Abstract
NUP98-HOXD13 (NHD13) and CALM-AF10 (CA10) are oncogenic fusion proteins produced by recurrent chromosomal translocations in patients with acute myeloid leukemia (AML). Transgenic mice that express these fusions develop AML with a long latency and incomplete penetrance, suggesting that collaborating genetic events are required for leukemic transformation. We employed genetic techniques to identify both pre-leukemic abnormalities in healthy transgenic mice as well as collaborating events leading to leukemic transformation. Candidate gene resequencing revealed that 6 out of 27 (22%) CA10 AMLs spontaneously acquired a Ras pathway mutation and 8 out of 27 (30%) acquired a Flt3 mutation. Two CA10 AMLs acquired a Flt3 internal-tandem duplication, demonstrating that these mutations can be acquired in murine as well as human AML. Gene expression profiles revealed a marked upregulation of Hox genes, particularly Hoxa5, Hoxa9, and Hoxa10 in both NHD13 and CA10 mice. Furthermore, mir196b, which is embedded within the Hoxa locus, was overexpressed in both CA10 and NHD13 samples. In contrast, the Hox cofactors Meis1 and Pbx3 were differentially expressed; Meis1 was increased in CA10 AMLs but not NHD13 AMLs, whereas Pbx3 was consistently increased in NHD13 but not CA10 AMLs. Silencing of Pbx3 in NHD13 cells led to decreased proliferation, increased apoptosis, and decreased colony formation in vitro, suggesting a previously unexpected role for Pbx3 in leukemic transformation.
Keywords: CALM, AF10, NUP98, HOXD13, Hoxa9, Meis1, Pbx3, acute myeloid leukemia (AML)
Introduction
Recurrent, balanced chromosomal translocations have been identified in a wide range of hematologic malignancies [1]. These translocations typically produce aberrant fusion proteins, the study of which has been a rich source of insight into leukemic transformation [2, 3]. Oncogenic fusion proteins resulting from balanced chromosomal translocations involving MLL, NUP98, or CALM produce aberrant transcription factors; whose expression typically leads to upregulation of Hoxa cluster genes and their co-factors, such as Meis1 [3–5]. Overexpression of Hoxa9 and Meis1 has been identified in approximately half of all patients with acute myeloid leukemia (AML) [6–11], with Hoxa9 overexpression as a marker for poor prognosis AML [6]. Several mouse models of AML have further highlighted the importance of Hoxa9 and Meis1 in leukemic transformation [12–15].
Expression of the oncogenic fusion proteins NUP98-HOXD13 (NHD13) or CALM-AF10 (CA10) in the hematopoietic compartment of mice results in the development of AML, with a long latency and incomplete penetrance [16, 17]. The AMLs that develop in these mouse models have an aggressive nature and features of B cell differentiation, including expression of the B-cell antigen B220 and clonal Igh gene rearrangements [16–18]. The NHD13 and CA10 leukemias share these features, as well as the overexpression of Hoxa9, with AML induced by MLL-fusions [3, 11, 14]. The late onset of leukemia in the NHD13 and CA10 murine models strongly suggests that expression of the oncogenic fusion proteins is required, but not sufficient, to promote leukemogenesis [16, 17]. This observation is consistent with the hypothesis that leukemic transformation requires complementary mutations that affect at least two different conserved pathways in hematopoietic stem/progenitor cells: one impairing maturation or self-renewal and the second promoting aberrant proliferation [19].
In this study we employed candidate gene resequencing and gene expression arrays (GEAs) to compare and contrast the biological pathways that are dysregulated in CA10 and NHD13 mice. We analyzed gene expression profiles (GEPs) from clinically healthy CA10 and NHD13 tissues to establish the direct effect of these transgenes on global gene expression in hematopoietic tissues. Subsequent analysis of GEPs from both clinically healthy and leukemic tissues highlighted important similarities and differences between the NHD13 and CA10 models including a similar deregulation of Hoxa cluster (Hoxa5, Hoxa9, and Hoxa10) genes and striking differences in expression of the Hoxc cluster genes and the TALE homeodomain proteins Meis1 and Pbx3.
Materials and Methods
Leukemia Diagnosis
Generation of CA10 and NHD13 mice and leukemia diagnosis has been described previously [16, 17]. Leukemia diagnoses were made according to the Bethesda proposals [20, 21]. Tissues obtained from CA10 or NHD13 mice in which no gross signs of leukemia were evident were described as clinically healthy.
Targeted Gene Resequencing and PCR analysis of Flt3 Length Mutations
Genomic DNA was amplified with the primer sets listed in Supplemental Table 1. PCR products were gel purified (Qiagen) and sequenced by the Sanger method. To confirm Flt3 length mutations, PCR products were sub-cloned into the pGemT-Easy Vector (Promega) and sequenced as above. Analysis for loss of the WT Flt3 allele in CA10 mice harboring an Flt3 length mutation was done by PCR analysis using primers flanking the region of the length mutations (Supplemental Table 1).
Isolation of Lineage Negative Bone Marrow (LNBM)
LNBM was isolated by negative selection using EasySep Mouse Hematopoietic Stem Cell enrichment kit (Stem Cell Technologies) according to the manufacturer’s protocol. LNBM was cultured in Iscove modified Dulbecco medium (IMDM) (Invitrogen) supplemented with 15% FBS, 100mM L-Glutamine and 10µg/ml Penicillin/Streptomycin (Invitrogen), 50ng/ml recombinant mouse Stem Cell Factor (SCF), 10ng/ml recombinant mouse interleukin 3 (Il3), and 10ng/ml recombinant human interleukin 6 (IL6) (R&D Systems) at 37°C at 5% CO2.
RNA isolation, cDNA synthesis and reverse transcriptase quantitative polymerase chain reaction (RQ-PCR)
RNA was isolated using Trizol (Invitrogen) reagent and the manufacturer’s protocol. 1µg of RNA was used to synthesize cDNA with Superscript III Reverse Transcriptase (Invitrogen) in a 20 uL reaction according to the manufacturer’s protocol. For RQ-PCR analysis, 1 µl of cDNA was amplified using Taqman primer and probe sets (Applied Biosystems) as indicated in figures or Supplemental Table 1. RQ-PCR analysis was performed on a 7500 Fast RT-PCR system (Applied Biosystems) using the default thermal cycling conditions. 18S ribosomal RNA was used as an internal control. ΔCT values were -log2 transformed and normalized to wild type (WT) bone marrow (BM).
GEAs and Gene Set Enrichment (GSEA)
RNA samples were hybridized to Affymetrix Mouse 430 2.0 arrays (Laboratory of Molecular Technology, Frederick, Maryland, USA). Gene expression data was analyzed using BRB Array Tools (see Acknowledgments). To generate tables of differentially expressed genes, genes with a 1.5 fold difference in expression and a parametric p-value less than 0.05 were tabulated. Probe sets with log2 transformed geometric mean intensities below 100 were not included in the analysis. For the analysis of leukemia samples, obtained from infiltrated spleens, genes increased 5.0 fold in normal spleens compared to normal BM were filtered out. For GSEA analysis, tissue comparisons listed in the figures were analyzed using the GSEA software [22, 23] and compared to AML gene signatures were taken from the Broad database. Leading Edge Analysis was performed comparing gene lists generated from CA10 AMLs/WT LNBM comparisons to previously described MLL-associated gene lists [24, 25].
microRNA Expression
10ng of RNA was reverse transcribed using the miRNA reverse-transcription kit and primer set for mir196b (Applied Biosystems) according to the manufacturer’s protocol. 1.3µl of cDNA was used for RT-PCR analysis under the following cycling conditions: 95°C for 10 minutes, 95°C for 15 seconds, and 60°C for 1 minute for 40 cycles. The U6 primer/probe set was used as an internal control. ΔCT values were -log2 transformed and normalized to WT BM.
