Abstract
1,3-Butadiene, an important petrochemical, is commonly burned off when excess amounts need to be destroyed. This combustion process produces butadiene soot (BDS), which is composed of a complex mixture of polyaromatic hydrocarbons in particulates ranging in size from <1μm to 1 mm. An organic extract of BDS is both cytotoxic and genotoxic to normal human bronchial epithelial (NHBE) cells. Based on the oxidizing potential of BDS, we hypothesized that an organic extract of this particulate matter would: 1) cause enzyme inactivation due to protein amino acid oxidation; and 2) induce oxidative DNA damage in NHBE cells. Thus, our aims were to determine the effect of butadiene soot ethanol extract (BSEE) on both enzyme activity and expression of proteins involved in the repair of oxidative DNA damage. Catalase was found to be sensitive to BDS as catalase activity was potently diminished in the presence of BSEE. Using Western analysis, both the alpha isoform of human 8-oxoguanine DNA glycosylase (α-hOGG1) and human apurinic/apyrimidinic endonuclease (APE-1) were shown to be significantly overexpressed as compared to untreated controls after exposure of NHBE cells to BSEE. Our results indicate that BSEE is capable of effectively inactivating the antioxidant enzyme catalase, presumably via oxidation of protein amino acids. The presence of oxidized proteins may partially explain the extranuclear fluorescence that is detected when NHBE cells are treated with an organic extract of BDS. Overexpression of both α-hOGG1 and APE-1 proteins following treatment of NHBE cells with BSEE suggests that this mixture causes oxidative DNA damage.
Keywords: APE-1, Butadiene soot, Catalase, OGG1, Oxidative DNA damage, Oxidative stress
Introduction
1,3-Butadiene is a high use industrial chemical present in numerous petrochemical mixtures and in cigarette smoke (Brunnemann et al. 1989). This compound is commonly burned off (i.e., flared) in areas with numerous refineries and chemical plants (e.g., Louisanna, TX) when excess amounts need to be destroyed. The combustion of 1,3-butadiene produces butadiene soot (BDS), which consists of particulates ranging in size from <1 μm to 1 mm; this particulate matter has been shown to contain complex mixtures of both aromatic and polycyclic aromatic hydrocarbons (PAHs), a number of which are known carcinogens (Catallo, 1998). Inhalation of fine particles of BDS has been shown to promote biotransformation, oxidative stress, and inflammation in murine lungs (Rouse et al. 2008).
A number of studies have utilized organic extracts of particles (e.g., ambient air particles, diesel exhaust particles) in order to measure the toxicity of he organic-soluble components on cultured epithelial cells. An organic extract of BDS has been previously shown to be a significant cytotoxic and genotoxic threat to human bronchial epithelial cells (Catallo et al. 2001). However, the mechanisms of action of BDS have yet to be identified.
Previous reports have suggested that BDS is highly reactive and may produce reactive oxygen species (ROS) in cultured normal human bronchial epithelia (NHBE) cells (Catallo et al. 2001). ROS can be produced in cells by either endogenous or exogenous processes and ROS have been shown to be involved in the development of cancer (Dreher and Junod 1996). Modification of DNA by ROS has led to the suggestion that oxidative stress may be an important factor in carcinogenesis. Cerutti (1994) stated that ROS possess three essential properties of carcinogens: 1) ROS elicit permanent structural changes in DNA (e.g., base-pair mutations and deletions); 2) ROS activate both cytoplasmic and nuclear signal transduction pathways; and 3) ROS modulate the activity of stress proteins and stress genes that function to regulate effector genes related to growth, differentiation, and cell death.
ROS cause several types of DNA damage including base modification, DNA-protein cross-links and strand breakage (Halliwell and Aruoma 1991). One of the major oxidatively modified pyrimidine bases in vivo is 8-oxo-2’-deoxyguanosine (Toyokuni et al. 1994). This product, which is also referred to as 7,8-dihydro-8-oxo-2’-deoxyguanosine or 8-oxo-dG, is pro-mutagenic and results in a G:C → T:A transversion mutation following DNA replication (Kuchino et al. 1987; Shibutani et al. 1991). Elevated levels of 8-oxo-dG have been detected in human cancers and persistent oxidative stress is a feature of carcinogenesis (Toyokuni et al. 1995). Fortunately, eukaryotic cells have a sophisticated repair system that both prevents the misincorporation of 8-oxo-2’-deoxyguanosine triphosphate (8-oxo-dGTP) into DNA and removes 8-oxo-dG residues present in DNA.
8-Oxo-dGTP can be formed by the reaction of dGTP in the free nucleotide pool with a hydroxyl radical, singlet oxygen or by direct photodynamic action. This oxidized nucleotide can be incorporated into DNA opposite adenine and cytosine (Maki and Sekiguchi 1992), resulting in both A:T → C:G and G:C → T:A transversions after replication (Cheng et al. 1992). The misincorporation of 8-oxo-dGTP into DNA is prevented by 7,8-dihydro-8-oxoguanine triphosphatase, the protein product of the human mutT homolog (hMTH1) gene; this enzyme hydrolyzes 8-oxo-dGTP to its corresponding monophosphate (Mo et al. 1992). Increased expression of either hMTH1 mRNA or the 7,8-dihydro-8-oxoguanine triphosphatase enzyme has been detected in human renal cell carcinomas (Okamoto et al. 1996), breast tumors (Wani et al. 1998), hepatocellular carcinomas (Zhou H et al. 2005), brain tumors (Iida et al. 2001), lung cancer cell lines (Hibi et al. 1998; Kennedy et al. 1998), and non-small cell lung carcinomas (Kennedy et al. 2003). Thus, there appears to be good evidence that both hMTH1 mRNA and 7,8-dihydro-8-oxoguanine triphosphatase are molecular markers of oxidative stress in carcinogenesis.
8-Oxo-dG residues present in DNA are removed by the action of 8-oxoguanine DNA glycosylase (OGG1) (Aburatani et al. 1997). In humans, the hOGG1 gene is located on chromosome 3 in a region (3p26.2) that is frequently lost in primary lung tumors (Wikman et al. 2000). Two forms of hOGG1 exist; the alpha form (α-hOGG1) is targeted both to the nucleus (90%) and mitochondria (~10%) while the beta form (β-hOGG1) is localized exclusively in the mitochondria (Takao et al. 1998; Nishioka et al. 1999). Although β-hOGG1 may be involved in base excision repair, this enzyme does not exhibit glycosylase activity (Hashiguchi et al. 2004). With respect to the effect of oxidative stress on OGG1 gene expression, OGG1 mRNA is induced in rat lungs after intratracheal administration of diesel exhaust particles (Tsurudome et al. 1999). Induction of hOGG1 mRNA has also been detected in human lung epithelial cells treated with crocidolite asbestos (Kim et al. 2001). Underexpression of both hOGG1 mRNA and hOGG1 protein has been associated with decreased repair of mitochondrial DNA damage in response to oxidative DNA damaging agents (Mambo et al. 2005).
Apurinic/apyrimidinic endonuclease (APE-1) is another enzyme involved in the excision of 8-oxo-dG residues from DNA (Demple et al. 1991). This protein is also a redox-modifying factor that acts to maintain a number of transcription factors in their active reduced states (Evans et al. 2000). Elevated levels of APE-1 gene products have been detected in human cervical, prostate and ovarian cancers (Evans et al. 2000) as well as in melanomas (Yang et al. 2005).
Human mutY homolog (hMYH) is also involved in the repair of oxidative DNA damage. This glycosylase acts to remove an adenine from an 8-oxo-dG:A base pair, thereby preventing a G:C → T:A transversion mutation following DNA replication (Shinmura et al. 2000). Overexpression of hMYH has been shown to increase cell survival in human alveolar epithelial cells (A549) exposed to either 95% oxygen or ionizing radiation (Kremer et al. 2004).
