudo-symmetric Homodimer
The two Plk4 PB1-PB2 molecules in the asymmetric unit form a homodimer. The two PB1-PB2 molecules are related by a pseudo-symmetric two-fold axis that runs parallel to the 1α1 helices (Figure 1D). A translational component along the pseudo two-fold axis shifts protomer A approximately 5 Å relative to protomer B. The homodimerization interface is mediated by PB1-PB1 contacts as well as PB2-PB2 contacts (Figure 2C,D). Due to the translational component, non-equivalent sets of residues are involved in the asymmetric dimerization. The PB1-PB1 interface primarily involves residues from 1α1 with additional contributions from neighboring residues in the β-sheet. The PB2-PB2 interface involves residues from 2β4, the 2β4-2β5 loop, 2β5, and 2α1. The PB1-PB1 and PB2-PB2 dimerization interfaces bury 1511 Å2 and 1065 Å2 of solvent accessible surface area respectively, collectively totaling 2576 Å2. While crystallographic interfaces between protomers exist (Figure S1A–C), the homodimer in the asymmetric unit buries the largest surface area and is the only interaction that involves both PB1 and PB2; thus it likely represents the biological dimer.
To verify that Plk4 PB1-PB2 formsan oligomeric species in solution, we analyzed the oligomeric state using light scattering. Size exclusion chromatography coupled to multi-angle static light scattering was conducted at pH 7.0. His6-Plk4 PB1-PB2 protein (MW 27.6 kDa) was injected at initial concentrations of 19 and 28 μM. Single elution peaks had experimentally determined molecular weights equal to 56.3 ± 4 kDa and 54.3 ± 2 kDa respectively (Figures 2E, S1D). We also investigated the oligomeric state using batch dynamic light scattering at pH 9.5 using 140 μM Plk4 PB1-PB2 (MW 25.7 kDa) which yielded an experimental mass of 66 ± 16 kDa (Table S1). Collectivley, the static and dynamic light scattering values indicate that Plk4 PB1-PB2 exists as a homodimer with potential higher order oligomers forming at elevated concentration and pH. This is supported by prior work showing that the mouse Plk4 PB1-PB2 region self-associates (Leung et al., 2002).
Unique Features of Plk4 PB1 and PB2 and Implications for Target Binding
While Plk4 PB1 and PB2 contain characteristic features of a PB domain, each diverges from PB structures determined to date, yielding implications for target binding. To highlight these differences, we compare Plk4’s PB1 and PB2 domains with Plk1’s PB1 domain bound to a phosphopeptide target (Figure 3A). The Plk1 PB1 1β1 strand forms key contacts with the phosphopeptide target. The Plk4 PB1 domain aligns to Plk1 PB1 with an rmsd of 2.0 Å across 76 aligned Cα atoms (Figure 3B). Structural divergence occurs at the Plk4 PB1 1β3-1β4 hairpin extension, the stirrup, the positioning of 1β6 and the elongated 1α1 N-terminal region. While there is structural divergence, the 1β1 strand, used in Plk1 PB1 to bind phosphopeptide targets, is accessible and may facilitate target binding as observed in Plk1 PB1 (Figure 3C).
Figure 3.
Plk4 PB1 and PB2 Diverge from Plk1 PB Domain Structures and Form a Unique Inter-domain Interaction. A) Structural alignment of Plk4 PB1 and Plk4 PB2 (colored as in Figure 1B) with human Plk1 PB1 bound to a phosphopeptide target (3FVH, β-strands in purple, α-helix in grey, peptide in blue). B) Matrix showing the rmsd (Å) between structures of human Plk1 PB1 and PB2, fly Plk4 PB1 and PB2, and mouse Plk4 PB3. C) Superposition of Plk4 PB1-PB2 homodimer and the Plk1 PB1-PB2-phosphopeptide structure, aligned over Plk4 PB1 and Plk1 PB1, highlighting the differential organization of Plk1 PB1-PB2 as compared to Plk4 PB1-PB2. The Plk4 PB1 stirrup overlaps with the corresponding binding site of Plk1 PB2. The location where Plk1 PB1 binds its phosphopeptide target is accessible on Plk4 PB1. D) Superposition of Plk4 PB1-PB2 and the Plk4 PB3 homodimer, aligned over single PB1 and PB3 domains. Insets show the orientation of each independent structure (C,D).
Plk4 PB2 diverges from the Plk1 PB1 structure, with differences in loops, a helix extension, and the positioning of 2β6. Plk4 PB2 aligns to Plk1 PB1 with a 2.3 Å rmsd over 72 structurally aligned Cα atoms (Figure 3B). Plk4 PB2 differs from Plk1 PB1 in the positioning of the 2β2-2β3 and 2β4-2β5 loops. Plk4 PB2 contains an ordered loop between 2β5 and 2α1 that substitutes for the Plk1 PB1 1β6 strand. Plk4 PB2 forms a C-terminal extension on the 2α1 helix. The Plk4 PB2 2β6 strand does not occur between 2β5 and 2α1, but resides C-terminal to 2α1 and runs parallel to 2β1, a site occupied by the phosphopeptide target in Plk1 PB1. Thus, 2β6 occludes the Plk4 PB2 domain from interacting with a phosphopeptide in a manner equivalent to Plk1 PB1.