Lentiviral Infection
pLKO.1-puro constructs containing three different short hairpin RNA (shRNA) inserts against either murine Pbx3 (shPbx3-1, shPbx3-2, shPbx3-3) or a scrambled non-targeting (NT) control (Sigma Aldrich) were co-transfected with pVSV-G and pΔR8.2 into the 293T cell line using calcium phosphate. 48 hours post-transfection, viral supernatants were used to infect 189MIG cells, a green fluorescent protein-tagged derivative of the Il3-dependent 189L2 cells that express the NHD13 protein [26]. 189MIG cells were incubated in 4mls of unconcentrated viral supernatants in the presence of 10ng/ml Il3 and 100µg/ml protamine sulfate for 24 hours at 37°C. Cells were washed and incubated for an additional 24 hours in complete media supplemented with Il3 before the addition of 1µg/ml of puromycin. To generate single cell clones of infected cells, cells were plated in a 96-well plate at 0.7 cells/well and grown in the presence of Il3 and puromycin for 10 days.
Cell Proliferation, Apoptosis and Methylcellulose Assays
Uninfected 189MIG cells or cells infected with shRNA particles were plated at a density of 5.0×104 in a 12-well dish and incubated in complete IMDM media supplemented with Il3. Viable cell number was determined by trypan blue exclusion. For methylcellulose cultures, uninfected or shRNA-infected 189MIG cells were plated at a density of 1×103 in M3434_methylcellulose (Stem Cell Technologies) and incubated at 37°C for 10–14 days. For cell cycle assays, cells were washed, fixed/permeablilized and incubated at 4°C for 30 minutes. Cells were washed and resuspended in PBS supplemented with 180 µg/ml propidium iodide and 15 µg/ml RNase for 30 minutes at room temperature and analyzed by flow cytometry for DNA content.
Results
Candidate gene resequencing identifies mutations in CA10 AMLs
We sequenced portions of Nras, Kras, Ptpn11, Flt3, Kit, Idh1, Idh2, and Wt1 from 27 CA10 leukemic spleens (Table 1 and Supplemental Table 2). No mutations were identified in Kit, Idh1, Idh2 or Wt1 (Supplemental Table 2), however, 3 of the 27 (11%) had acquired an Nras mutation and 2 of 27 (7%) acquired a Kras mutation in codons 12, 13, or 61. Activating mutations of Ptpn11, a downstream effector of Ras signaling, have been identified in patients with juvenile myelomonocytic leukemia (JMML), myelodysplastic syndrome (MDS), and AML [27]. We sequenced exons 3 and 13 of Ptpn11, corresponding to the regions most frequently mutated in leukemic patients, and identified one activating mutation in exon 3 of Ptpn11. In sum, 6 of 27 (22%) CA10 leukemic samples had acquired activating mutations of the Ras pathway; this frequency was comparable to that previously reported for NHD13 leukemias (32%) [28].
Table 1.
Targeted gene resequencing of CALM-AF10 leukemic mice
| Mouse | IgH rgmt | Nras | Kras | Ptpn11 | Flt3 |
|---|---|---|---|---|---|
| 2953 | yes | GL | codon 12 GGT>GCT (G>A) | GL | codon 822 GTC>GCC (V>A) |
| 7026 | no | GL | GL | GL | GL |
| 7081 | no | GL | GL | GL | codon 841 GAC>GGC (D>G) |
| 7088 | no | codon 12 GGT>GAT (G>D) | GL | GL | codon 841 GAC>GGC (D>G) |
| 7092 | yes | GL | GL | codon 76 GAA>AAA (E>K) | GL |
| 7095 | yes | GL | GL | GL | GL |
| 7160 | yes | GL | GL | GL | codon 841 GAC>GGC (D>G) |
| 7196 | ND | GL | GL | GL | 12 bp duplication |
| 7214 | yes | GL | GL | GL | GL |
| 7223 | no | GL | GL | GL | GL |
| 7249 | ND | GL | GL | GL | GL |
| 7280 | ND | GL | GL | GL | GL |
| 7339 | ND | GL | GL | GL | GL |
| 7408 | ND | GL | GL | GL | GL |
| 7412 | ND | GL | GL | GL | GL |
| 7415 | ND | GL | GL | GL | GL |
| 7438 | ND | GL | GL | GL | GL |
| 7468 | yes | GL | GL | GL | 39 bp duplication |
| 7520 | ND | GL | GL | GL | GL |
| 7541 | ND | codon 61 CAA>CAT (Q>H) | GL | GL | GL |
| 9001 | yes | GL | GL | GL | GL |
| 9010 | yes | codon 13 GGT>GAT (G>D) | GL | GL | GL |
| 9014 | no | GL | GL | GL | GL |
| 9043 | yes | GL | GL | GL | codon 837 in frame deletion (D) |
| 9068 | yes | GL | GL | GL | codon 841 GAC>GGC (D>G) |
| 9120 | ND | GL | GL | GL | GL |
| 9139 | yes | GL | codon 61 CAA>CGA (Q>R) | GL | GL |
ND- Not done
Flt3 is a receptor tyrosine kinase frequently mutated in AML patients; these mutations most commonly take the form of single nucleotide substitutions within the tyrosine kinase domain (TKD) or internal tandem duplications (ITDs) within the juxtamembrane domain (JM) [29]. We analyzed CA10 leukemias for ITD mutations in exons 14–15 or for TKD mutations in exon 19, and identified acquired mutations in 8 of 27 (30%) CA10 samples (Table 1). Six mice had acquired TKD mutations; four had an identical GAC>GCC transversion at codon 841, resulting in a D>G change, one had a GTC>GCC transition at codon 822, resulting in a V>A change, and one had an in frame deletion of codon 837, which encodes aspartic acid. Although mutations similar to the D>G missense substitution and the D deletion have been reported in AML patients [30, 31], the V>A substitution has not been reported and it is unclear whether this V>A substitution is oncogenic. Of note, this mouse also has an oncogenic Kras mutation (Table 1).