In this study, we examined intracellular fluorescence induced by butadiene soot DMSO extract (BSDE) or butadiene soot ethanol extract (BSEE) in BEAS-2B cells, an immortalized nontumorigenic human brochial epithelial cell line (Reddel et al. 1988), to determine whether BSEE induces extranuclear fluorescence in the same manner as that seen previously with BSDE (Catallo et al. 2001). Therefore, the oxidizing potential of BDS may be reduced when DMSO is used as a vehicle. The ability of BSEE to cause enzyme inactivation was addressed in an attempt to ascertain whether inhibition of antioxidant enzymes could be involved in the development of oxidative stress when BEAS-2B cells are treated with BDS (Rouse et al. 2008). Catalase was chosen for this experiment because diesel exhaust particles and extracts thereof have been shown to inhibit the activity of this enzyme (Mori et al. 1996). The expression of enzymes involved in the prevention and repair of oxidative DNA damage after exposure of NHBE cells to BSEE was assessed to determine the ability of BSEE to cause oxidative DNA damage in these cells.
Materials and Methods
Chemicals
Nonfat dry milk was purchased from Safeway (Pleasanton, CA). Phenylmethylsulfonyl fluoride (PMSF) was obtained from Boehringer Mannheim (Mannheim, Germany). Phosphate-buffered saline (PBS, 1X solution, pH 7.4), sodium chloride (NaCl, 5M solution) and tris(hydroxymethyl)aminomethane (Tris) buffer (1M solution, pH 7.4) were purchased from Digene (Beltsville, MD). Cellgo Tris-HCl buffer (1M solution, pH 8.0) was obtained from Mediatech, Inc. (Herndon, VA). Protease inhibitor cocktail tablets were from Roche Diagnostics (Indianapolis, IN). Sodium dodecyl sulfate (SDS, 20% w/v solution) was from Quality Biological, Inc. (Gaithersburg, MD). Tween 20 was purchased from BioRad (Hercules, CA). 1,3-Butadiene, catalase, nonidet P-40 and sodium orthovanadate were from Sigma-Aldrich, Inc. (St. Louis, MO).
Antibodies for Western Analysis
Primary antibodies for human MutT homologue 1 (hMTH1), human 8-oxoguanine DNA-glycosylase (α-hOGG1), and human mitochondrial-8-oxoguanine DNA-glycosylase (β-hOGG1) were purchased from Novus Biologicals (Littleton, CO). Anti-human MutY homologue (hMYH) was obtained from Alpha Diagnostic International (San Antonio, TX). Anti-human AP endonuclease was purchased from Trevigen® (Gaithersburg, MD). Anti-human actin, anti-human cytochrome c, and secondary antibodies (anti-mouse IgG-HRP and anti-rabbit IgG-HRP) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Preparation of Butadiene Soot Extracts
Butadiene soot was produced by introducing room temperature gas-phase 1,3-butadiene into a 20 cm steel burner at slow feed rates (0.5 – 5 ml/s) and collected on solvent-clean glass fiber filters (Catallo 1998). Organic extracts of BDS were prepared by adding either absolute ethanol or DMSO to BDS in a glass vial and then rapidly shaking the capped vial for ~5 min. Each resulting extract was filtered through a Millex®-SR 0.5 μm filter unit (Millipore, Solvent Resistant, PTFE Membrane) to yield a stock solution (25 mg soot/ml DMSO or ethanol) that was stored at 4° C prior to use. BDS extracted with DMSO is termed butadiene soot DMSO extract, while BDS extracted with EtOH is termed butadiene soot ethanol extract.
Cell Culture
Normal human bronchial epithelial cells from a never smoker (Lot # 1F1811) and bronchial epithelial growth medium (BEGM) were purchased from BioWhittaker, Inc. (Walkersville, MD). The cryopreserved cells (passage 1) were plated on Corning 100 mm plastic tissue culture dishes (Fisher Scientific, Springfield, NJ) pre-coated with FNC coating mix (Athena Environmental Sciences, Inc., Baltimore, MD), expanded to ~90% confluence and subcultured. BEAS-2B cells, immortalized nontumorigenic human bronchial epithelial cells (Reddel 1988), were purchased from the American Type Culture Collection (Mannasas, VA) and cultured in the same manner as NEBE cells.
Measurement of Intracellular Fluorescence
BEAS-2B cells (passage 2) were cultured and expanded to ~50% confluence. BSDE or BSEE was added to BEGM just prior to exposure at a ratio of 1:1,000 to yield a final concentration of 25 ng soot/ml of media, and a final DMSO or EtOH concentration of 0.1 % (v/v). Five ml of BEGM containing BSDE or BSEE was added to each T25 flask of BEAS-2B cells. Controls received 5 ml of BEGM containing DMSO or EtOH (0.1% v/v). The flasks were placed in a humidified incubator at 37°C for up to 72 hr. During the incubation period, the cells were visualized with an Olympus CKX41 inverted phase-contrast fluorescent microscope using an LCAchN20XPhP fluorescence objective lens (Olympus America Inc., Garden City, NJ). The cells were photographed using a DP71 digital camera (Olympus) with DB2-BSW software (Olympus) under either fluorescent light (emitted by a U-RFL-T power supply unit through a BP460–490C excitation filter) or visible light using phase contrast. Fluorescence excitation wavelength range=460–490 nm. Emission wavelength range=510–580 nm. Magnification =x200. Fluorescence spectroscopy measurements of cell extracts. BEAS-2B cells (passage 2) were cultured and expanded to ~50% confluence. Freshly prepared BSDE (or BSEE) was added to BEGM just prior to exposure at a ratio of 1:1,000 to yield a final concentration of 25 ng soot/ml of media and a final DMSO (or EtOH) concentration of 0.1% (v/v). Five milliliters of BEGM containing BSDE or BSEE was added to each flask of BEAS-2B cells. Controls received 5 ml of BEGM containing DMSO or EtOH (0.1% v/v). The flasks were placed in a humidified incubator at 37°C for 24 h. The media was then aspirated off and 3.0 ml of DMSO was added. After swirling for several minutes, the DMSO was transferred into an organic-safe PE syringe barrel equipped with a Millex®-SR 0.45 μm filter unit (Millipore, Solvent Resistant, PTFE Membrane). The samples were filtered and then analyzed by fluorescence spectroscopy using a RF5000U spectrofluorophotometer (Shimadzu, Columbia, MD) set to an excitation wavelength of 365 nm. Sample emission was measured between 400 and 600 nm.
Effect of BSEE on Catalase Activity
The method of Cohen et al. (1996) was utilized to measure the effect of BSEE on catalase activity. Briefly, 1 ml reaction mixtures were prepared containing catalase (10 U), and either BSEE (25 μg soot/μl) or ethanol (0.1 % v/v as a vehicle control) in 10 mM sodium phosphate buffer (pH 7.0). After inverting the tubes several times to mix the components, the tubes were incubated at 4°C for 4 hr. A 1 μl aliquot was then removed from each reaction mixture and added to tubes containing H2O2 (6 mM) in 10 mM sodium phosphate buffer (pH 7.0) at 4° C. Aliquots of these solutions (100 μl) were removed at 2, 5 and 10 min after initiation of the catalytic reaction. These aliquots were added to tubes containing 4 ml of H2SO4 (0.6 M in dH2O) and 1 ml of ferrous sulfate (10 mM in dH2O). After mixing, 0.4 ml of potassium thiocyanate (2.5 M in dH2O) was added to each tube, and this final reaction mixture was inverted several times to mix the components. The absorbance of these solutions was measured at 471 nm, the experimentally determined λmax for ferrithiocyanate, using an ultraviolet/visible spectrophotometer. Semi-logarithmic plots were prepared by plotting the natural logarithm of the absorbance versus reaction time in order to visualize the effect of BSEE on catalase activity. The first order rate constant for each reaction (i.e., ± BSEE) was determined as described (Cohen et al. 1996). Reactions were performed in triplicate to ensure reproducibility of the results.
