Abstract
Gene expression profiling using microarray technologies provides a powerful approach to understand complex biological systems and the pathogenesis of diseases. In the field of liver cancer research, a number of genome-wide profiling studies have been published. These studies have provided gene sets, that is, signature, which could classify tumors and predict clinical outcomes such as survival, recurrence, and metastasis. More recently, the application of genomic profiling has been extended to identify molecular targets, pathways, and the cellular origins of the tumors. Systemic and integrative analyses of multiple data sets and emerging new technologies also accelerate the progress of the cancer genomic studies. Here, we review the genomic signatures identified from the genomic profiling studies of hepatocellular carcinoma (HCC), and categorize and characterize them into prediction, phenotype, function, and molecular target signatures according to their utilities and properties. Our classification of the signatures would be helpful to understand and design studies with extended application of genomic profiles.
Keywords: signature, microarray, integrative analysis, hepatocellular carcinoma
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most common cancers with dismal clinical outcome accounting for the third cause of cancer-related death in the world [1], and its incidence has continued to increase in the United States and Western Europe [2]. A variety of etiological and risk factors including hepatitis virus, dietary exposure of aflatoxin B1, alcohol, diabetes, or parasites have associated with the development of HCC. Moreover, multi-step processes including genetic and epigenetic alterations are thought to be accumulated during progression of HCC [3,4]. Such complexity of the pathophysiology leads to a daunting heterogeneity of the tumors precluding unveiling disease mechanism.
In the last decade, numerous genome-wide profiling studies have been performed to delineate the complexity of the cancers. Based on gene expression patterns, heterogeneous HCC has been classified into homogeneous tumors with correlation of clinical outcomes. Molecular-based subgrouping of patients allowed the development of new targeted therapies [5,6]. This success could be accomplished by substantial progress on data analysis. Using gene set (i.e., signature)-based rather than single gene-based approach, data normalization, clustering, classification, and validation workflows are now becoming standardized [7–9]. Those developments of analysis strategies and tools have been accelerated by publicly open statistical computing resources such as bioconductor (http://www.bioconductor.-org), gene set enrichment analysis (GSEA) [10], genetic network analysis, and other analogous approaches.
More recently, research interest has moved toward identifying molecular determinants of disease pathogenesis and novel therapeutic targets. For this purpose, systematic and integrative analyses using multiple and multi-level data resources are thought as a state-of-art strategy to identify pivotal aberrations of genetic events or signaling pathways [11]. Accordingly, the study design including analytical strategy and reporting might be different according to the study purposes. With this reason, the gene sets identified by genome-wide profiling studies may have different biological meanings and usage.
Here, we categorized the previously identified gene signatures from gene expression profiling studies of liver cancer into four signature classes of prediction, phenotype, function, and molecular targets (Table 1), and discuss their distinct properties in study design, analysis, and validation strategies.
Table 1.
Classification of Gene Signatures Identified by Gene Expression Profiling
| Category | Description | Clinical correlations | External validation methoda |
|---|---|---|---|
| Prediction signature: predict clinical outcomes | |||
| Iizuka et al. [12] | Predict early recurrence by 12 gene signature | Recurrence | Same platform |
| Ye et al. [13] | Predict metastasis in HBV-related HCC | Metastasis | Same platform |
| Lee et al. [14] | Predict survival by 406 gene signature | Survival | Same platform |
| Budhu et al. [15] | Predict metastasis by stromal tissue signature | Metastasis, survival | Same platform |
| Wang et al. [16] | Predict recurrence by 57 gene signature | Recurrence | Same platform |
| Woo et al. [17] | Predict early recurrence in HBV-related HCC CD24 was identified as biomarker for early recurrence | Recurrence | Cross platform |
| Hoshida et al. [18] | Predict late recurrence using formalin-fixed, paraffin-embedded (FFPE) stromal tissues | Recurrence | Same platform |
| Budhu et al. [19] | Predict metastasis by microRNA signature | Metastasis, survival | Same platform |
| Ji et al. [20] | Identify mir-26 as predictor for survival and response to IFN therapy | Survival, response to IFN therapy | Same platform |
| Poon et al. [21] | Predict survival by DNA copy numbers | Survival | Not determined |
| Hernandez-Vargas et al. [22] | Predict survival by 58 DNA methylation | Survival | Not determined |
| Phenotype signature: define characteristics of cancer subtype | |||
| Nam et al. [31] | Identify classifiers for low-to high-grade dysplasia and HCCs | Not determined | Not determined |
| Wurmbach et al. [32] | Identify dysregulated pathways for 4 neoplastic stages of HCV-related HCC | Not determined | Not determined |
| Ura et al. [33] | Identify a microRNA signature which is differentially expressed between HBV- and HCV-related HCC | Not determined | siRNA-mediated silencing |