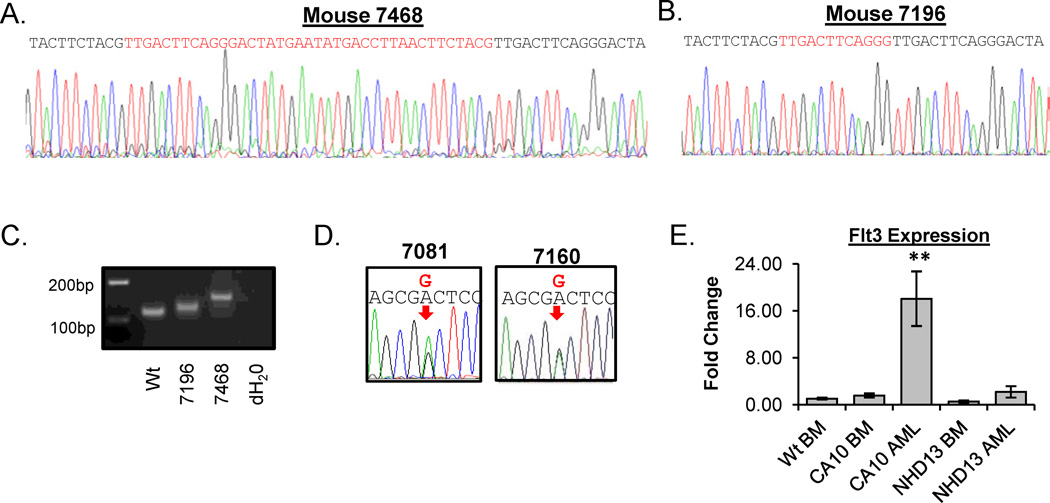
Interestingly, we identified two CA10 leukemias that harbored Flt3-ITD mutations in the JM domain. These samples had 12 or 39 bp in-frame insertion which occurred at the identical nucleotide within exons 14 of Flt3 (nucleotide 1778, codon 593 of NM_010229 coding sequence) (Figure 1). Since some AML patients with Flt3-ITD mutations lose the remaining WT Flt3 allele [29–31] and the sequence electropherograms of the PCR products did not show two overlying tracings, we hypothesized that the wild-type allele had been lost. To test this, we amplified the region containing the Flt3 length mutations. PCR analysis showed only the mutant allele, with no evidence for retention of the WT allele, consistent with the electropherogram tracing (Figure 1C). In contrast to the Flt3-ITD mutations, loss of the WT allele in AML patients with an Flt3-TKD mutation is a rare event, reported in approximately 4% [30, 31]. Consistent with that observation, analysis of sequence tracings from CA10 leukemias with Flt3-TKD mutations did not show loss of the WT allele (Figure 1D).
Figure 1. CALM-AF10 AMLs acquire Flt3 mutations.
(A) and (B) Sequence tracings from CA10 AML samples 7468 and 7196 harboring 39 bp and 12 bp insertions in Flt3 exon 19. Red sequences represent the inserted sequences and the black sequences represent the WT sequences. (C) PCR analysis of genomic DNA from WT and CA10 infiltrated spleens using primers that flanked the ITD. Note the absence of the WT allele in samples 7196 and 7468. (D) Sequence tracings from two CA10 mice with a Flt3-TKD mutation. Note the presence of two peaks (mutant and WT) for the TKD mutations (indicated with a red arrow). (E) RQ-PCR analysis of Flt3 expression in WT BM (n=4), CA10 clinically healthy BM (n=3), CA10 AML (n=5), NHD13 clinically healthy BM (n=4) and NHD13 AML (n=16). 18S ribosomal RNA was used as an internal control and samples were normalized to WT BM.
To determine if Flt3 expression was increased in the CA10 mice with a Flt3 mutation, we performed RQ-PCR assays. Flt3 expression was modestly increased in BM from clinically healthy CA10 mice compared to WT mice, but was significantly up-regulated (18.0 fold) in CA10 AML samples (Figure 1E). In contrast, there was no difference in expression of Flt3 in AML samples or BM from clinically healthy NHD13 mice compared to WT BM (Figure 1E). This is consistent with our prior study which showed that none of the 22 AML sample from NHD13 mice had acquired Flt3 mutations [28], therefore we concluded that overexpression or mutation of Flt3 is not a frequent oncogenic event in the NHD13 mouse model. However, for the CA10 mouse model, our data strongly suggests that mutation of Flt3 is a frequent acquired oncogenic event associated with leukemic transformation in vivo (Table 1 and Figure 1A–E). Fisher’s Exact analysis of Flt3 mutations from CA10 (8/27) and NHD13 (0/22) AMLs showed a statistically significant difference (p=0.0056).
Hoxa9 and Meis1 are differentially expressed in hematopoietic tissue from CA10 mice
To determine the effect of enforced expression of CA10, we compared the gene expression profiles (GEP) of hematopoietic tissues (BM, LNBM, spleen, and thymus) from clinically healthy CA10 mice to WT mice (Supplemental Table 3). We purposely chose hematopoietic tissues from non-leukemic CA10 mice for this study, since we anticipated that any gene expression changes in leukemic CA10 mice would reflect a combination of changes caused by the primary mutation (the CA10 transgene), as well as those caused by additional acquired mutations (such as Nras, Kras, or Flt3). Analysis of GEP revealed a relatively small number of genes whose expression in CA10 BM differed from WT BM by more than 1.5 fold (Supplemental Table 3). Remarkably, in this unbiased survey, the top two upregulated genes were Hoxa9 and Meis1, two genes that we had previously suspected to be increased in CA10 leukemias based on published reports [16, 32]. In addition, we previously showed that T and B lymphocyte maturation are abnormal in CA10 mice [16]; therefore, we compared the GEP’s of CA10 thymus and spleen to that of WT thymus and spleen (Supplemental Table 3). Similar to findings in BM, Hoxa cluster genes (Hoxa9 and Hoxa5) and Meis1 were among the most highly upregulated genes in the CA10 thymus and spleen. Finally, to determine the effect of enforced expression of CA10 on hematopoietic stem/progenitor cells, we compared the GEP of immature cells from CA10 and WT LNBM (Supplemental Table 3). Again, Hoxa9, Hoxa5, and Meis1 were amongst the most significantly increased genes.
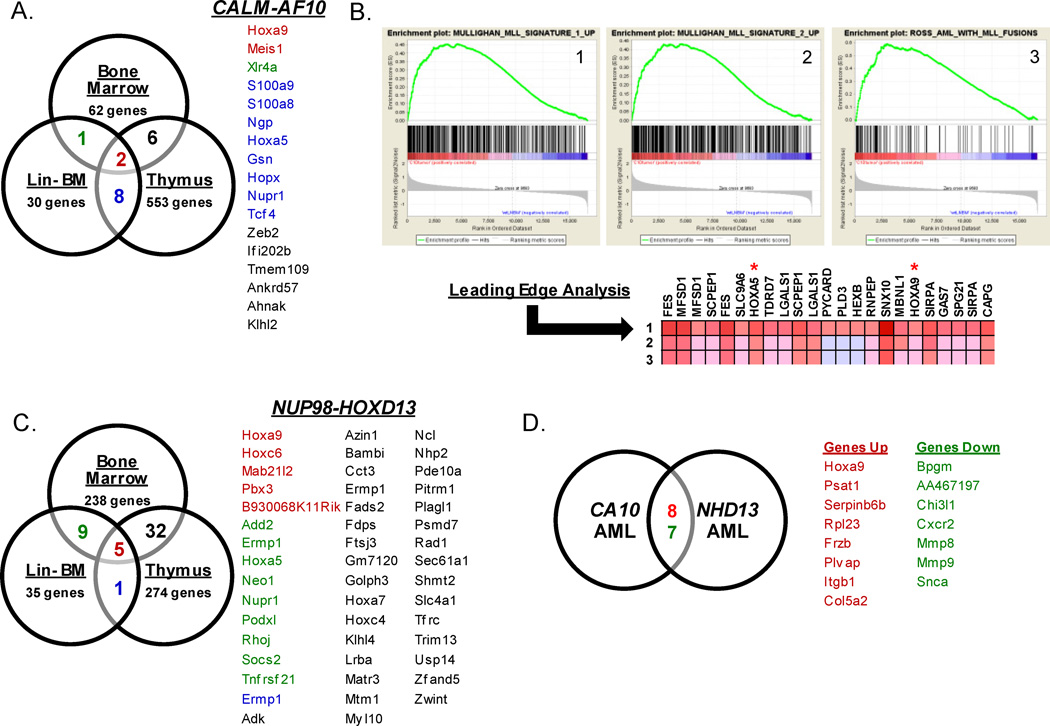
To determine a gene signature generally associated with the expression of CA10, we analyzed the sets of genes that were increased in CA10 BM, thymus, and LNBM (Figure 2A). We found that Hoxa9 and Meis1 were the only genes consistently upregulated in all CA10 hematopoietic tissues (Figure 2A) suggesting that their upregulation is an important downstream genetic event in tissues expressing CA10.