Exposure of NHBE cells to BSEE for protein preparation
NHBE cells (passage 3) were cultured and expanded to <50% confluence. Freshly prepared BSEE (25 mg soot/ml) was added to BEGM just prior to exposure at a ratio of 1:1,000 to yield a final concentration of 25 ng soot/ml of media and a final ethanol concentration of 0.1% (v/v). Ten milliliters of BEGM containing BSEE was added to each dish of NHBE cells. The dishes were placed in a humidified incubator at 37°C and then removed at various time points after exposure (6, 12, 18, 24, and 48 h) and harvested. Controls were either untreated (BEGM only) or vehicle controls (BEGM plus 0.1% v/v ethanol). Vehicle controls were incubated 24 h prior to harvesting. Untreated controls (fresh BEGM only) were harvested after placing the plates of BSEE treated cells and vehicle controls in the incubator. Cells were harvested by the following steps: (1) aspirating off the media, (2) washing with ice-cold Dulbecco's phosphate-buffered saline (DPBS), (3) aspirating off the supernatant, (4) scraping the cells with a rubber cell scraper in 10 ml of ice-cold DPBS, (5) pelleting the cells in a centrifuge at 4°C for 5 min at 500 g, (6) aspirating off the DPBS, (7) washing the pellet with 10 ml ice-cold DPBS and centrifuging again, and (8) aspirating off the wash solution.
Preparation of Cell Lysates
Harvested cells were solubilized by adding two volumes of ice-cold lysis buffer (1X PBS, 1% Nonidet P-40, 0.1% SDS, 1 mM sodium orthovanadate, 10 mg/ml PMSF) containing a protease inhibitor cocktail to each cell pellet and pipeting up and down until the cells were lysed. The lysates were then spun at 10,000g for 15 min at 4° C. The supernatants were removed and spun again at 10,000g for an additional 15 min at 4° C. Protein concentrations of the resulting supernatants were determined by the Bradford assay using a kit (Bio Rad, Hercules, CA). Protein samples were stored at –80°C prior to Western analysis.
Western Analysis
For immunoblotting, protein samples (50 μg per lane) were separated on 4-20% Tris-Glycine gels (10 well, 1.0 mm, Invitrogen, Carlsbad, CA) for ~2 hr at 125 V in 1X Tris-Glycine SDS running buffer (Invitrogen). Lane one contained 10 μl See Blue Plus 2 Protein Standard (Invitrogen) to facilitate determination of protein molecular weights. Following separation, proteins were transferred to nitrocellulose membranes (0.45 μm pore size, Invitrogen) using an X Cell II™ Blot Module (Invitrogen) at 25 V for ~1.5 hr in 1X Tris-Glycine SDS transfer buffer (Invitrogen) containing 20% methanol. The blots were then cut into strips to facilitate probing several proteins per blot. The membrane strips were placed in 50 ml disposable centrifuge tubes containing 10 ml Blotto (0.15 M NaCl, 10 mM Tris, pH 8.0, 0.05% v/v Tween 20, 0.2% v/v Nonidet P-40, 0.02% w/v SDS, and 5% w/v nonfat dried milk). The tubes were placed horizontally in a 24-tube plastic tube rack on its side. The rack was then placed in a wooden holder (which was constructed at the NCI according to a design by Dr. Angelo Russo) attached to a Dual Action Shaker (Lab Line Instruments, Inc., Melrose Park, IL). The membranes were blocked for 1 hr at room temperature with gentle agitation. Primary antibody was then added to the designated tubes at the following dilutions: anti-actin, 1:1000; anti-APE-1, 1:1000; anti-hMTH1, 1:1000; anti-hMYH, 1:2000; anti-α-hOGG1, 1:1000, anti-β-hOGG1, 1:1000; and anti-cytochrome C, 1:5000. The membrane strips were incubated with the primary antibodies overnight on a Red Rocker (Hoeffer Scientific Instruments, San Francisco, CA) at 4° C at a speed of 10 rev/min. The membranes were then washed three times in 15 ml of 1X TBST (50 mM Tris, 150 mM NaCl, 0.1 % Tween 20, pH 7.4) for 20 min per wash at 4° C. The membranes were blocked again by incubating them at room temperature for 60 min in Blotto on a Dual Action Shaker with gentle agitation. HRP-conjugated secondary antibodies were added to the tubes at a dilution of 1:10,000 followed by incubation at room temperature for 30 min on a Dual Action Shaker with gentle agitation. Blots were washed three times in 15 ml of 1X TBST for 20 min per wash at room temperature. Proteins were detected using the Western Lightning chemiluminescence system (NEN Life Science Products, Inc, Boston, MA). Membranes were exposed to autoradiography film (Biomax ML and MR, Eastman Kodak Company, Rochester, NY) for various times. Signal intensity of protein expression was quantified by scanning densitometry using QuantiScan software (Biosoft, Ferguson, MO). Western blotting was performed in duplicate. In each case, a set of protein samples from an individual exposure of NHBE cells to BSEE was used for Western analysis to confirm the reproducibility of the results.
Statistical Analysis of Protein Expression
Peak areas of individual proteins at specific time points were quantified by scanning densitometry and divided by the corresponding peak areas of cytochrome c to adjust for differences in protein loading prior to electrophoresis. Student's t-test was utilized to determine significant changes in protein expression when comparing protein levels at various times after BSEE exposure to that of time zero controls.
Results