Plk4 PB1-PB2 has a Novel Inter-domain and Homodimeric Arrangement
PB structures determined to date show diverse intra- and inter-molecular PB-PB interactions. Here we compare Plk4 PB1-PB2 with Plk1 PB1-PB2 and Plk4 PB3. Plk1 PB1-PB2 is monomeric, with PB1 and PB2 positioned to form a collective β-sandwich. In contrast, Plk4 PB3 is homodimeric and dimerizes across a symmetric β-sandwich (Leung et al., 2002). Like Plk1 PB1-PB2, the Plk4 PB3 β-sheets are sandwiched orthogonal to one another; however, PB3 dimerizes across the opposite face of its PB βsheets. Plk4 PB1-PB2 adopts a third, unique PB domain arrangement (Figure 3C,D). To highlight the differential inter-domain arrangements across these paired PB domain structures, we superimposed the Plk4 PB1-PB2 structure and the Plk1 PB1-PB2 structure after a least squares fit of their respective PB1 domains (Figure 3C). Relative to PB1, the Plk4 PB2 domain is positioned dramatically different than Plk1 PB2, each engaging a distinct, non-overlapping face on their respective PB1 partner. In Figure 3D, the Plk4 PB1-PB2 structure is superimposed on the Plk4 PB3-PB3 structure after a least squares fit of individual Plk4 PB1 and PB3 domains. The position of the PB3 dimeric mate does not correlate with the relative positioning of PB2, or the PB1 dimeric mate. Overall, all PB-PB structures exhibit non-homologous domain arrangements. One consistent feature across Plk4 PB structures is homodimerization of the individual PB domains. However, while the PB3-PB3 interface is symmetric, the PB1-PB1 and PB2-PB2 interfaces are asymmetric.
The Conserved Plk4 PB1-PB2 Inter-domain Groove Corresponds to the Plk1 PB1 Target Binding Site
To highlight conserved determinants across the Plk4 PB1-PB2 structure, we generated an alignment of PB1-PB2 across ten diverse species and contoured identity at 100% and 80% (Figure 4A, green and yellow, respectively). When mapped onto the Plk4 PB1-PB2 homodimer structure, the prime cluster of invariant residues occurs at a composite site formed at the PB1-PB2 junction, with contributions by PB1 1β1-1β2 and PB2 2β1-2β4 (Figure 4B,C, green arrows). The majority of conserved sequences in PB1 1β2-1β5 are occluded by the stirrup and accessibility would thus require a dramatic rearrangement of the stirrup which we do not rule out. The PB1-PB2 junction is formed on a single protomer, thereby constituting two independent sites on the homodimer. The PB1-PB2 junction is concave with both hydrophobic and basic determinants (Figure 4B,C, blue arrows). When Plk1 PB1 with bound phosphopeptide is aligned with Plk4 PB1, the phosphopeptide is positioned at the Plk4 PB1-PB2 conserved junction (Figures 3C; 4B,C). Whether this site on Plk4 binds targets remains to be determined.
Figure 4.
Plk4 PB1 and PB2 Form a Composite Inter-domain Groove Delineated by Conserved and Basic Residues. A) Plk4 sequence alignment across ten species. Protomer A solvent accessible surface area (ASA) (Å2) is indicated. 100% identity is highlighted in green, 80% identity in yellow (homologous residues also highlighted in yellow where the 80% identity criteria is met). Human Plk1 PB1 sequence is aligned against Plk4 PB1 and PB2, based on structural alignment. Residues involved in homodimerization are indicated below the alignment, colored as in Fig 2C,D. B) Plk4 PB1-PB2 homodimer structure shown in sphere format with conserved residues colored as in A (left) and in surface representation (right) showing electrostatics contoured from −2.0 to +2.0 kT/e. C) Protomer A rotated 45° relative to the orientation shown in B showing conservation and electrostatics as in B. The phosphopeptide from the Plk1 PB1-PB2 structure (3FVH) shown in stick format and colored blue, is docked onto the Plk4 structure in both B and C, based on the structural alignment of Plk1 PB1 and Plk4 PB1 shown in Fig 3C. D–E) Anti-GFP immunoprecipitates from S2 cell lysates transiently-expressing N-terminal Asl-V5 and the indicated Plk4-GFP construct or control GFP, probed for GFP and V5.