Figure 2. Analysis of gene expression profiles from CA10 and NHD13 mice.
(A) Venn diagram of CA10 BM, thymus and LNBM. List of genes common to tissues are colored according to the tissues where they overlap. Genes labeled in red correspond to genes common in all CA10 tissues analyzed. (B) Gene Set Enrichment Analysis (GSEA) Leading Edge Analysis of CA10 AML versus WT LNBM compared to AML gene signatures [24, 25]. (C) Venn diagram of NHD13 BM, thymus and LNBM. (D) Venn diagram of gene expression of profiles from CA10 and NHD13 AMLs compared to WT LNBM. Genes labeled in red correspond to genes upregulated in both gene sets and genes labeled in green correspond to genes down-regulated in both gene sets.
Murine CA10 AMLs display gene expression patterns similar to human leukemias expressing CA10 or MLL fusions
To identify a gene signature that is important for malignant transformation of hematopoietic cells that expressed a CA10 fusion, we compared the GEP of CA10 leukemia samples to that of WT BM and WT LNBM cells (Supplemental Table 4). The comparison of CA10 AMLs to WT BM again confirmed the up-regulation of Hoxa9 and Meis1 at a 17.7 and 17.2 fold, respectively.
We used Gene Set Enrichment Analysis (GSEA) [22, 23] to determine if the GEPs from CA10-induced AMLs were similar to that of patients with AML. We found that the CA10 AML gene signature shared a high degree of similarity to gene signatures from AML patients harboring MLL fusions [24, 25] (Figure 2B). Leading edge analysis of genes that contribute significantly to the signature revealed that Hoxa5 and Hoxa9 were two of the most prominent and significant genes in the signature. Taken together, we conclude that the CA10 mouse AMLs share disruption of similar biological pathways to those seen in patients with AML caused by MLL fusions.
Although patients with CA10 positive leukemias are most commonly gamma/delta T-cell acute lymphoid leukemia (ALL), these leukemias often display myeloid features, such as CD33 or CD34 expression [33]. Using a publicly available data-set [34], we identified 264 genes that were over-expressed at least 1.5-fold in CA10 positive T-ALL’s compared to CA10 negative T-ALLs and compared that list to the set of genes that was 1.5-fold increased in the murine CA10 AMLs compared to WT LNBM. We found 79 genes that were up-regulated in both the human CA10 positive leukemias and the murine CA10 AML samples (Supplemental Figure 1A), including Hoxa5, Hoxa9, Meis1, and Brd4. Furthermore, comparison of our CA10 associated upregulated genes to the top 100 genes associated with CA10 in patient samples identified in Mulaw et al. [35], identified 27 genes in commonly upregulated including Hoxa9, Meis1, and Flt3 (Supplemental Figure 1B) suggesting that expression of CA10 in transgenic mice dysregulates pathways similar to those disrupted in human CA10 leukemias.
Hox genes and Pbx3 are over-expressed in hematopoietic tissues from NHD13 mice
We previously showed that mice which express a NHD13 transgene also upregulate Hoxa cluster genes [36]. To expand these findings, we compared the GEP of hematopoietic tissues from NHD13 mice to WT mice. Similar to findings with the CA10 mice, NHD13 tissues demonstrated increased expression of Hox genes (Supplemental Table 5). Hoxa9 (9.9 Fold), Hoxa5 (6.5 fold), Hoxc4 (4.8 fold) and Hoxc6 (3.1 fold) were significantly increased in the NHD13 BM compared to WT; similar trends were detected in LNBM and thymus. The TALE homeodomain family member Pbx3 was consistently increased in all NHD13 clinically healthy tissues (4.9–17.4 fold) compared to their WT counterparts (Supplemental Table 5). Of note, Meis1, was not identified as a differentially expressed gene in clinically healthy tissue from NHD13 mice. Similar to the analysis of CA10 mice, we identified 5 genes that were significantly increased in all NHD13 BM, LNBM, and thymus; these genes included Hoxa9, Hoxc6, Mab21l2, Pbx3, and B930068K11Rik (Figure 2C).
To determine which genes and pathways might be important for NHD13-mediated leukemic transformation, we analyzed changes in gene expression in the NHD13 leukemias as above. There were 3073 differentially expressed genes in the NHD13 leukemias compared to WT BM (Supplemental Table 6). Similar to the findings with NHD13 BM from clinically healthy mice, NHD13 AML samples displayed significantly increased levels of several Hox genes (Hoxa5, Hoxa7, Hoxa9, and Hoxc4) and Pbx3.
To identify genes and pathways that are commonly dysregulated in both CA10 and NHD13 leukemias, we identified genes that were differentially expressed in CA10 and NHD13 AML compared to WT BM (Figure 2D). We found 8 genes that were significantly up-regulated at least 4 fold in both the CA10 and NHD13 AMLs: Hoxa9, Psat1, Serpinb6b, Rpl23, Fzrb, Plvap, and Itgb1, and 7 genes down-regulated at least 4 fold in CA10 and NHD13 AMLs: Bpgm, AA467197, Chi3l1, Cxcr2, Mmp8, Mmp9, and Scna.
Verification of Hoxa gene locus overexpression in CA10 and NHD13 tissues
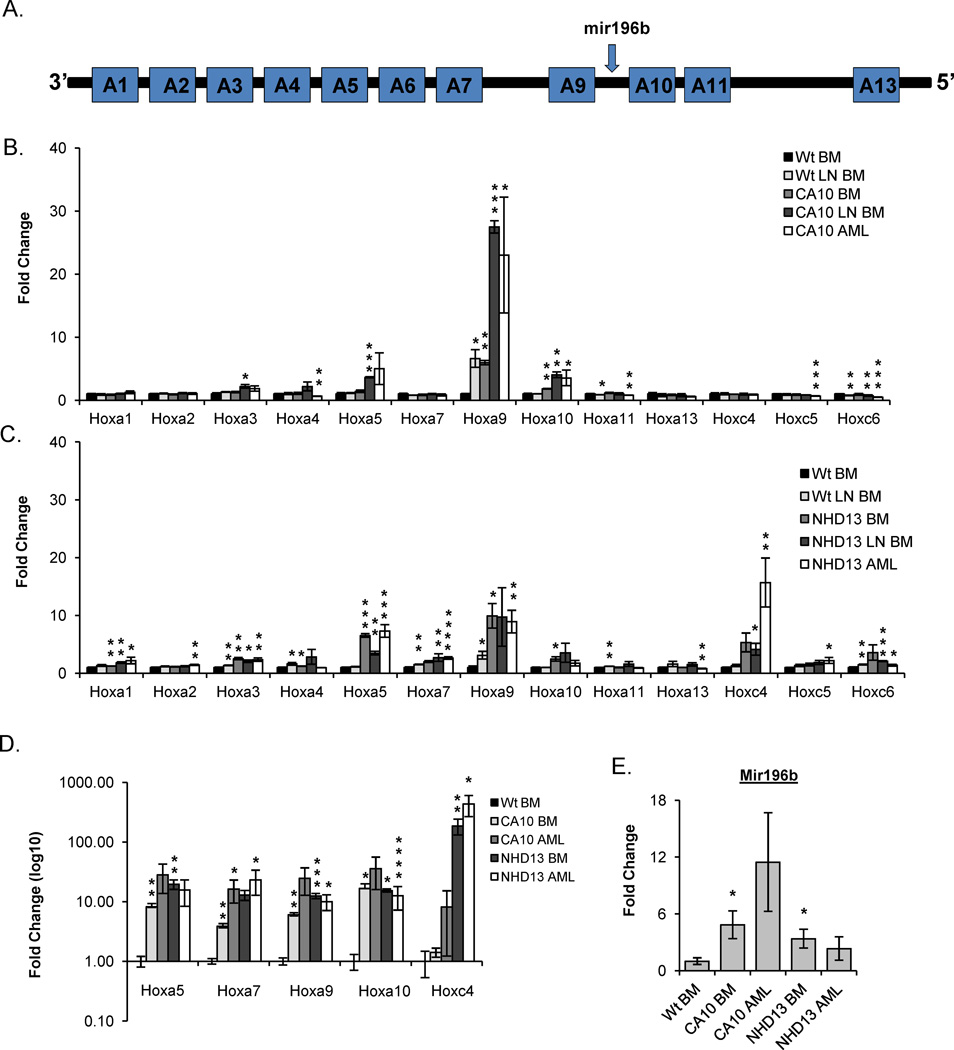
To further evaluate the expression levels of the Hoxa locus (Figure 3A), and portions of the Hoxc cluster, we compared the log2 transformed values from the GEAs for CA10 (Fig 3B) and NHD13 (Fig 3C) to WT BM samples. The Hoxa cluster genes consistently upregulated in both the CA10 and NHD13 leukemias were Hoxa5, Hoxa9, and Hoxa10, with Hoxa9 showing the most dramatic increase compared to WT BM (Figure 3B and 3C). Although expression of the CA10 fusion led to relatively minor (<1.5 fold) changes in the Hoxc cluster genes examined, Hoxc4 was significantly increased in BM (5.3 fold), LNBM (4.1 fold) and AML (15.7 fold) from the NHD13 mice compared to WT BM (Figure 3C). Expression of Hoxa5, Hoxa7, Hoxa9, Hoxa10 and Hoxc4 was confirmed by RQ-PCR analysis for both CA10 and NHD13 tissues (Figure 3D).