Intracellular fluorescence in HBE cells exposed to BSDE or BSEE
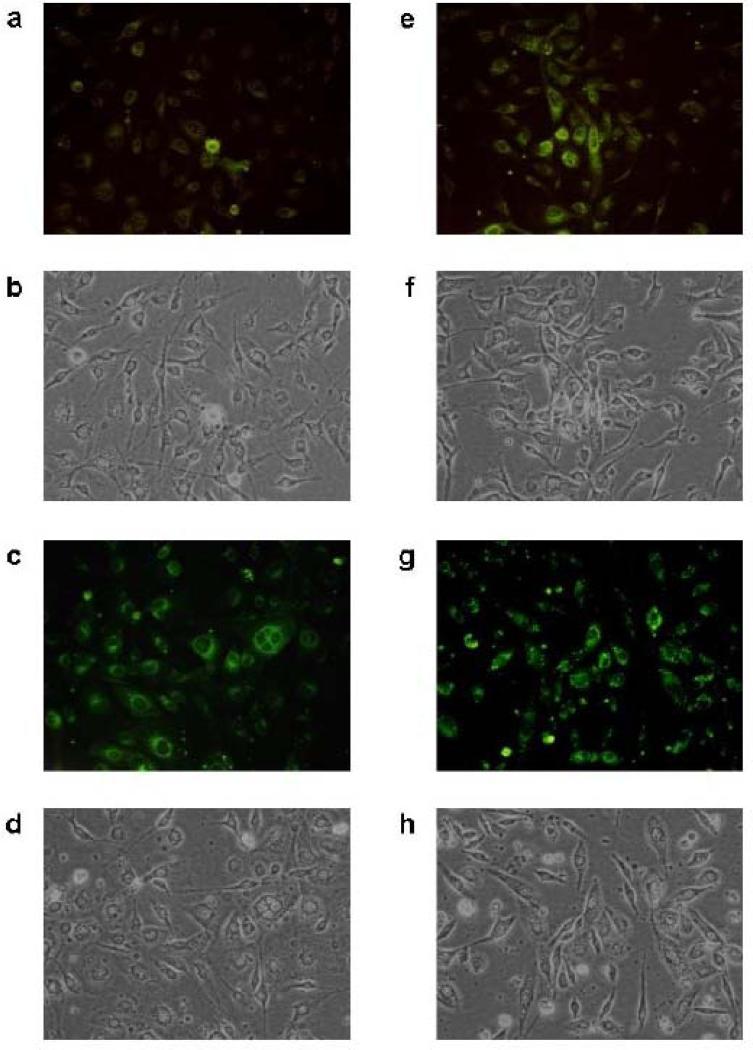
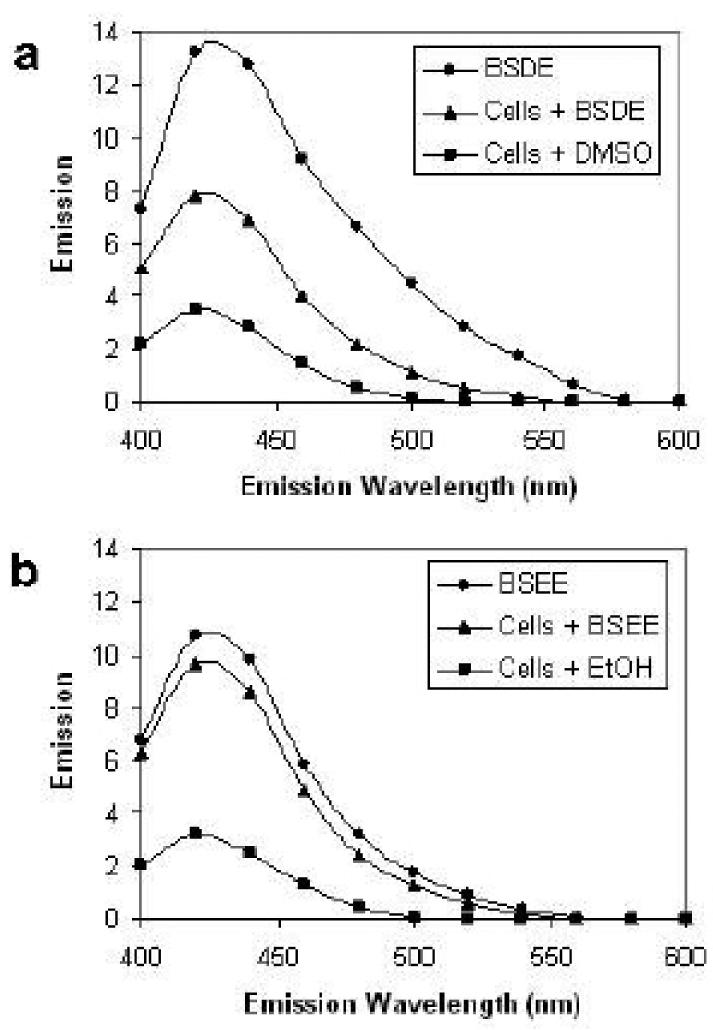
Previously, we reported that extranuclear fluorescence was detected within 4 h after exposure of NHBE cells to BSDE and that the fluorescence intensified up to 24 h (Catallo et al. 2001). In order to establish that BSEE also causes extranuclear fluorescence, BEAS-2B cells exposed to either a vehicle control (DMSO or EtOH) or toxicant mixture (BSDE or BSEE) were visualized 6, 24, 48, and 72 h after exposure. No intracellular fluorescence was detected in either vehicle controls (at all time points) or toxicant-treated cells 6 h after exposure (data not shown). Figure 1 shows photomicrographs of BEAS-2B cells treated with either BSDE or BSEE. Very little extranuclear fluorescence is visible in cells 24 h after exposure to BSDE (Fig. 1a). By comparing this image with a phase-contrast image of the same cells (Fig. 1b), it can be seen that many BEAS-2B cells do not exhibit fluorescence 24 h after exposure to BSDE. However, significant extranuclear fluorescence is observed in these same cells 72 h after exposure (Fig. 1c). By comparing this image with a phase-contrast image of the same cells (Fig. 1d), it can be seen that the majority of BEAS-2B cells do exhibit fluorescence 72 h after exposure to BSDE. However, at this time point, the extranuclear fluorescence is still diffuse in the cytoplasm. In contrast, extranuclear fluorescence is readily visible in BEAS-2B cells 24 h after treatment of cells with BSEE (Fig. 1e). By comparing this image with a phase-contrast image of the same cells (Fig. 1f), it can be seen that a significant fraction of BEAS-2B cells do exhibit extranuclear fluorescence 24 h after exposure to BSEE. Both the number of cells exhibiting fluorescence and the intensity of this fluorescence is dramatically increased in BEAS-2B cells 72 h after exposure to BSEE (Fig. 1g). By comparing this image with a phase-contrast image of the same cells (Fig. 1h), it is observed that the majority of these cells exhibit fluorescence. Further, the extranuclear fluorescence in BSEE-treated BEAS-2B cells 72 after exposure (Fig. 1g) is not as diffuse as that seen in BSDE-treated cells (Fig. 1c) and is, in part, in the form of punctiform bodies which correspond to black circular formations in the phase contrast image (Fig. 1h). Fluorescence of DMSO extracts of HBE cells after treatment with either BSDE or BSEE Fluorescence spectroscopy was performed using DMSO extracts of BEAS-2B cells treated with either BSDE or BSEE in order to better determine the nature of the species responsible for the extranuclear fluorescence shown in Fig. 1. BEAS-2B cells exposed to either a vehicle control (DMSO or EtOH) or toxicant mixture (BSDE or BSEE) were extracted with DMSO 24 h after exposure and analyzed by fluorescence spectroscopy. Figure 2a shows the fluorescence emission spectra of BSDE (at the same concentration as that used to treat cells), BEAS-2B cells treated with BSDE, and BEAS-2B cells treated with DMSO (vehicle control). The fluorescence emission spectrum of BSDE is significantly different than DMSO extracts of BEAS-2B cells treated with either BSDE or DMSO. A DMSO extract of BEAS-2B cells treated with DMSO (vehicle control) exhibits a similar fluorescence emission spectrum to that of BEAS-2B cells treated with BSDE. Figure 2b shows the fluorescence emission spectra of BSEE (at the same concentration as that used to treat cells), BEAS-2B cells treated with BSEE, and BEAS-2B cells treated with EtOH (vehicle control). The fluorescence emission spectrum of BSEE is essentially the same as that of DMSO extracts of BEAS-2B cells treated with either BSEE or EtOH. Finally, these results also show that BSEE causes a greater increase in cellular fluorescence than does BSDE (Fig. 2a,b), which is consistent with the images shown in Fig. 1.
Fig 1.
Extranuclear cellular fluorescence in HBE cells after a single acute exposure to BSDE or BSEE. BEAS2B cells were treated with either BSDE or BSEE and incubated for up to 72 h at 37°C. The cells were photographed at various time points after exposure under either fluorescent light (a, c, e, g) or visible light using phase contrast (b, d, f, h). a, b Cells 24 h after treatment with BSDE; c, d cells 72 h after treatment with BSDE; e, f cells 24 h after treatment with BSEE; and g, h cells 72 h after treatment with BSEE. Excitation wavelength range=460–490 nm. Emission wavelength range=510–580 nm. Magnification = x200
Fig 2.
Fluorescence emission spectra of HBE cell extracts after a single acute exposure to BSDE or BSEE. BEAS-2B cells were treated with BSDE, BSEE, DMSO, or EtOH and incubated for 24 h at 37°C prior to being extracted with DMSO. a Fluorescence emission spectrum of BSDE (diluted in DMSO at the concentration used for cell treatments), cells treated with BSDE and cells treated with DMSO (vehicle control). b Fluorescence emission spectrum of BSEE (diluted in DMSO at the concentration used for cell treatments), cells treated with BSEE and cells treated with EtOH (vehicle control). Fluorescence of the extracts was analyzed by using a spectrofluorophotometer. Excitation wavelength = 365 nm. Emission wavelength range = 400–600 nm.