| Chiang et al. [34] | Classify phenotypes of proliferation, CTNNB1, IFN-related, polysomy 7, and unannotated | Recurrence | Not determined |
| Katoh et al. [35] | Identify subclasses, e.g., c-Myc-induced HCC, 6p/1q-amplified HCC, and 17q-amplified HCC | Survival | Rapamycin sensitivity in 17q gained tumor cell lines |
| Boyault et al. [36] | Identify 6 classes based on gene expression, mutation, methylation, and LOH | Survival | Not determined |
| Lee et al. [37] | Identify hepatoblast-like class | Survival, recurrence | Same platform |
| Yamashita et al. [38] | Identify progenitor-like class using EpCAM and AFP | Survival | Same platform, immunohistochemistry |
| Andersen et al. [39] | Identify progenitor-like class using CK19 expression | Survival, recurrence | Not determined |
| Cairo et al. [40] | Identify child hepatoblastoma-derived signature | Survival | qPCR |
| Woo et al. [41] | Identify cholangiocarcinoma-like class | Survival, recurrence | Cross-platform |
| Function signature: functionally related gene sets (pathways) | |||
| Kaposi-Novak et al. [44] | Met knock-out signature | Survival | Cross platform |
| Coulouarn et al. [45] | TGF-β knock-out signature | Survival | Cross platform |
| Kaposi-Novak et al. [46] | Myc signature expression during malignant conversion | Not determined | Cross platform |
| Mayhew et al. [47] | RB knock-out signature | Survival | Not determined |
| Lee et al. [48] | Signatures from mouse transgenics of E2F, MYC, MYC-E2F1, and MYC-TGFA | Survival group | Not determined |
| Molecular target signature: candidate gene sets for molecular therapies | |||
| Zender et al. [55] | cIAP1 and Yap are identified in DNA amplicons | Not determined | In vivo overexpression |
| Zender et al. [56] | 13 genes including XPO4 are identified by shRNA screening of the overlapped deleted genomic regions in human and mouse tumors | Not determined | shRNA-mediated silencing |
| Woo et al. [57] | 50 potential driver genes are identified by combined analysis of array CGH and gene expression profiles | Survival | siRNA-mediated silencing |
Functional validations by experiments and external validations using independent data sets were considered.
CLASSIFICATION OF SIGNATURES
Prediction Signature
The majority of the gene signatures in previous studies are the prediction signature which predicts clinical outcomes. It includes the signatures generated from gene expression [12–18], microRNA [19,20], DNA copy numbers [21], and epigenetic regulation [22]. The gene signatures are usually developed and trained by the clinical information (e.g., survival, recurrence, or metastasis) to be predicted.
Major concern for the prediction signatures is its clinical applicability. For clinical use, the prediction signatures should be validated in a concrete manner. Because the predictability of the signatures could be affected by many confounding factors including experimental conditions (e.g., sample preparation and hybridization conditions) or population characteristics (e.g., postoperative treatment, patient’s health, and etiological differences), rigorous validation with appropriate sample size is required. External validation with independent data sets is also indispensable. Simon and coworkers have suggested helpful guidelines for study design, analysis, and validation methods for the development of predictors [23,24]. In general, the prediction signatures do not necessarily have functional roles [25], because they merely reflect the general condition of liver functions or host responses without causal relations. Therefore, the prediction signature genes may not appropriate candidates for therapeutic targets.
There are several issues to be resolved for the clinical use of the prediction signatures. Cross-platform difference of microarray is thought to be one of major obstacle for the clinical use of microarrays. Although the Microarray Quality Control (MAQC) project launched by US Food and Drug Administration (FDA) has addressed the technical reproducibility of microarray data across different sites and platforms [26], many of microarray studies generated only a small number of overlapping genes [24,27]. This discrepant result might be derived from the use of different platforms, probe sets, data normalization, and processing as well as differences in the study populations [28]. To resolve this issue, we have shown cross-platform consistency and robustness of tumor recurrence predictors by comparing two independent data sets [17]. Such validation by cross-platform comparison would be a strong evidence for the robustness of the clinical utility of prediction signatures.
Sample preparation is also one of critical drawback for the clinical use of microarray, because handling of the RNA samples from frozen tissues is not easy in clinical setting. Recently, Hoshida et al. [18] have shown the clinical utility of RNAs prepared from formalin fixed, paraffin-embedded (FFPE) tissues for the prediction of prognostic outcomes using a cDNA-mediated annealing selection extension and ligation (DASL) assay. This study has brought the microarray technology one step closer to clinical use, because the FFPE samples can be easily assessed in clinic [29]. They showed prognostic significance of the prediction signature generated from nontumoral tissues, but failed to show prognostic significance of the signatures from tumoral tissues. This finding is discrepant to the previous studies [12–14,16,17] which have consistently demonstrated the prognostic values of the signatures from tumoral tissue, therefore, further elaboration might be required [30].