The Full Plk4 PB1-PB2 Cassette is Required for Asterless Binding
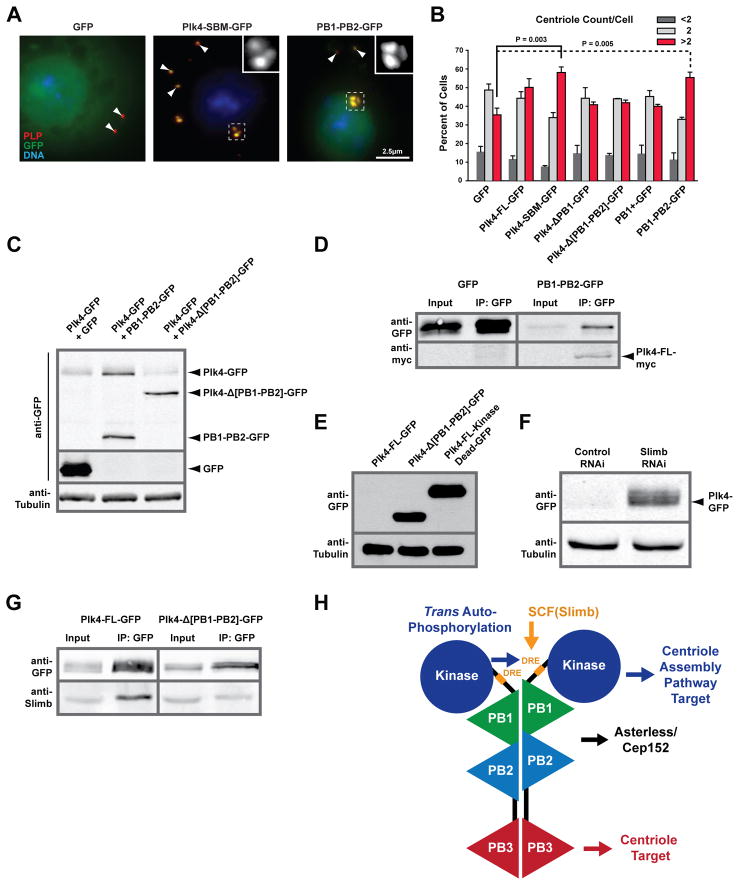
A fragment of Plk4’s PB1-PB2 region has been shown to bind the centriole component Asl in vitro (Dzhindzhev et al., 2010). The Asl-binding region spans Plk4 residues 376–525, while a shorter fragment spanning residues 376–500 (encompassing only PB1), lacks Asl-binding activity. The Plk4 PB1-PB2 structure spans residues 382–602, with proline P500 defining the PB1-PB2 bridge and residues 501–525 encoding the first three contiguous anti-parallel β-strands in PB2 (2β1-2β3). While 2β1-2β3 is conserved and contributes to the composite PB1-PB2 conserved patch, it is unlikely that 2β1-2β3 would fold into an ordered β-sheet in the absence of 2α1, though it may fold upon Asl binding. To test the ability of Plk4 PB1-PB2 to bind Asl, taking structural insight into construct design, we immunoprecipitated various Plk4-GFP constructs from S2 cell lysates transiently co-expressing the Asl Plk4-binding domain, V5-Asl (residues 1–300), and immunoblotted for these proteins. As expected, full-length Plk4 did not express at high levels, due to its ubiquitin-mediated degradation, and thus did not co-immunoprecipitate detectable levels of Asl (Figure 4D). To ensure that Plk4 was capable of immunoprecipitating Asl, we examined a full-length Plk4 Slimb Binding Mutant (SBM), S293A/T297A, that prevents phospho-dependent Slimb binding and concomitant Plk4 ubiquitination, yielding stable Plk4 (Rogers et al., 2009). Plk4-SBM-GFP was stably expressed and co-immunoprecipitate Asl (Figure 4E). Intriguingly, Plk4 lacking PB1 (Plk4 ΔPB1) expressed to a high level, implicating a possible role for PB1 in Plk4 degradation (Figure 4D). However, Plk4 ΔPB1 failed to co-immunoprecipitate Asl. Strikingly, expression of only PB1-PB2 (residues 382–602) robustly co-immunoprecipitated Asl. In contrast, a construct containing the previously described Plk4 Asl-binding domain, Plk4 PB1+ (residues 382–525), exhibited low expression and failed to co-immunoprecipitate detectable levels of Asl, suggesting that the full PB1-PB2 structural cassette is required for robust Asl binding in vivo.