Effect of BSEE on catalase activity
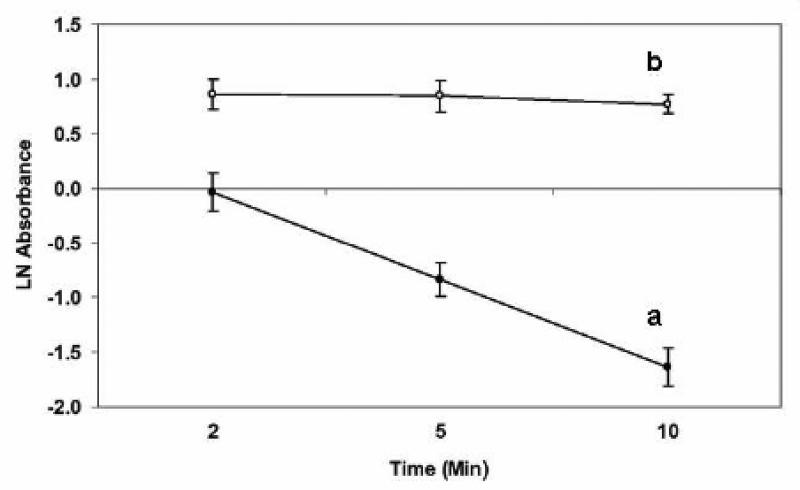
Based on the oxidizing potential of BDS (Catallo et al. 2001), we believed that these particles would oxidize proteins in cells. If this is the case, then BDS would be expected to inhibit the activity of enzymes. It has been shown that both diesel exhaust particles and extracts of these particles inhibit catalase activity in vitro (Mori et al. 1996). Based on the results of this study, we hypothesized that an organic extract of BDS would also inhibit catalase activity. Since butadiene soot caused oxidation of the DMSO used for extraction of BDS in our previous study (Catallo et al. 2001), ethanol was used to extract BDS to study the effects of this complex mixture on catalase activity. A standard catalase assay (Cohen et al. 1996) was used to determine enzymatic activity after treatment of catalase with either ethanol (vehicle control) or BSEE for 4 h at 4°C. Figure 3 shows the absorbance of ferrithiocyanate (A471) as a function of time after the addition of hydrogen peroxide to the reaction mixtures. A linear decrease in absorbance is indicative of high catalase activity after treatment of this enzyme with EtOH (Fig. 3a). Conversely, treatment of catalase with BSEE caused almost complete inhibition of enzymatic activity (Fig. 3b).
Fig 3.
Effect of BSEE on the enzymatic activity of catalase. The activity of this enzyme was measured after a 4-h incubation with either EtOH (vehicle control—line a) or BSEE (line b). Hydrogen peroxide (HP) was added after the incubation period, and aliquots were removed from the reaction mixture at various time points after the addition of HP. The absorbance of ferrithiocyanate was measured at 471 nm at 2, 5, and 10 min after the addition of HP. A decrease in absorbance of the ferrithiocyanate complex is indicative of the disappearance of HP due to its conversion to oxygen and water by catalase.
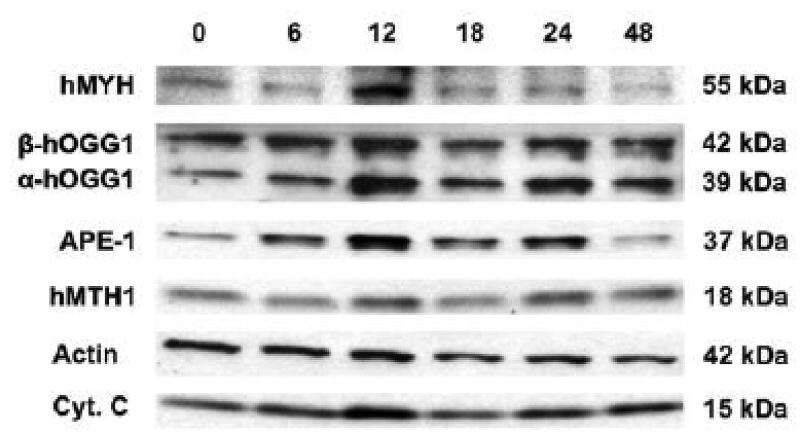
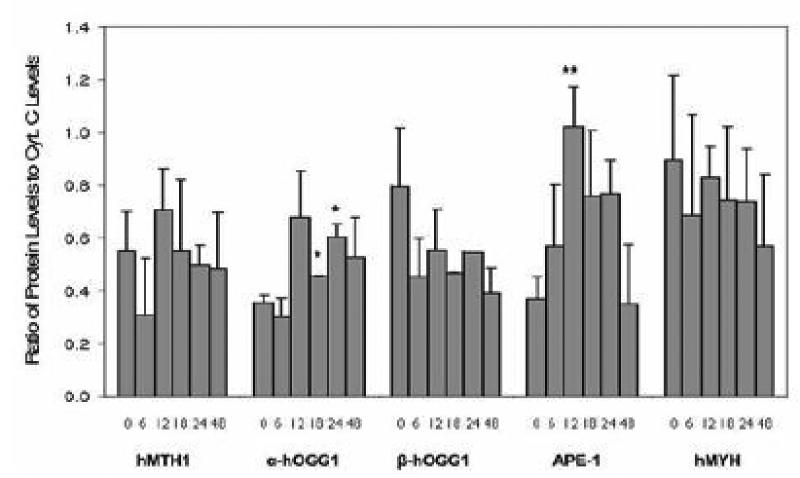
Effect of BSEE on protein expression of oxidative DNA damage repair enzymes Previously, expression of hMTH1 mRNA was shown to exhibit an inverse linear relationship with levels of 8-oxo-deoxyguanosine, a marker of oxidative DNA damage, in NHBE cells, virus immortalized HBE (BEAS-2B) cells, and lung cancer cell lines (Kennedy et al. 1998). This suggested that cells undergoing oxidative stress (i.e., BEAS-2B cells and lung cancer cell lines) overexpress hMTH1, an 8-oxo-dGTPase, in order to protect their DNA from further oxidative damage. These results led us to believe that oxidative DNA damage repair enzymes would be upregulated in response to BSEE in NHBE cells. To test this hypothesis, NHBE cells were treated with BSEE and harvested at various time points after exposure. Protein from these cells was isolated and Western analyses were performed to determine protein levels of the following DNA damage repair enzymes: APE-1, hMTH1, hMYH, α-hOGG1, and β-hOGG1. The results of Western analyses are shown in Fig. 4. Two different loading controls were used (actin and cytochrome c), as actin, the most commonly used loading control for Western blots, was shown to reproducibly exhibit a decrease in protein expression at times greater than 12 h after exposure of NHBE cells to BSEE. This decrease is readily observable in Fig. 4. Levels of all of the proteins examined, with the exception of hMTH1 and β-hOGG1, appeared to increase at various times after exposure of NHBE cells to BSEE. However, when densitometry was performed on the Western blots and protein expression was expressed as the ratio of individual proteins to cytochrome c levels, only the nuclear form of hOGG1 (α-hOGG1) and APE-1 were found to be significantly overexpressed compared to time zero controls (Fig. 5). Although there appears to be a decrease in expression of the mitochondrial form of hOGG1 (β-hOGG1), this decrease was not found to be statistically significant.
Fig 4.
Effect of BSEE on expression of oxidative DNA damage repair enzymes in NHBE cells. NHBE cells were treated with a single dose of BSEE and then harvested at various time points (h) after exposure as indicated in the figure. Protein expression of oxidative DNA damage repair enzymes and loading controls (β-actin, cytochrome c) were measured by Western analysis. The molecular weight of each protein is expressed in kilodaltons.
Fig 5.
Quantification of protein levels of oxidative DNA damage repair enzymes in NHBE cells at various time points after acute exposure to BSEE. Cells were treated with a single dose of BSEE and harvested at various times after exposure. Values shown represent the area of bands detected by Western analysis divided by the area of bands of a loading control (cytochrome c) relative to unexposed controls. Values are the mean ± SD. Student's t-test was utilized to determine significant changes in protein expression when comparing protein levels at various times after BSEE exposure to that of time zero (i.e., unexposed) controls. *p<0.05; **p<0.01.
Discussion
The addition of either BSDE or BSEE to BEAS-2B cells shows that weak intracellular fluorescence is detected 24 h after exposure and intensifies up to 72 h after exposure (Fig. 1). A previous study (Catallo et al. 2001) determined that this fluorescence is detectable as early as 4 h after exposure of NHBE cells to BSDE, continues to increase up to 24 h after exposure, and is still present 72 h after exposure. In the current study, no fluorescence could be detected in BEAS-2B cells exposed to either BSDE or BSEE 6 h after exposure (data not shown). Further, the formation of punctiform fluorescent bodies could not be detected until 72 h after exposure of these cells to BSEE (Fig. 1g) and were barely present in cells exposed to BSDE (Fig. 1c). The disparity between the results of the previous study (Catallo et al. 2001) and the results described herein may be due to the fact that BEAS-2B cells are a pretumorigenic cell line (Reddel et al. 1993). Therefore, the results shown in Fig. 1 could reflect the actions of butadiene soot components on human bronchial epithelial cells in the progression phase of lung carcinogenesis.