Figure 3. CA10 and NHD13 mice up-regulate Hoxa cluster genes.
(A) Schematic of Hoxa locus including the microRNA mir196b. (B) Hoxa and Hoxc cluster gene expression data from expression arrays for WT LNBM (n=3), CA10 BM (n=3), CA10 LNBM (n=3) and CA10 AMLs (n=6) normalized to WT BM (n=3). (C) Hoxa and Hoxc cluster gene expression data from expression arrays for WT LNBM (n=3), NHD13 BM (n=3), NHD13 LNBM (n=3) and NHD13 AMLs (n=6) normalized to WT BM (n=3). (D) RQ-PCR confirmation of the up-regulation of Hoxa5, Hoxa7, Hoxa9, Hoxa10 and Hoxc4 in CA10 BM (n=3), CA10 AML (n=5), NHD13 BM (n=5), and NHD13 AMLs (n=5) normalized to WT BM (n=4). (E) RQ-PCR analysis of mir196b expression in CA10 BM (n=3), CA10 AML (n=6), NHD13 BM (n=3) and NHD13 AML (n=6) normalized to WT BM (n=3). * , **, and *** indicate p<0.05, 0.01, and 0.001 respectively.
mir196b is a microRNA (miRNA) situated within the Hoxa locus between Hoxa9 and Hoxa10 (Fig 3A), and has been shown to be overexpressed in some patients with AML, especially those with MLL or CA10 gene fusions [37–40]. Given its location and the observation that Hoxa9 and Hoxa10 are increased in CA10 and NHD13 leukemias (Figure 3B–C), we suspected that mir196b expression might also be increased in these tissues. RQ-PCR analysis of CA10 BM and AML samples demonstrated an increase in mir196b expression of 4–11 fold, whereas NHD13 BM and AML samples showed a more modest 2–3 fold increase (Figure 3E). Taken together, we conclude that dysregulation of the Hoxa locus is a common feature in both NHD13 and CA10 mice.
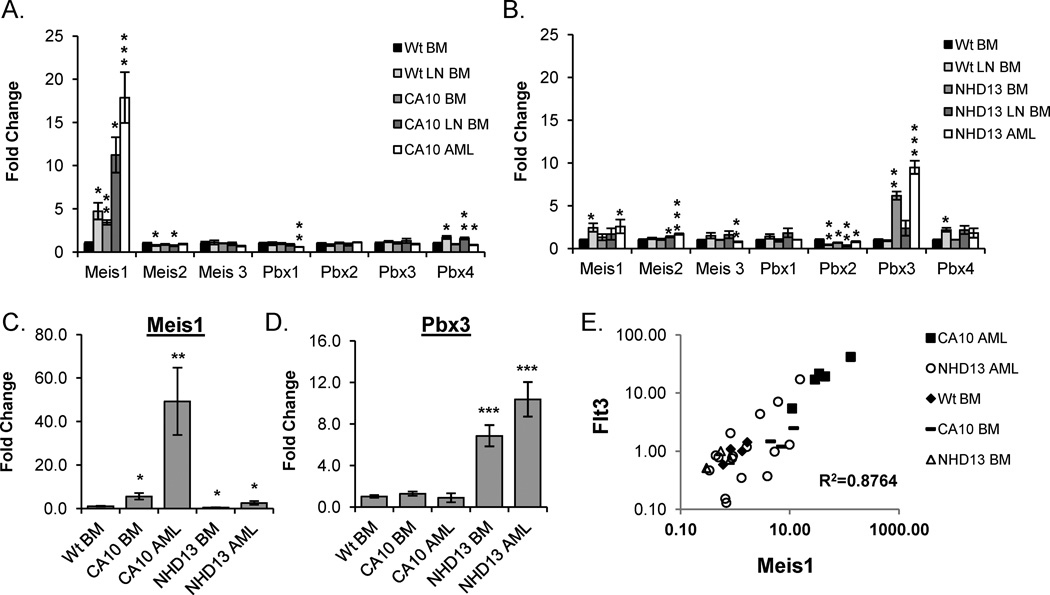
TALE homeodomain family members Meis1 and Pbx3 are differentially expressed in CA10 and NHD13 tissues
TALE homeodomain family members, including Meis1, Pbx1, Pbx2, and Pbx3 are transcriptional co-factors important for Hox function [41]; of these, Meis1 is most commonly overexpressed in human and murine AML samples [8, 11, 13, 16, 42]. Log2 transformed expression values from GEAs normalized to WT BM is shown in Figure 4A and 4B. Meis1 is the only TALE gene markedly (>2 fold) upregulated in the CA10 samples; this upregulation is seen in CA10 BM samples from clinically healthy and leukemic mice, suggesting that Meis1 upregulation may be an early or initiating event in -CA10-mediated leukemic transformation. In contrast, although a subset of NHD13 AML samples had clear upregulation of Meis1 there was not a consistent increase in Meis1 in tissues from NHD13 mice, suggesting that Meis1 overexpression was not required for NHD13-mediated leukemic transformation (Fig 4B and 4C). Interestingly, Pbx3 was consistently upregulated in NHD13 hematopoietic tissues (Figure 4B and 4D), and was the only TALE mRNA overexpressed in all NHD13 samples.
Figure 4. Differential expression of TALE homeodomain family genes in CA10 and NHD13 tissues.