The PB1-PB2 Cassette is Necessary and Sufficient for Robust Centriole Targeting
In light of our finding that the “cryptic polo box” is composed of two bona fide PB domains, we set out to determine whether individual PBs could mediate centriole localization or if the full PB1-PB2 structure was required for centriole targeting. Localization experiments were conducted in interphase S2 cells containing endogenous Plk4, as Plk4 depletion causes dramatic centriole loss. We transiently expressed a series of inducible Plk4 PB-GFP constructs (Figure 5A). After transgene induction, cells were immunostained for pericentrin-like protein (D-PLP) to mark centrioles, and co-localization was scored as strong, weak, or no centriole localization (Figure 5B-5I). Only two constructs, PB1-PB3 (aa 382–741) and PB1-PB2 (aa 382–602), containing the entire PB1-PB2 cassette defined in our crystal structure, displayed robust centriole localization. PB1-PB3 and PB1-PB2 strongly co-localized with centrioles in 88% and 96% of cells assayed respectively (Figure 5B–D). Examination of constructs lacking the full PB1-PB2 cassette showed significantly reduced centriole co-localization. Expression of the single PB domains that compose the PB1-PB2 cassette, individually displayed no centriole localization in the majority of cells scored (Figure 5B,E,F). We also scored the centriole targeting activity of the previously described Plk4 Asl-binding region (PB1+) (Dzhindzhev et al., 2010). PB1+ displayed a dramatic reduction in centriole co-localization as compared to PB1-PB2 (Figure 5B, G), suggesting that the full PB1-PB2 cassette is necessary to bind Asl and target Plk4 to centrioles. To determine if the PB1-PB2 cassette is necessary for centriole targeting, we designed a full length, SBM construct with the PB1-PB2 cassette deleted (SBM Δ[PB1-PB2]). SBM Δ[PB1-PB2] showed strong centriole co-localization in only 6% of cells and weak co-localization in 49% of cells (Figure 5B,H). [PB1-PB2]-independent centriole association is likely conferred by PB3 as mouse PB3 has weak centriole targeting activity (Leung at al., 2002). In agreement, we found that a Linker-PB3 construct (L-PB3; aa 602–741) showed some level of centriole co-localization in 49% of cells (Figure 5B,I).
Figure 5.
The Plk4 PB1-PB2 Cassette is Required for Robust Centriole Localization.A) Schematic of PB-GFP containing constructs assayed for centriole localization. B) S2 cells were transiently-transfected with the constructs shown in A (green), induced to express for 24 hours, and immunostained with anti D-PLP antibody to mark centrioles (red). Centriole co-localization was classified as strong, weak, or no co-localization. Total number of cells analyzed and independent experiments performed noted at right. C, D) Expression of PB1-PB3 (B) or the PB1-PB2 cassette (D) results in strong centriole co-localization. Cell margins are indicated (white dashed lines). E–I) Single or incomplete PBs primarily display weak or no centriole co-localization. Representative images of differential localization are shown. Boxed regions are magnified in the lower panels. Scale, 5μm.
PB1-PB2 Scaffolds Plk4 Trans-autophosphorylation to Limit Centriole Duplication
Previous work has revealed an auto-regulatory mechanism in Plk1 whereby its PB1-PB2 cassette binds the kinase domain in trans and inhibits kinase activity in vitro. It has also been shown that the mouse Plk4 central region (encompassing PB1-PB2) can bind the Plk4 kinase domain in trans (Leung et al., 2002). To determine whether inhibition of kinase activity is a conserved PB feature, we incubated a fly Plk4 construct containing the kinase domain + DRE (Kin-DRE) with increasing molar ratios of the PB1-PB2 cassette or the PB3 domain and assayed Plk4 autophosphorylation. No change in autophosphorylation was detected upon titration with PB1-PB2 or PB3 in trans (Figure S2).
Endogenous Plk4 levels are tightly regulated through trans-autophosphorylation of the DRE. Trans-autophosphorylation should be promoted by Plk4 oligomerization. Given that our PB1-PB2 structure is homodimeric, we tested whether expression of the PB1-PB2 cassette could inhibit Plk4 degradation by heterodimerizing with full-length Plk4 and preventing trans-autophosphorylation. We first analyzed Plk4 stabilization by assessing centriole number, as increased Plk4 stabilization promotes centriole amplification. Cells transfected with full-length Plk4 or non-degradable Plk4-SBM resulted in centriole amplification, with Plk4-SBM generating a stronger effect (Rogers et al., 2009). Strikingly, cells transfected with PB1-PB2 displayed centriole amplification on par with Plk4-SBM (Figures 6A–B, S3). Other Plk4 constructs including Plk4-ΔPB1, Plk4-Δ[PB1-PB2], and PB1+ did not alter the centriole count from the GFP control, indicating that the entire PB1-PB2 cassette is required to stimulate centriole amplification, presumably by hyper-stabilizing endogenous Plk4. While low PB1-PB2 induction levels caused centriole amplification, this effect varied slightly at higher induction levels, potentially due to PB1-PB2 homodimers saturating centriole binding sites (Figure S4).
Figure 6.