A recent study (Penn et al. 2005) has shown that exposure of BEAS-2B cells to BDS causes the induction of fluorescence that can be detected as early as 30 min after exposure. Further, the fluorescence detected following treatment of BEAS-2B cells with BDS has been localized to lipid droplets within the cytoplasm of these cells and concentration of PAHs within these droplets is ascribed as the source of the fluorescence (Murphy et al. 2007). Based on the fact that the fluorescence emission spectrum of a DMSO extract of BSDE-treated cells differed from that of BSDE alone (Fig. 2a), we suggest that the fluorescence was not due merely to components of BSDE but instead resulted, at least in part, from the oxidation of cellular biomolecules. The fluorescence emission spectrum of BSEE is essentially the same as that of DMSO extracts of BEAS-2B cells treated with either BSEE or EtOH (Fig. 2b). These results also show that BSEE causes a greater increase in cellular fluorescence than does BSDE (Fig. 2a, b), which is consistent with the images shown in Fig. 1. Previous work (Catallo et al. 2001) has shown that that BSDE contains strong oxidants (e.g., a component reactive enough to generate methyl radicals from DMSO). Thus, it is likely that BSDE (or BSEE) oxidizes cellular components to generate modified biomolecules that undergo fluorescence when excited with either 365 nm (Fig. 2) or 460–490 nm radiation (Fig. 1).
Previously, diesel exhaust particles and extracts of these particles have been shown to inhibit catalase activity (Mori et al. 1996). Therefore, we hypothesized that an extract of BDS (BSEE) would also inhibit the activity of this protein. To test this hypothesis, the enzymatic activity of catalase was measured in the presence and absence of BSEE. The results of this assay showed that incubation of catalase with BSEE for 4 h (i.e., the time necessary to observe fluorescence in NHBE cells treated with BSDE) caused almost complete loss of catalase activity (Fig. 3). Based on the oxidizing potential of BDS (Catallo et al. 2001), inactivation of catalase by BSEE may be due to oxidative denaturation of this protein. It has been reported that the selective degradation of oxidized proteins in living cells requires proteasomes (Grune et al. 1995). However, it has been noted that mildly oxidized proteins are the best substrates for proteolysis by proteosomes; conversely, highly oxidized proteins are resistant to degradation and undergo aggregation and cross-linking (Grune et al. 1997). Thus, although the mechanism of catalase inactivation by BSEE remains to be elucidated, we believe that this protein is highly oxidized by both BDS and BSEE, resulting in cross-linking and aggregation. Despite this, we cannot preclude the possibility that the absence of catalase activity in the presence of BSEE could be caused by active site occupation or any simple nonoxidative conformational changes in this enzyme.
Microarray analysis performed in a recent study (Murphy et al. 2007) has shown that BDS upregulates genes involved in NF-E2-related factor 2-mediated oxidative stress responses. Due to the inherent oxidizing potential of BDS particles (Catallo et al. 2001) and its apparent ability to induce oxidative stress, we reasoned that exposure of NHBE cells to BSEE would also result in the oxidation of DNA. In order to test this hypothesis, we measured the expression of enzymes involved in the prevention and repair of oxidative DNA damage at various time points after the treatment of NHBE cells with BSEE. Overexpression of these proteins was utilized as an indirect measure of oxidative DNA damage.
The hMTH1 gene encodes 7,8-dihydro-8-oxoguanine triphosphatase, an enzyme that acts to inhibit the formation of 8-oxo-dG residues in DNA by preventing the misincorporation of the oxidized nucleotide 8-oxo-dGTP by DNA polymerase (Mo et al. 1992). With respect to the lung, hMTH1 mRNA is overexpressed in lung cancer cell lines relative to NHBE cells (Hibi et al. 1998; Kennedy et al. 1998). Further, levels of the protein product of hMTH1 are elevated in non-small cell lung carcinomas compared to adjacent histologically normal lung tissue (Kennedy et al. 2003). These results suggest that overexpression of hMTH1 gene products constitutes a molecular marker of oxidative stress in lung cancer. In our study, we reasoned that BSEE would cause oxidation of dGTP to yield 8-oxo-dGTP based on the oxidizing potential of this complex mixture. This, in turn, should cause an upregulation of hMTH1 gene expression.
Although we detected a slight increase in hMTH1 protein levels 12 h after the treatment of NHBE cells with BSEE (Figs. 4 and 5), this change was not statistically significant. Lack of induced hMTH1 gene expression by BSEE may indicate that endogenous levels of 7,8-dihydro-8-oxoguanine triphosphatase are sufficient to hydrolyze 8-oxo-dGTP formed in the free nucleotide pool. However, it is also possible that BSEE may interfere with upregulation of hMTH1 gene expression. Since the mechanism of control of hMTH1 gene expression has not been reported, it is not clear by what means BSEE might interfere with this process. The hOGG1 protein participates in repair of oxidative DNA damage by excising 8-oxo-dG residues present in DNA (Aburatani et al. 1997). The hOGG1 gene encodes two isoforms of this protein: α-hOGG1 is targeted to both the nucleus (<90%) and mitochondria (<10%) while β-hOGG1 is localized exclusively in the mitochondria (Takao et al. 1998; Nishioka et al. 1999). Cancer cell lines which exhibit higher than normal levels of hOGG1 protein have been shown to be proficient in the removal of 8-oxodG from DNA; conversely, cell lines which exhibit lower than normal levels of hOGG1 gene products have been shown to have decreased ability to repair oxidatively damaged DNA with respect to both nuclear DNA and mitochondrial DNA (Mambo et al. 2005). Reduced hOGG1 enzyme activity has been shown to be a risk factor in non-small cell lung cancer based on the results of a functional DNA repair blood test; the ability of hOGG1 to remove 8-oxo-dG from DNA is suggested to be a potential biomarker for lung cancer risk (Paz Elizur et al. 2005). In the present study, the expression of α-hOGG1 was found to significantly increase within 18 h following exposure of NHBE cells to BSEE (Figs. 4 and 5). These results suggest that BSEE causes both nuclear and mitochondrial oxidative DNA damage and that expression of α-hOGG1 is increased in response to this damage. Conversely, no significant change in protein levels of β-hOGG1 was observed in treated NHBE cells (Figs. 4 and 5). Since β-hOGG1 does not exhibit glycosylase activity and its role in base excision repair is unknown (Hashiguchi et al. 2004), upregulation of α-hOGG1 may also reflect its importance in the repair of BSEE-induced mitochondrial oxidative DNA damage. Another putative explanation for the lack of upregulation of β-hOGG1 is that BSEE exposure interferes with the post-transcriptional formation of this protein from the corresponding full length mRNA transcript. However, the molecular mechanisms underlying the induction of both α- hOGG1 and β-hOGG1 are not presently established.
In concert with hOGG1, the enzyme APE-1 is also involved in the excision of 8-oxo-guanine residues from DNA (Demple et al. 1991). The subcellular localization of APE-1 has been shown to be of prognostic significance with respect to nonsmall cell lung carcinomas; cytoplasmic localization, as opposed to nuclear expression, is associated with poor outcome of patients (Puglisi et al. 2001). Expression of the yeast homolog of APE-1 in lung epithelial (A549) cells has been shown to significantly decrease both DNA damage and cytotoxicity caused by exposure of these cells to bleomycin (He et al. 2001). Intratracheal installation of rats with respirable quartz, a toxicant known to induce oxidative stress, has been shown to induce APE-1 expression in rat lungs (Albrecht et al. 2005). In our study, significant overexpression of APE-1 was observed 12 h following the exposure of NHBE cells to BSEE (Figs. 4 and 5). These results provide further evidence that BSEE causes oxidative DNA damage in the nucleus and that expression of APE-1 is increased in response to this damage.