(A) Expression of TALE homeodomain genes from expression arrays for WT LNBM (n=3), CA10 BM (n=3), CA10 LNBM (n=3) and CA10 AML (n=6), normalized to WT BM (n=3). (B) Expression of the TALE homeodomain genes from expression arrays for WT LNBM (n=3), NHD13 BM (n=3), NHD13 LNBM (n=3) and NHD13 AML (n=6), normalized to WT BM (n=3). (C and D) Expression of Meis1 and Pbx3 measured by RQ-PCR in CA10 BM (n=4), CA10 AML (n=7), NHD13 BM (n=5), and NHD13 AML (n=16) normalized to WT BM (n=4). (E) Meis1 and Flt3 expression values for CA10 BM (n=3), CA10 AML (n=5), NHD13 BM (n=4) and NHD13 AML (n=16) were normalized to WT BM (n=4), plotted, and the correlation coefficient calculated. * , **, and *** indicate p<0.05, 0.01, and 0.001 respectively.
Several studies have suggested that Flt3 gene expression is regulated, at least in part, by Hoxa9 and Meis1 [43, 44]. To determine if there was a correlation between Meis1 and Flt3 expression in CA10 and NHD13 transgenic mice, we compared Meis1 and Flt3 gene expression data, as measured by RQ-PCR, from several different sample sets (Figure 4E). There was a strong correlation between Meis1 and Flt3 expression (R2=0.8764) suggesting that Meis1 may have a direct role in the regulation of Flt3 expression. Analysis of a publicly available dataset [34] demonstrated a significant correlation between FLT3 and MEIS1 expression in the CA10 positive leukemias but not in the CA10 negative leukemias, supporting the assertion that this CA10 mouse model shares a similar biology to CA10 leukemias in humans (Supplemental Figure 2).
Knockdown of Pbx3 in an NHD13 cell line inhibits proliferation and promotes apoptosis
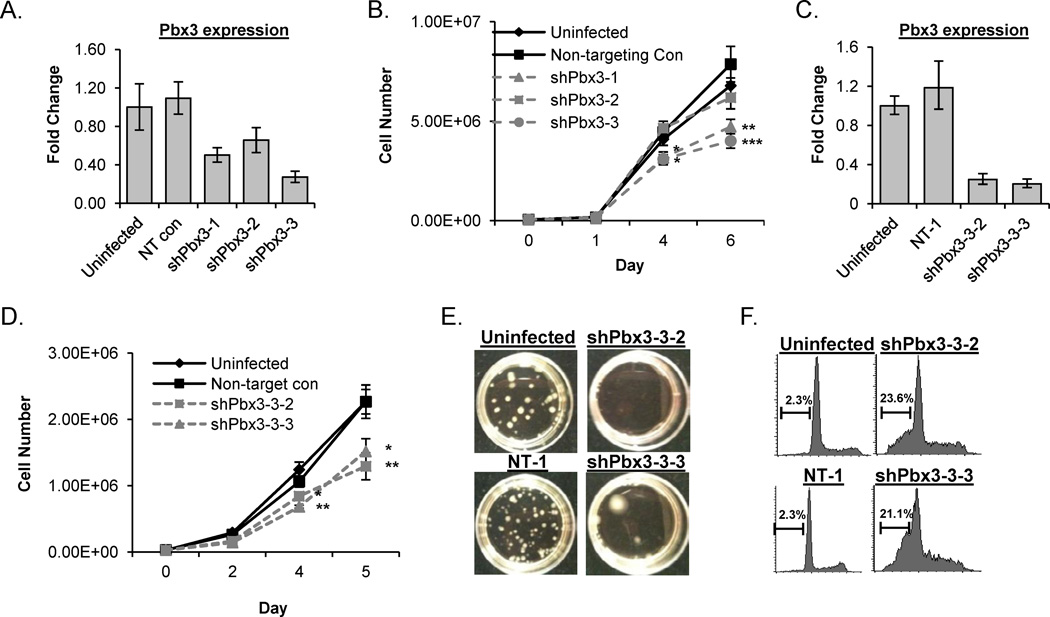
To determine if Pbx3 is important for leukemic transformation in the context of the NHD13 fusion, we utilized 189MIG cells, a non-transformed, Il3-dependent cell line that was generated by in vitro differentiation of murine embryonic stem cells that express an NHD13 fusion [26]. 189MIG cells were infected with lentiviral shRNA particles directed against Pbx3 or a non-targeting (NT) shRNA control and selected with puromycin. Expression of each Pbx3 shRNA inhibited Pbx3 expression to a variable degree (Figure 5A). Cells infected with shPbx3-1 or shPbx3-3 showed a significant decrease in proliferation, whereas cells infected with the shPbx3-2 construct, which had the least degree of Pbx3 knockdown, did not (Figure 5B).
Figure 5. Knockdown of Pbx3 inhibits proliferation and promotes apoptosis in a NHD13 cell line.
(A) RQ-PCR analysis for Pbx3 expression from pools of 189MIG cells infected with shRNA against a non-targeting (NT) control or Pbx3 (shPbx3-1, shPbx3-2, and shPbx3-3) compared to uninfected 189 MIG cells. (B) Proliferation curves of pooled clones from uninfected, NT control, shPbx3-1, shPbx3-2, and shPbx3-3 infected cells. (C) RQ-PCR analysis of Pbx3 expression from single cell clones of 189MIG cells infected with the NT control (NT-1) and two clones infected with shPbx3-3 (designated shPbx3-3-2 and shPbx3-3-3), compared to uninfected 189MIG cells. (D) Proliferation curves from single cell clones from NT-1 and shPbx3-3-2 and shPbx3-3-3 compared to uninfected 189MIG cells. (E) Analysis of uninfected, NT-1, shPbx3-3-2 and shPbx3-3-3 in methylcellulose CFU assay (F) Measurement of DNA content from uninfected 189MIG, NT-1 shRNA control, shPbx3-3-2 and shPbx3-3-3 at day 5 in liquid culture. Apoptotic events are those with less than 2N DNA. * , **, and *** indicate p<0.05, 0.01, and 0.001 respectively.
Since pools of cells expressing a shPbx3 may have variable levels of knockdown within individual clones, we isolated pure single cell clones infected with the shPbx3-3 lentivirus by limiting dilution. RQ-PCR demonstrated that expression of Pbx3 was decreased by over 75% compared to uninfected 189MIG cells and NT-1 controls in two independent clones transduced with shPbx3-3 (shPbx3-3-2 and shPbx3-3-3) (Figure 5C). Figure 5D demonstrates that cells with decreased Pbx3 expression proliferated at a significantly lower rate than uninfected and NT-1 controls. In addition, inhibition of Pbx3 led to a dramatic decrease in the number of colony forming units (CFU) produced by the 189MIG cells in methylcellulose. The shPbx3-3-2 clone gave rise to no colonies and the shPbx3-3-3 gave rise to only 7 colonies, a 23-fold decrease compared to the NT1 control (Figure 5E).
To determine if the decrease in proliferation was due to differentiation of shPbx3-3 infected 189MIG cells, we analyzed the shPbx3-3 clones by flow cytometry for expression of differentiation markers including Mac1, Gr1, and Ter119, but saw no evidence of differentiation (data not shown). Analysis of PI stained cells did not indicate a G1 or G2 arrest, but did show a dramatic (10-fold) increase in sub-G1, apoptotic cells in shPbx3-3 subclones (Figure 5F). Taken together, these results suggest that continued Pbx3 upregulation is important for the survival of cells that express the NHD13 fusion, and therefore may be an important co-regulator of NHD13-mediated survival and transformation.