Plk4 PB1-PB2 Promotes Centriole Amplification and Protects Full-length Plk4 in trans. A) S2 cells were transiently-transfected with either inducible GFP, Plk4 PB1-PB2-GFP, or non-degradable Plk4-SBM-GFP (green), induced for 3 days, fixed, and stained for centrioles (PLP, red) and DNA (blue). Arrowheads mark centrioles. Boxed regions are magnified in the insets and highlight centriole clusters not observed in controls. B) Histograms of centriole counts were measured from S2 cells transiently expressing the indicated constructs after 3 days of induction (see Figures S3,4). The percentage of cells with a centriole count per cell <2, 2, and >2 is indicated. Error bars indicate standard deviation. C) Ectopic Plk4 PB1-PB2-GFP expression is sufficient to stabilize full-length Plk4-GFP. Immunoblots of S2 cell lysates showing that overexpression of PB1-PB2-GFP (but not Plk4-Δ[PB1-PB2]-GFP or GFP) stabilizes wild-type full-length Plk4-GFP. Tubulin, loading control. D) PB1-PB2-GFP co-immunoprecipitates Plk4-FL-myc. Immunoblots of anti-GFP immunoprecipitates from S2 cells co-transfeted with Plk4-FL-myc and PB1-PB2-GFP or GFP (control). E) Anti-GFP immunoblot of Plk4-GFP constructs transiently expressed in S2 cells showing differential stability. Tubulin, loading control. F) Plk4-FL-GFP is expressed but rapidly degraded by Slimb-mediated ubiquitination. Cell lysates from S2 cells transfected with Plk4-FL-GFP and treated with control or Slimb dsRNA. Tubulin, loading control. G) Plk4 lacking the PB1-PB2 cassette shows reduced auto-phosphorylation of the DRE as assayed by Slimb binding. Immunoblots of anti-GFP immunoprecipitates from S2 cells transfected with Plk4-FL-GFP or Plk4-Δ[PB1-PB2]-GFP and blotted for GFP and Slimb. H) Model of the Plk4 homodimer. PB1 and PB2 mediate homodimerization. PB1 and PB2 form a composite Asl/Cep152 binding site, recruiting Plk4 to the centriole. PB1-PB2 homodimerization scaffolds Plk4 trans-autophosphorylation, priming the DRE for SCFSlimb binding and ubiquitination.
To directly test if PB1-PB2 promotes centriole amplification through stabilization of Plk4, we co-transfected full-length Plk4-GFP with either PB1-PB2-GFP, Plk4-Δ[PB1-PB2]-GFP or GFP and analyzed Plk4-GFP levels. We found that full length Plk4-GFP levels increased dramatically when co-transfected with PB1-PB2-GFP but not with Plk4-Δ[PB1-PB2]-GFP or GFP alone, demonstrating that PB1-PB2 promotes Plk4 stability in trans (Figure 6C). To test if PB1-PB2-GFP could bind Plk4-FL-myc in trans, we co-transfected these constructs, immunoprecipitated PB1-PB2-GFP and probed for Plk4-FL-myc. We found that PB1-PB2-GFP was able to immunoprecipitate Plk4-FL-myc, indicative of an interaction mediated by PB1-PB2 (Figure 6D). Both PB1-PB2 and PB3 independently confer dimerization. We asked whether Plk4 homodimerization, as mediated by PB1-PB2, is required for efficient trans-autophosphorylation and degradation or whether PB3-mediated dimerization suffices. To test this, we expressed a construct containing PB3 but lacking PB1-PB2 (Plk4-Δ[PB1-PB2]-GFP) and compared its levels to wild-type Plk4-GFP and a stable, full-length Plk4 kinase-dead mutant (Figure 6E) (Brownlee et al., 2011). Wild-type Plk4-GFP protein levels were extremely low. In contrast, Plk4-Δ[PB1-PB2] protein levels were dramatically stabilized and on par with kinase-dead Plk4. To ensure that Plk4-GFP was expressed in these cells and could be compared to Plk4-Δ[PB1-PB2], we depleted Slimb via RNAi and immunoblotted for GFP. Slimb depletion yielded detectable Plk4-GFP, confirming Plk4 expression and its Slimb-mediated degradation (Figure 6F). These findings indicate that PB1-PB2 plays a key role in Plk4 degradation beyond homodimerization and may extend to scaffolding the kinase for trans-autophosphorylation (Guderian et al., 2010). To test if PB1-PB2 plays a role in auto-phosphorylation of the DRE, we examined Slimb-binding as a read-out for DRE phosphorylation. Plk4-FL-GFP ran as a broad band on SDS PAGE and was able to co-immunoprecipitate Slimb, however Plk4-Δ[PB1-PB2]-GFP ran as a tight doublet and co-immunoprecipitated comparatively reduced levels of Slimb, indicative that the PB1-PB2 cassette enhances auto-phosphorylation, priming the DRE for Slimb binding (Figure 6G).
DISCUSSION
The Plk4 CPB is Two Architecturally Distinct Polo Box Domains
Plk4’s essential role in centriole duplication is well established, but a fundamental understanding of its mechanism has been lacking. Here, we have determined the structure of the Plk4 CPB and found that it is actually composed of two PB domains, PB1-PB2. This finding recalibrates the number of PB domains in Plk4 from one to three. Both the Plk4 PB1 and PB2 domains have unique structural features that distinguish them from Plk1 PB1, PB2 and Plk4 PB3.