Another enzyme involved in the repair of oxidative DNA damage is hMYH, a glycosylase which acts to remove an adenine from an 8-oxo-dG:A base pair (Shinmura et al. 2000). With respect to the lung, overexpression of the hMYH gene (via retroviral vector transfection) has been shown to increase cell survival in human alveolar epithelial cells (A549) exposed to either 95% oxygen or ionizing radiation, but not H2O2 (Kremer et al. 2004). In our study, treatment of NHBE cells with BSEE did not result in any significant changes in hMYH protein levels (Figs. 4 and 5), suggesting that hMYH gene expression is not induced by BSEE and may be of less consequence than α-hOGG1 and APE-1.
In summary, the results of our study indicate that BSEE causes the induction of intracellular fluorescence, enzyme inactivation (i.e., catalase), and overexpression of certain genes involved in the repair of oxidative DNA damage (i.e., α-hOGG1 and APE-1). The fact that BSEE caused inactivation of catalase, an important antioxidant enzyme, suggests that part of the mechanism of cytotoxicity of this mixture of toxicants may be via inhibition of the cell's antioxidant response. The selective induction of α-hOGG1 and APE-1 (but not hMTH1, β-hOGG1, or hMYH) may be specific to the organic compounds in BDS since cigarette smoke condensate induced hMTH1 gene expression in treated NHBE cells (data not shown). The fact that treatment of NHBE cells with BSEE resulted in overexpression of both α-hOGG1 and APE-1 strongly suggests that nuclear oxidative DNA damage occurs following exposure to this mixture of toxicants. This, in turn, provides evidence for the induction of oxidative stress in NHBE cells exposed to BSEE. However, it should be noted that the results of this study only apply to one strain of NHBE cells from a never smoker. Thus, the effects of BSEE on the induction of enzymes involved in the repair of oxidative DNA damage observed in this study could be specific to the strain of NHBE cells utilized. Further studies need to be performed using multiple NHBE strains to further define the effects of BDS on gene expression and DNA repair pathways.
List of Abbreviations
- APE-1
apurinic/apyrimidinic endonuclease
- BDS
butadiene soot
- BEAS-2B
immortalized nontumorigenic human bronchial epithelial cells
- BEGM
bronchial epithelial growth medium
- BSDE
butadiene soot DMSO extract
- BSEE
butadiene soot ethanol extract
- DMSO
dimethyl sulfoxide
- EtOH
ethanol
- HBE cells
human bronchial epithelial cells
- hMTH1
human mutT homolog
- hMYH
human mutY homolog
- hOGG1
human 8-oxoguanine DNA glycosylase
- NHBE cells
normal human bronchial epithelial cells
- 8-oxo-dG
8-oxo-2’-deoxyguanosine
- 8-oxo-dGTP
8-oxo-2’-deoxyguanosine triphosphate
- PAHS
polycyclic aromatic hydrocarbons
- ROS
reactive oxygen species
Footnotes
This work was performed while CHK was a Senior Research Fellow in the Radiation Biology Branch, National Cancer Institute, Bldg. 10, Rm. B3MB69, Bethesda, MD 20892.
References
- Aburatani H, Hippo Y, Ishida T, Takashima R, Matsuba C, Kodama T, Takao M, Yasui A, Yamamoto K, Asano M. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997;57:2151–2156. [PubMed] [Google Scholar]
- Albrecht C, Knaapen AM, Becker A, Hohr D, Haberzettl P, van Schooten FJ, Borm PJ, Schins RP. The crucial role of particle surface reactivity in respirable quartz-induced reactive oxygen/nitrogen species formation and APE/Ref-1 induction in rat lung. Respir Res. 2005;2:129. doi: 10.1186/1465-9921-6-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunnemann KD, Kagan MR, Cox JE, Hoffmann D. Determination of benzene, toluene and 1,3-butadiene in cigarette smoke by GC-MDS. Exp Pathol. 1989;37:108–113. doi: 10.1016/s0232-1513(89)80026-x. [DOI] [PubMed] [Google Scholar]
- Catallo WJ. Polycyclic aromatic hydrocarbons in combustion residues from 1,3-butadiene. Chemosphere. 1998;37:108–13. [Google Scholar]
- Catallo WJ, Kennedy CH, Henk W, Barker SA, Grace SC, Penn A. Combustion products of 1,3-butadiene are cytotoxic and genotoxic to human bronchial epithelial cells. Environ Health Perspect. 2001;109:965–971. doi: 10.1289/ehp.01109965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti PA. Oxy-radicals and cancer. Lancet. 1994;344:862–863. doi: 10.1016/s0140-6736(94)92832-0. [DOI] [PubMed] [Google Scholar]
- Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G→T and A→C substitutions. J Biol Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- Cohen G, Kim M, Ogwu V. A modified catalase assay suitable for a plate reader and for the analysis of brain cell cultures. J Neurosci Methods. 1996;67:53–56. [PubMed] [Google Scholar]
- Demple B, Herman T, Chen DS. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc Natl Acad Sci USA. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher D, Junod AF. Role of oxygen free radicals in cancer development. Eur J Cancer. 1996;32A:30–38. doi: 10.1016/0959-8049(95)00531-5. [DOI] [PubMed] [Google Scholar]
- Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutat Res. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- Grune T, Reinheckel T, Joshi M, Davies KJA. Protein degradation in cultured liver epithelial cells during oxidative stress. J Biol Chem. 1995;270:2344–51. doi: 10.1074/jbc.270.5.2344. [DOI] [PubMed] [Google Scholar]
- Grune T, Reinheckel T, Davies KJA. Degradation of oxidized proteins in mammalian cells. FASEB J. 1997;11:526–34. [PubMed] [Google Scholar]
- Halliwell B, Aruoma OI. DNA damage by oxygen-derived species. Its mechanism and measurement in mammalian systems. FEBS Lett. 1991;28:9–19. doi: 10.1016/0014-5793(91)80347-6. [DOI] [PubMed] [Google Scholar]
- Hashiguchi K, Stuart JA, de Souza-Pinto NC, Bohr VA. The C-terminal αO helix of human Ogg1 is essential for 8-oxoguanine DNA glycosylase activity: the mitochondrial β-Ogg1 lacks this domain and does not have glycosylase activity. Nucleic Acids Res. 2004;32:5596–608. doi: 10.1093/nar/gkh863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YH, Wu M, Kobune M, Xu Y, Kelley MR, Martin WJ. Expression of yeast apurinic/apyrimidinic endonuclease (APN1) protects lung epithelial cells from bleomycin toxicity. Am J Respir Cell Mol Biol. (2nd) 2001;25:692–698. doi: 10.1165/ajrcmb.25.6.4550. [DOI] [PubMed] [Google Scholar]
- Hibi K, Liu Q, Beaudry GA, Madden SL, Westra WH, Wehage SL, Yang SC, Heitmiller RF, Bertelsen AH, Sidransky D, Jen J. Serial analysis of gene expression in non-small cell lung cancer. Cancer Res. 1998;58:5690–5694. [PubMed] [Google Scholar]
- Iida T, Furuta A, Kawashima M, Nishida J, Nakabeppu Y, Iwaki T. Accumulation of 8-oxo-2’- deoxyguanosine and increased expression of hMTH1 protein in brain tumors. Neurooncology. 2001;3:73–81. doi: 10.1093/neuonc/3.2.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy CH, Cueto R, Belinsky SA, Lechner JF, Pryor WA. Overexpression of hMTH1 mRNA: a molecular marker of oxidative stress in lung cancer cells. FEBS Lett. 1998;429:17–20. doi: 10.1016/s0014-5793(98)00505-5. [DOI] [PubMed] [Google Scholar]