Discussion
Through a targeted gene resequencing approach we demonstrated that both CA10 and NHD13 mice have acquired Ras pathway mutations (Table 1), at a frequency similar to that seen in human AMLs [45]. Interestingly, we identified Flt3 mutations as a frequent collaborating event in CA10 but not NHD13-mediated leukemogenesis (Table 1) [28]. Although the most common Flt3 mutations in the CA10 mice were point mutations in the TKD, we documented for the first time a spontaneous Flt3-ITD in a mouse model of leukemia (Table 1). These CA10 leukemias harboring Flt3-ITD mutations lost the WT Flt3 allele in the leukemic clones (Figure 1 A–C), similar to human AML [31].
Gene expression profiling of CA10 hematopoietic tissue and leukemias identified Hoxa9 and Meis1 as the two most consistent and significantly overexpressed genes in CA10 transgenic mice, suggesting that an important downstream effect of CA10 in the hematopoietic compartment is to promote dysregulation of Hoxa9 and Meis1. This is consistent with GEPs obtained from leukemic patients harboring a MLL-rearrangement as well as CA10 positive leukemias [34]. Utilizing GSEA, we show that the GEPs from CA10 AMLs were similar to those from human MLL-rearranged leukemias (Figure 2B) [24, 25]. An additional comparison between the murine CA10 leukemias and GEPs from patients with CA10 T-ALL showed a significant overlap in genes that have been up-regulated (Supplemental Figure 1) [34, 35]. This data indicates that the CA10 mouse model shares a similar biology to human leukema and that targeted therapies used to treat CA10 might also be applicable to the treatment of MLL-rearranged leukemias.
MicroRNAs (miRNA) are small non-coding RNA molecules that regulate the stability and translation of mRNAs involved in wide range of biological processes including leukemogenesis [46]. mir196b is a miRNA embedded within the Hoxa locus in both humans and mice, and is overexpressed in a subset of AML patients [47]. Although the upregulation of mir196b was initially noted in MLL-fusion leukemic samples [37, 47], subsequent studies demonstrated that samples from non MLL-fusion leukemias, including CA10 and SET-NUP214, also showed overexpression of mir196b [38], suggesting that up-regulation of mir196b is associated with deregulation of the Hoxa locus by oncogenic fusion proteins. Therefore, in addition to the increase in Hoxa cluster gene expression, both the CA10 and NHD13 leukemias show upregulation of mir196b, underscoring the similarity of these leukemias to each other as well as MLL-fusion leukemias.
An important distinction between the CA10 and NHD13 murine leukemias was differential gene expression of TALE homeodomain family genes Meis1 and Pbx3. Meis1 was identified as a critical, rate limiting factor in MLL-induced leukemic transformation, although loss of both Pbx2 and Pbx3 could partially impair MLL-induced transformation [13]. Our findings suggest that, similar to MLL-fusions, up-regulation of Meis1 as an important event in CA10-mediated transformation. However, Meis1 expression was only modestly increased in the AML samples from NHD13 mice and, in some cases, was decreased relative to WT BM (Supplemental Figure 1). Analysis of GEPs from NHD13 thymus, BM, and LNBM revealed a consistent up-regulation of Pbx3 (Figure 4B–D). In addition, knockdown of Pbx3 expression in an NHD13 cell line resulted in decreased proliferation, increased apoptosis, and a dramatic decrease in the ability to form colonies in methylcellulose (Figure 5) suggesting that loss of Pbx3 is important to the maintenance and survival of cells overexpressing an NHD13 fusion. This data is supported by a recent clinical report demonstrating that the co-expression of Hoxa9 and Pbx3 was an important predictor of poor outcome in a large subset of AML patients [48].
Through the use of several genetic approaches, we have identified genes and pathways that are similar or unique in leukemias triggered by expression of CA10 or NHD13. Both fusion genes produce an AML that is associated with B-lineage features, and both overexpress Hoxa cluster genes similar to AMLs associated with MLL fusions. Although spontaneous Flt3 mutations were not identified in the NHD13-induced AML [28], spontaneous Flt3 mutations were common in the CA10-induced AMLs. We speculate that this may be due to the fact that the CA10 mice overexpress Flt3, whereas NHD13 mice do not, as opposed to the possibility that the NHD13 fusion can not collaborate with Flt3 to transform cells. This hypothesis is supported by the observation that the NHD13 fusion does in fact collaborate with a Flt3 ITD mutation to generate AML in an experimental mouse model [49]. We have identified reciprocal expression of Meis1 and Pbx3 in the CA10 and NHD13 murine models and have also shown a functional role for Pbx3 expression in NHD13-mediated transformation. Taken together, these murine models of AML provide tools to study pathways that cooperate with oncogenic fusions proteins and identify potential targets for AML treatment.
Supplementary Material
Acknowledgements
The authors thank Drs. Yang Jo Chung, Sheryl Gough, Masahiro Onoazawa, Hyunkyung Kim and Zhenhua Zhang for insightful discussions and Maria Jorge for expert technical assistance with the mouse colonies. This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute. Gene expression analyses were performed using BRB-ArrayTools developed by Dr. Richard Simon and BRB-ArrayTools Development Team.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Disclosure:
P.D.A. and C.S. have received royalties from the NIH Division of Technology Transfer for the invention of NUP98-HOXD13 mice.
References
- 1.Rabbitts TH. Chromosomal translocations in human cancer. Nature. 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 2.Rowley JD. The role of chromosome translocations in leukemogenesis. Semin Hematol. 1999;36:59–72. [PubMed] [Google Scholar]
- 3.Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- 4.Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood. 2011;118:6247–6257. doi: 10.1182/blood-2011-07-328880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caudell D, Aplan PD. The role of CALM-AF10 gene fusion in acute leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2008;22:678–685. doi: 10.1038/sj.leu.2405074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 7.Palmqvist L, Pineault N, Wasslavik C, Humphries RK. Candidate genes for expansion and transformation of hematopoietic stem cells by NUP98-HOX fusion genes. PLoS One. 2007;2:e768. doi: 10.1371/journal.pone.0000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawagoe H, Humphries RK, Blair A, Sutherland HJ, Hogge DE. Expression of HOX genes, HOX cofactors, and MLL in phenotypically and functionally defined subpopulations of leukemic and normal human hematopoietic cells. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 1999;13:687–698. doi: 10.1038/sj.leu.2401410. [DOI] [PubMed] [Google Scholar]
- 9.Lawrence HJ, Christensen J, Fong S, et al. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005;106:3988–3994. doi: 10.1182/blood-2005-05-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Afonja O, Smith JE, Jr, Cheng DM, et al. MEIS1 and HOXA7 genes in human acute myeloid leukemia. Leuk Res. 2000;24:849–855. doi: 10.1016/s0145-2126(00)00059-x. [DOI] [PubMed] [Google Scholar]
- 11.Drabkin HA, Parsy C, Ferguson K, et al. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2002;16:186–195. doi: 10.1038/sj.leu.2402354. [DOI] [PubMed] [Google Scholar]
- 12.Iwasaki M, Kuwata T, Yamazaki Y, et al. Identification of cooperative genes for NUP98-HOXA9 in myeloid leukemogenesis using a mouse model. Blood. 2005;105:784–793. doi: 10.1182/blood-2004-04-1508. [DOI] [PubMed] [Google Scholar]