Plk4 PB1-PB2 has a Novel Inter-domain Arrangement and Dimerization Interface
The spatial arrangement of Plk4 PB1 and PB2 differs from the arrangement observed in Plk1 PB1-PB2 structures. Plk4 PB1 and PB2 pack end-to-end, linked by a short, ordered threonine-proline segment, flanked by hydrophobic residues that anchor PB1 on PB2. The PBs are rotated relative to their β-sheet planes and translated, placing the 1α1 helix axis in line with the PB2 2β2 strand. In contrast, Plk1’s tandem PBs are packed around a pseudo 2-fold axis, forming a β-sandwich. Thus, Plk4 and Plk1 each have distinct, non-homologous arrangements of their PB1-PB2 domains. The Plk1 PB1-PB2 structure is monomeric in contrast to the homodimeric Plk4 PB1-PB2 structure. Plk4 PB1-PB2 has extensive pseudo-symmetric dimerization interfaces across PB1-PB1 and PB2-PB2. Our light scattering data confirm a Plk4 PB1-PB2 homodimeric state, in agreement with previous work showing that the CPB confers self-association (Leung et al., 2002). The Plk4 PB3 structure is also homodimeric (Leung et al., 2002), indicating that full-length Plk4 has at least three dimerization interfaces mediated by its three PB domains. Overall, the three Plk4 PBs are unique among themselves and distinct from Plk1 PB1 and PB2 in their structure, multi-domain arrangement, and oligomeric state.
The PB1-PB2 Cassette Mediates Centriole Targeting and Asterless Binding
Plk4 PB1-PB2 localizes robustly to centrioles in S2 cells while the individual PB1 and PB2 domains display only weak centriole co-localization, indicating that the full PB1-PB2 cassette collectively confers strong centriole targeting. Previous work examining Plk4’s interaction with Asl in vitro mapped the Plk4 determinants to a segment embodying PB1 and the first three β-strands of PB2 (PB1+) (Dzhindzhev et al., 2010). In the same study, the Plk4 PB1 region failed to bind Asl in vitro. This maps key Plk4 Asl-binding determinants to the conserved inter-domain, concave junction defined by PB1 1β1 and PB2 2β1-2β3 (Figure 4B,C). We found that while PB1+ enhances centriole localization over PB1 alone, it is not as effective as the full PB1-PB2 cassette. In support we found that PB1-PB2 co-immunoprecipitates Asl while PB1+ does not. Collectively, our work and previous work can be interpreted in light of our PB1-PB2 structure. While PB1+ contains prime Asl-binding determinants, the remaining PB2 elements are likely required to complete the domain fold and stabilize the determinants that bind Asl and afford robust centriole targeting.
The PB1-PB2 Inter-domain Groove and Implications for Asterless Binding
The conserved PB1-PB2 inter-domain groove contains the PB1 1β1 strand, which in Plk1 PB1 is primarily responsible for phosphopeptide binding. It is possible that the Plk4 PB1 1β1 strand is used to bind Asl, but Plk1 and Plk4 target binding will have significant differences. First, bacterially expressed Asl binds Plk4 in vitro, indicative that the interaction, in contrast to Plk1, is not phospho-dependent. Second, Plk4 binds to a minimal, 300 residue N-terminal segment of Asl (Dzhindzhev et al., 2010). This is in contrast to the short, phosphorylated motifs Plk1 recognizes and suggests that Plk4 binds a domain in Asl rather than a short motif. In support, Plk4 PB1-PB2 forms a large, concave surface that could accommodate domain binding. This contrasts with Plk1, where PB1 and PB2 clamp around a phosphopeptide target.
The PB1-PB2 Cassette Regulates Plk4 Stability and Concomitant Centriole Number
Expression of the PB1-PB2 cassette caused interesting dominant effects. First, PB1-PB2 amplified centriole levels on par with the non-degradable Plk4-SBM construct. Second, we found that full-length Plk4 was stabilized in trans by the PB1-PB2 construct. Plk4 down-regulates its own protein level by trans-autophosphorylating its DRE to promote Slimb binding (Holland et al., 2010; Guderian et al., 2010). We found that the PB1-PB2 construct heterodimerizes with endogenous Plk4 and protects it from trans-autophosphorylation. Trans-autophosphorylation is not simply mediated by oligomerization because a Plk4 construct lacking the PB1-PB2 cassette but retaining the PB3 homodimerization domain was itself dramatically stabilized. This indicates that PB1-PB2 dimerization positions the kinase domains and DREs optimally for trans-autophosphorylation in order to restrict centriole duplication to once and only once per cell cycle (Figure 6H).
Conclusion
Our results reveal unique structural and functional determinants in Plk4 (Figure 6H). In contrast to Plk1-3, Plk4 is a unique polo kinase member, containing three PBs that mediate centriole localization as well as homodimerization. The PB1-PB2 cassette collectively binds Asl and affords robust centriole localization, optimally positioning the kinase domain for trans-autophosphorylation. While PB1-PB2 affords centriole localization via Asl, we do not rule out the possibility that PB1-PB2 interacts with additional centriole factors. While PB3 does not bind Asl in vitro (Dzhindzhev et al., 2010), it does mediate centriole localization, albeit weakly, implicating a non-Asl PB3-binding factor at the centriole. Further structural studies are needed to illuminate the Plk4/Asl interaction, understand how the PB1-PB2 cassette regulates trans-autophosphorylation, and identify additional Plk4 centriole targets.