- Kennedy CH, Pass HI, Mitchell JB. Expression of human MutT homologue (hMTH1) protein in primary non-small-cell lung carcinomas and histologically normal surrounding tissue. Free Radic Biol Med. 2003;34:1447–1457. doi: 10.1016/s0891-5849(03)00176-x. [DOI] [PubMed] [Google Scholar]
- Kim HN, Morimoto Y, Tsuda T, Ootsuyama Y, Hirohashi M, Hirano T, Tanaka I, Lim Y, Yun IG, Kasai H. Changes in DNA 8-hydroxyguanine levels, 8-hydroxyguanine repair activity, and hOGG1 and hMTH1 mRNA expression in human lung alveolar epithelial cells induced by crocidolite asbestos. Carcinogenesis. 2001;22:265–269. doi: 10.1093/carcin/22.2.265. [DOI] [PubMed] [Google Scholar]
- Kremer TM, Rinne ML, Xu Y, Chen XM, Kelley MR. Protection of pulmonary epithelial cells from oxidative stress by hMYH adenine glycosylase. Respir Res. 2004;5:16. doi: 10.1186/1465-9921-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y, Mori F, Kasai H, Inoue H, Iwai S, Miura K, Ohtsuka E, Nishimura S. Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature. 1987;327:77–79. doi: 10.1038/327077a0. [DOI] [PubMed] [Google Scholar]
- Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- Mambo E, Chatterjee A, de Souza-Pinto NC, Mayard S, Hogue BA, Hoque MO, Dizdaroglu M, Bohr VA, Sidransky D. Oxidized guanine lesions and hOgg1 activity in lung cancer. Oncogene. 2005;24:4496–4508. doi: 10.1038/sj.onc.1208669. [DOI] [PubMed] [Google Scholar]
- Mo JY, Maki H, Sekiguchi M. Hydrolytic elimination of a mutagenic nucleotide, 8-oxodGTP, by human 18-kilodalton protein: sanitization of nucleotide pool. Proc Natl Acad Sci USA. 1992;89:11021–11025. doi: 10.1073/pnas.89.22.11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y, Murakami S, Sagae T, Hayashi H, Sakata M, Sagai M, et al. Inhibition of catalase activity in vitro by diesel exhaust particles. J Toxicol Environ Health. 1996;47:125–34. doi: 10.1080/009841096161834. [DOI] [PubMed] [Google Scholar]
- Murphy G, Jr, Rouse RL, Polk WW, Henk WG, Barker SA, Boudreaux J, et al. Combustion-derived hydrocarbons localize to lipid droplets in respiratory cells. Am J Respir Cell Mol Biol. 2007;38:532–540. doi: 10.1165/rcmb.2007-0204OC. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Ohtsubo T, Oda H, Fujiwara T, Kang D, Sugimachi K, Nakabeppu Y. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol Biol Cell. 1999;10:1637–1652. doi: 10.1091/mbc.10.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Toyokuni S, Kim WJ, Ogawa O, Kakehi Y, Arao S, Hiai H, Yoshida O. Overexpression of human mutT homologue gene messenger RNA in renal-cell carcinoma: evidence of persistent oxidative stress in cancer. Int J Cancer. 1996;65:437–441. doi: 10.1002/(SICI)1097-0215(19960208)65:4<437::AID-IJC7>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Paz-Elizur T, Krupsky M, Elinger D, Schechtman E, Livneh Z. Repair of the oxidative DNA damage 8-oxoguanine as a biomarker for lung cancer risk. Cancer Biomark. 2005;1:201–205. doi: 10.3233/cbm-2005-12-308. [DOI] [PubMed] [Google Scholar]
- Penn A, Murphy G, Barker S, Henk W, Penn L. Combustion-derived ultrafine particles transport organic toxicants to target respiratory cells. Environ Health Perspect. 2005;113:956–963. doi: 10.1289/ehp.7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi F, Aprile G, Minisini AM, Barbone F, Cataldi P, Tell G, Kelley MR, Damante G, Beltrami CA, Di Loreto C. Prognostic significance of Ape1/ref-1 subcellular localization in non-small cell lung carcinomas. Anticancer Res. 2001;21:4041–4049. [PubMed] [Google Scholar]
- Reddel RR, Ke Y, Gerwin BI, McMenamin MG, Lechner JF, Su RT, et al. Transformation of human bronchial epithelial cells by infection with SV40 or adenovirus-12 SV40 hybrid virus, or transfection via strontium phosphate coprecipitation with a plasmid containing SV40 early region genes. Cancer Res. 1988;48:1904–9. [PubMed] [Google Scholar]
- Reddel RR, Salghetti SE, Willey JC, Ohnuki Y, Ke Y, Gerwin BI, et al. Development of tumorigenicity in simian virus 40-immortalized human bronchial epithelial cell lines. Cancer Res. 1993;53:985–91. [PubMed] [Google Scholar]
- Rouse RL, Murphy G, Boudreux MJ, Paulsen DB, Penn AL. Soot nanoparticles promote biotransformation, oxidative stress, and inflammation in murine lungs. Am J Respir Cell Mol Biol. 2008 doi: 10.1165/rcmb.2008-0057OC. [DOI] [PubMed] [Google Scholar]
- Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- Shinmura K, Yamaguchi S, Saitoh T, Takeuchi-Sasaki M, Kim SR, Nohmi T, Yokota J. Adenine excisional repair function of MYH protein on the adenine:8-hydroxyguanine base pair in double-stranded DNA. Nucleic Acids Res. 2000;28:4912–4918. doi: 10.1093/nar/28.24.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao M, Aburatani H, Kobayashi K, Yasui A. Mitochondrial targeting of human DNA glycosylases for repair of oxidative DNA damage. Nucleic Acids Res. 1998;26:2917–22. doi: 10.1093/nar/26.12.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyokuni S, Mori T, Dizdaroglu M. DNA base modifications in renal chromatin of Wistar rats treated with a renal carcinogen, ferric nitrilotriacetate. Int J Cancer. 1994;57:123–128. doi: 10.1002/ijc.2910570122. [DOI] [PubMed] [Google Scholar]
- Toyokuni S, Okamoto K, Yodoi J, Hiai H. Persistent oxidative stress in cancer. FEBS Lett. 1995;358:1–3. doi: 10.1016/0014-5793(94)01368-b. [DOI] [PubMed] [Google Scholar]
- Tsurudome Y, Hirano T, Yamato H, Tanaka I, Sagai M, Hirano H, Nagata N, Itoh H, Kasai H. Changes in levels of 8-hydroxyguanine in DNA, its repair and OGG1 mRNA in rat lungs after intratracheal administration of diesel exhaust particles. Carcinogenesis. 1999;20:1573–1576. doi: 10.1093/carcin/20.8.1573. [DOI] [PubMed] [Google Scholar]
- Wani G, Milo GE, D'Ambrosio SM. Enhanced expression of the 8-oxo-7,8-dihydrodeoxyguanosine triphosphatase gene in human breast tumor cells. Cancer Lett. 1998;125:123–130. doi: 10.1016/s0304-3835(97)00507-7. [DOI] [PubMed] [Google Scholar]
- Wikman H, Risch A, Klimek F, Schmezer P, Spigelhalder B, Dienemann H, Kayser k, Schulz V, Drings P, Bartsch H. hOGG1 polymorphism and loss of heterozygosity (LOH): significance for lung cancer susceptibility in a caucasian population. Int J Cancer. 2000;15:932–937. doi: 10.1002/1097-0215(20001215)88:6<932::aid-ijc15>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Yang S, Irani K, Heffron SE, Jurnak F, Meyskens FL., Jr Alterations in the expression of the apurinic/apyrimidinic endonuclease-1/redox factor-1 (APE/Ref-1) in human melanoma and identification of the therapeutic potential of resveratrol as an APE/Ref-1 inhibitor. Mol Cancer Ther. 2005;4:1923–1935. doi: 10.1158/1535-7163.MCT-05-0229. [DOI] [PubMed] [Google Scholar]
- Zhou H, Cheng B, Lin J. Expression of DNA repair enzyme hMTH1 mRNA and protein in hepatocellular carcinoma. J Huazhong Univ Sci Technolog Med Sci. 2005;25:389–392. doi: 10.1007/BF02828204. [DOI] [PubMed] [Google Scholar]