- 13.Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faber J, Krivtsov AV, Stubbs MC, et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113:2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroon E, Thorsteinsdottir U, Mayotte N, Nakamura T, Sauvageau G. NUP98-HOXA9 expression in hemopoietic stem cells induces chronic and acute myeloid leukemias in mice. EMBO J. 2001;20:350–361. doi: 10.1093/emboj/20.3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caudell D, Zhang Z, Chung YJ, Aplan PD. Expression of a CALM-AF10 fusion gene leads to Hoxa cluster overexpression and acute leukemia in transgenic mice. Cancer Res. 2007;67:8022–8031. doi: 10.1158/0008-5472.CAN-06-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin YW, Slape C, Zhang Z, Aplan PD. NUP98-HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood. 2005;106:287–295. doi: 10.1182/blood-2004-12-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung YJ, Choi CW, Slape C, Fry T, Aplan PD. Transplantation of a myelodysplastic syndrome by a long-term repopulating hematopoietic cell. Proc Natl Acad Sci U S A. 2008;105:14088–14093. doi: 10.1073/pnas.0804507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly LM, Gilliland DG. Genetics of myeloid leukemias. Annu Rev Genomics Hum Genet. 2002;3:179–198. doi: 10.1146/annurev.genom.3.032802.115046. [DOI] [PubMed] [Google Scholar]
- 20.Morse HC, 3rd, Anver MR, Fredrickson TN, et al. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood. 2002;100:246–258. doi: 10.1182/blood.v100.1.246. [DOI] [PubMed] [Google Scholar]
- 21.Kogan SC, Ward JM, Anver MR, et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100:238–245. doi: 10.1182/blood.v100.1.238. [DOI] [PubMed] [Google Scholar]
- 22.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 24.Mullighan CG, Kennedy A, Zhou X, et al. Pediatric acute myeloid leukemia with NPM1 mutations is characterized by a gene expression profile with dysregulated HOX gene expression distinct from MLL-rearranged leukemias. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2007;21:2000–2009. doi: 10.1038/sj.leu.2404808. [DOI] [PubMed] [Google Scholar]
- 25.Ross ME, Mahfouz R, Onciu M, et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004;104:3679–3687. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- 26.Slape C, Chung YJ, Soloway PD, Tessarollo L, Aplan PD. Mouse embryonic stem cells that express a NUP98-HOXD13 fusion protein are impaired in their ability to differentiate and can be complemented by BCR-ABL. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2007;21:1239–1248. doi: 10.1038/sj.leu.2404648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matozaki T, Murata Y, Saito Y, Okazawa H, Ohnishi H. Protein tyrosine phosphatase SHP-2: a proto-oncogene product that promotes Ras activation. Cancer Sci. 2009;100:1786–1793. doi: 10.1111/j.1349-7006.2009.01257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slape C, Liu LY, Beachy S, Aplan PD. Leukemic transformation in mice expressing a NUP98-HOXD13 transgene is accompanied by spontaneous mutations in Nras, Kras, and Cbl. Blood. 2008;112:2017–2019. doi: 10.1182/blood-2008-01-135186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Small D. FLT3 mutations: biology and treatment. Hematology Am Soc Hematol Educ Program. 2006:178–184. doi: 10.1182/asheducation-2006.1.178. [DOI] [PubMed] [Google Scholar]
- 30.Bacher U, Haferlach C, Kern W, Haferlach T, Schnittger S. Prognostic relevance of FLT3-TKD mutations in AML: the combination matters--an analysis of 3082 patients. Blood. 2008;111:2527–2537. doi: 10.1182/blood-2007-05-091215. [DOI] [PubMed] [Google Scholar]
- 31.Thiede C, Steudel C, Mohr B, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 32.Dik WA, Brahim W, Braun C, et al. CALM-AF10+ T-ALL expression profiles are characterized by overexpression of HOXA and BMI1 oncogenes. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2005;19:1948–1957. doi: 10.1038/sj.leu.2403891. [DOI] [PubMed] [Google Scholar]
- 33.Asnafi V, Radford-Weiss I, Dastugue N, et al. CALM-AF10 is a common fusion transcript in T-ALL and is specific to the TCRgammadelta lineage. Blood. 2003;102:1000–1006. doi: 10.1182/blood-2002-09-2913. [DOI] [PubMed] [Google Scholar]
- 34.Grossmann V, Bacher U, Kohlmann A, et al. EZH2 mutations and their association with PICALM-MLLT10 positive acute leukaemia. British journal of haematology. 2012;157:387–390. doi: 10.1111/j.1365-2141.2011.08986.x. [DOI] [PubMed] [Google Scholar]
- 35.Mulaw MA, Krause AJ, Deshpande AJ, et al. CALM/AF10-positive leukemias show upregulation of genes involved in chromatin assembly and DNA repair processes and of genes adjacent to the breakpoint at 10p12. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2012;26:1012–1019. doi: 10.1038/leu.2011.307. [DOI] [PubMed] [Google Scholar]
- 36.Choi CW, Chung YJ, Slape C, Aplan PD. A NUP98-HOXD13 fusion gene impairs differentiation of B and T lymphocytes and leads to expansion of thymocytes with partial TCRB gene rearrangement. J Immunol. 2009;183:6227–6235. doi: 10.4049/jimmunol.0901121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Popovic R, Riesbeck LE, Velu CS, et al. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;113:3314–3322. doi: 10.1182/blood-2008-04-154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schotte D, Lange-Turenhout EA, Stumpel DJ, et al. Expression of miR-196b is not exclusively MLL-driven but is especially linked to activation of HOXA genes in pediatric acute lymphoblastic leukemia. Haematologica. 2010;95:1675–1682. doi: 10.3324/haematol.2010.023481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Li Z, He C, et al. MicroRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol Dis. 2010;44:191–197. doi: 10.1016/j.bcmd.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cammarata G, Augugliaro L, Salemi D, et al. Differential expression of specific microRNA and their targets in acute myeloid leukemia. Am J Hematol. 2010;85:331–339. doi: 10.1002/ajh.21667. [DOI] [PubMed] [Google Scholar]
- 41.Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 42.Roche J, Zeng C, Baron A, et al. Hox expression in AML identifies a distinct subset of patients with intermediate cytogenetics. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2004;18:1059–1063. doi: 10.1038/sj.leu.2403366. [DOI] [PubMed] [Google Scholar]
- 43.Wang GG, Pasillas MP, Kamps MP. Meis1 programs transcription of FLT3 and cancer stem cell character, using a mechanism that requires interaction with Pbx and a novel function of the Meis1 C-terminus. Blood. 2005;106:254–264. doi: 10.1182/blood-2004-12-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmqvist L, Argiropoulos B, Pineault N, et al. The Flt3 receptor tyrosine kinase collaborates with NUP98-HOX fusions in acute myeloid leukemia. Blood. 2006;108:1030–1036. doi: 10.1182/blood-2005-12-007005. [DOI] [PubMed] [Google Scholar]
- 45.Bowen DT, Frew ME, Hills R, et al. RAS mutation in acute myeloid leukemia is associated with distinct cytogenetic subgroups but does not influence outcome in patients younger than 60 years. Blood. 2005;106:2113–2119. doi: 10.1182/blood-2005-03-0867. [DOI] [PubMed] [Google Scholar]
- 46.Marcucci G, Mrozek K, Radmacher MD, Garzon R, Bloomfield CD. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood. 2011;117:1121–1129. doi: 10.1182/blood-2010-09-191312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schotte D, Chau JC, Sylvester G, et al. Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2009;23:313–322. doi: 10.1038/leu.2008.286. [DOI] [PubMed] [Google Scholar]
- 48.Li Z, Huang H, Li Y, et al. Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood. 2012;119:2314–2324. doi: 10.1182/blood-2011-10-386235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greenblatt S, Li L, Slape C, et al. Knock-in of a FLT3/ITD mutation cooperates with a NUP98-HOXD13 fusion to generate acute myeloid leukemia in a mouse model. Blood. 2012;119:2883–2894. doi: 10.1182/blood-2011-10-382283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.