EXPERIMENTAL PROCEDURES
Cloning and Protein Purification
D.m. Plk4 PB1-PB2 (residues 382–602) was subcloned into pET28 (Novagen), expressed in BL21 DE3 E. coli and induced with 0.1 mM IPTG for 16 hr at 20°C. Protein was purified using successive Ni2+-NTA (Qiagen) and SP-sepharose (GE Healthcare) chromatography. The His6 tag was proteolytically removed. Protein was exchanged into 50 mM Tris pH 9.5, 300 mM NaCl, 0.1% β-ME, 10% glycerol and concentrated to 7.2 mg/ml. The SeMet-M517A mutant was generated using standard protocols (Leahy et al., 1994).
Structure Determination and Analysis
Native Plk4 was crystallized by the hanging drop method using a mother liquor (1 ml) containing 1.2 M Li2SO4, 100 mM Hepes, pH 7.5 and a drop containing 2 μl 7.2 mg/ml protein stock and 2 μl mother liquor. Native crystals were transferred to Fomblin oil (Sigma) and frozen in liquid nitrogen. SeMet-M517A Plk4 was crystallized using a mother liquor containing 1.4 M Li2SO4, 100 mM Hepes pH 7.5 and a drop containing 2 μl 3.2 mg/ml protein stock and 1 μl mother liquor. Diffraction data were collected at the Advanced Photon Source SER-CAT beamlines 22-ID and 22-BM. HKL2000 (Otwinowski and Minor, 1997), Phenix (Adams et al., 2010) and Coot (Emsley et al., 2010) were used to integrate and scale diffraction data, solve and refine, and build the structure respectively.
Centriole Localization and Count Assays
S2 cell culture was performed as described in Rogers and Rogers (2008). Primary rabbit anti-D-PLP antibody (1:3000) and Cy3 labeled 2° antibody (Jackson ImmunoResearch Laboratories, 1:500) were used to label centrioles for colocalization studies. To count centriole numbers, we co-transfected S2 cells with Plk4 constructs and Nlp-EGFP (a constitutively expressed nuclear protein) (Rogers et al., 2009), and analyzed cells with GFP-positive nuclei. Centrioles were labeled with anti-D-PLP and a 2° antibody (conjugated with Cy2, Rhodamine Red-X, or Cy5 [Jackson ImmunoResearch Laboratories]). At least 200 cells were counted per construct. Statistical analyses of centriole counts were performed using two-tailed two-sample t-tests, assuming equal variances.
Supplementary Material
HIGHLIGHTS.
The Plk4 CPB is a structurally unique, tandem polo box array: PB1-PB2.
Full length Plk4 comprises three polo box domains, a unique polo kinase architecture.
The Plk4 PB1-PB2 cassette is required for Asterless binding and centriole targeting.
The Plk4 PB1-PB2 homodimer potentiates Plk4 degradation.
Acknowledgments
The authors thank S. Rogers and M. Peifer for insightful discussion, A. Tripathy for technical support, and D. Roberts for a custom vector. We are grateful for support from the National Cancer Institute P30 CA23074, the American Cancer Society IRG 74-001-31, the GI SPORE (NCI/NIH P50 CA95060), the Arizona Biomedical Research Commission (ABRC #1210), the March of Dimes Basil O’Connor Award #5-FY10-44 (to G.C.R.), National Institutes of Health Grants 5R03HD064881 and 1R01GM094415 (to K.C.S.), and the National Science Foundation Graduate Research Fellowship under Grant No. DGE-0646083 (to L.K.S.).
Footnotes
ACCESSION NUMBERS
Coordinates have been deposited in the Protein Data Bank (4G7N).
Supplemental Information includes Extended Experimental Procedures and four figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66(Part A):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10:265–275. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- Azimzadeh J, Marshall WF. Building the centriole. Curr Biol. 2010;20:816–825. doi: 10.1016/j.cub.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends Genet. 2011;27:307–315. doi: 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Bornens M. The centrosome in cells and organisms. Science. 2012;335:422–426. doi: 10.1126/science.1209037. [DOI] [PubMed] [Google Scholar]
- Brownlee CW, Klebba JE, Buster DW, Rogers GC. The protein phosphatase 2A regulatory subunit Twins stabilizes Plk4 to induce centriole amplification. J Cell Biol. 2011;195:231–43. doi: 10.1083/jcb.201107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng KY, Lowe ED, Sinclair J, Nigg EA, Johnson LN. The crystal structure of the human polo-like kinase-1 polo box domain and its phospho-peptide complex. EMBO J. 2003;22:5757–5768. doi: 10.1093/emboj/cdg558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cizmecioglu O, Arnold M, Bahtz R, Settele F, Ehret L, Haselmann-Weiss U, Antony C, Hoffmann I. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J Cell Biol. 2010;191:731–739. doi: 10.1083/jcb.201007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Ferreira I, Rodrigues-Martins A, Bento I, Riparbelli M, Zhang W, Laue E, Callaini G, Glover DM, Bettencourt-Dias M. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr Biol. 2009;19:43–49. doi: 10.1016/j.cub.2008.11.037. [DOI] [PubMed] [Google Scholar]
- Dzhindzhev NS, Yu QD, Weiskopf K, Tzolovsky G, Cunha-Ferreira I, Riparbelli M, Rodrigues-Martins A, Bettencourt-Dias M, Callaini G, Glover DM. Asterless is a scaffold for the onset of centriole assembly. Nature. 2010;467:714–718. doi: 10.1038/nature09445. [DOI] [PubMed] [Google Scholar]
- Eckerdt F, Yamamoto TM, Lewellyn AL, Maller JL. Identification of a polo-like kinase 4-dependent pathway for de novo centriole formation. Curr Biol. 2011;21:428–432. doi: 10.1016/j.cub.2011.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia AEH, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-Box Domain. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Cryst. 2010;D66(Part 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guderian G, Westendorf J, Uldschmid A, Nigg EA. Plk4 trans-autophosphorylation regulates centriole number by controlling βTrCP-mediated degradation. J Cell Science. 2010;123:2163–2169. doi: 10.1242/jcs.068502. [DOI] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. Cep152 interacts with Plk4 and is required for centriole duplication. J Cell Biol. 2010;191:721–729. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J Cell Biol. 2010;188:191–198. doi: 10.1083/jcb.200911102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa D, Busso C, Flückiger I, Gönczy P. Phosphorylation of SAS-6 by ZYG-1 is critical for centriole formation in C. elegans embryos. Dev Cell. 2009;17:900–907. doi: 10.1016/j.devcel.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Vakonakis I, Olieric N, Hilbert M, Keller D, Olieric V, Bortfeld M, Erat MC, Flückiger I, Gönczy P, Steinmetz MO. Structural Basis of the 9-Fold symmetry of centrioles. Cell. 2011;144:364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Leahy DJ, Erickson HP, Aukhil I, Joshi P, Hendrickson WA. Crystallization of a fragment of human fibronectin: introduction of methionine by site-directed mutagenesis to allow phasing via selenomethionine. Proteins. 1994;19:48–54. doi: 10.1002/prot.340190107. [DOI] [PubMed] [Google Scholar]
- Leung GC, Hudson JW, Kozarova A, Davidson A, Dennis JW, Sicheri F. The Sak polo-box comprises a structural domain sufficient for mitotic subcellular localization. Nat Struc Biol. 2002;9:719–724. doi: 10.1038/nsb848. [DOI] [PubMed] [Google Scholar]
- Lowery DM, Lim D, Yaffe MB. Structure and function of Polo-like kinases. Oncogene. 2005;24:248–259. doi: 10.1038/sj.onc.1208280. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139(4):663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Stearns T. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology 276: Macromolecular Crystallography Part A. 1997:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L, O’Toole E, Schwager A, Hyman AA, Müller-Reichert T. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444:619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- Puklowski A, Homsi Y, Keller D, May M, Chauhan S, Kossatz U, Grünwald V, Kubicka S, Pich A, Manns, et al. The SCF-FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nat Cell Biol. 2011;13:1004–1009. doi: 10.1038/ncb2282. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. Revisiting the role of the mother centriole in centriole biogenesis. Science. 2007;316:1046–1050. doi: 10.1126/science.1142950. [DOI] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC. Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nature Protocols. 2008;3:606–611. doi: 10.1038/nprot.2008.18. [DOI] [PubMed] [Google Scholar]
- Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL. The SCFSlimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J Cell Biol. 2009;184:225–239. doi: 10.1083/jcb.200808049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario CO, Ko MA, Haffani YZ, Gladdy RA, Paderova J, Pollett A, Squire JA, Dennis JW, Swallow CJ. Plk4 is required for cytokinesis and maintenance of chromosomal stability. Proc Natl Acad Sci. 2010;107:6888–6893. doi: 10.1073/pnas.0910941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillibourne JE, Bornens M. Polo-like kinase 4: the odd one out of the family. Cell Div. 2010;5(25):1–9. doi: 10.1186/1747-1028-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MH, Miliaras NB, Peel N, O’Connell KF. Centrioles: some self-assembly required. Curr Opin Cell Biol. 2008;20:688–693. doi: 10.1016/j.ceb.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swallow CJ, Ko MA, Siddiqui NU, Hudson JW, Dennis JW. Sak/Plk4 and mitotic fidelity. Oncogene. 2005;24:306–312. doi: 10.1038/sj.onc.1208275. [DOI] [PubMed] [Google Scholar]
- Tsou MFB, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- Tsou MFB, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell. 2009;17:344–354. doi: 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breugel M, Hirono M, Andreeva A, Yanagisawa HA, Yamaguchi S, Nakazawa Y, Morgner N, Petrovich M, Ebong IO, Robinson CV, et al. Structures of SAS-6 suggest its organization in centrioles. Science. 2011;331:1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- Varmark H, Llamazares S, Rebollo E, Lange B, Reina J, Schwarz H, Gonzalez C. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr Biol. 2007;17:1735–1745. doi: 10.1016/j.cub.2007.09.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.