Phenotype Signature
While the prediction signatures are generally obtained from the parameters to be predicted, the phenotype signatures represent distinct class phenotype regardless of the prediction parameters. For example, clinical and pathological features such as tumor grade [31], clinical stage [32], etiology (e.g., HBV or HCV) [33], drug responses, or the presence of certain biomarkers can be used for sample classification labels. Also, genetic events such as gene mutations or DNA copy number variations can be used as phenotype classifiers [34–36].
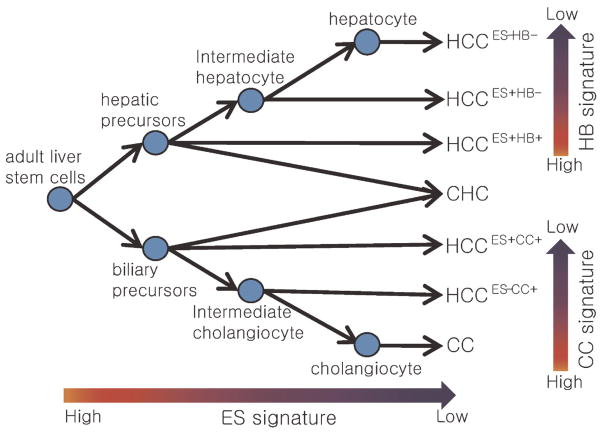
Remarkably, recent identification of the expression traits of tumor cellular origins could represent distinct tumor phenotypes. The identification of hepatoblast-like signature has stratified a novel HCC subtype harboring hepatic progenitor cell-like trait [37]. This study has demonstrated a strong correlation of the cell-type traits with patients’ prognosis, suggesting that the cellular origin of the tumors is a critical determinant for cancer progression. EpCam [38] and CK19 [39] were also identified as classifier for progenitor-derived HCC. Similarly, the hepato-blastoma-derived signature has been identified from childhood hepatoblastomas predicting clinical outcome [40]. However, the progenitors might be heterogeneous according to their differentiation status. With this concern, we have recently identified a cholangiocarcinoma (CC)-like gene expression trait (i.e., CC signature) representing a biliary differentiation trait in HCC [41]. In this study, we have shown functional and prognostic implications of the CC signature in HCC progression, suggesting distinct property of differentiation trait compared with those of stem-like trait. Taken these findings together, we could suggest that the different cellular origins from distinct developmental stage of primitive hepatic progenitor cells to differentiated hepatic or biliary precursors can give rise to heterogeneous HCC phenotypes (Figure 1).
Figure 1.
Postulated diagram of the cellular origin of HCC. The postulated cellular origins of HCC based on the expression status of cholangiocarcinoma (CC), embryonic stem (ES) cell, and hepatoblast (HB) signatures were illustrated. Details for cellular origin of CC were omitted. + The expression of cellular origin signatures. CHC represents combined hepatocellular cholangiocarcinoma. Reprinted from Ref. [41].
As like the prediction signature, the phenotype signatures have correlated with clinical outcomes (Table 1) [37,40,41]. However, these phenotype signatures do not necessarily have strong prediction power for clinical outcomes, because their primary goal is to address the functional characteristics of a certain tumor subtype. For example, the HCC class identified by a certain genetic phenotypes such as 17q amplification was not strongly associated with patient’s survival, but functionally liked to drug sensitivity to rapamycin [35]. Thus, the major concern for the phenotype signature is class characterization rather than prediction of clinical outcomes. Proper classification with well-defined phenotype signatures and linking them with functional and clinical utilities such as chemical or drug responses would be a promising strategy to develop novel cancer treatment modality for a certain tumor subtype, opening new era of personalized medicine [42,43].
Function Signature
The gene signatures representing distinct cellular processes or molecular pathways are referred as function signature. For example, the target gene signatures for MET [44], TGFB [45], MYC [46], and RB [47] have been identified from knock-out or transgenic animal models. As an application of function signatures, comparative analysis using different mouse models has demonstrated that the MYC-TGFA transgenics had similar expression patterns with HCCs of the poorer survival group, suggesting the regulatory role of TGFA signaling in aggressive progression of HCC [48]. In addition to such experimental models, computationally predicted or manually curated databases can be categorized as function signatures such as Gene Ontology (http://www.geneontology.org), microRNA targets (e.g., MirBase, http://microrna.sanger.ac.uk, or TargetScan, http://www.targetscan.org), protein–protein interactions (e.g., IntAct, http://www.ebi. ac.uk/intact), or transcription factor-binding (e.g., TRASNFAC, http://www.biobase-international.com).
As like phenotype signature, function signatures can represent phenotypes, but they have more causal or functional relevance. Thus, they are useful in functional interpretation of tumor classification. However, since gene expression profile is only a “snap-shot” of gene-to-gene interaction, they inevitably include many of nonspecific “by-stander” genes which are not functionally associated. Therefore, functional roles of the genes in function signature need to be addressed more carefully in detail. Particularly, proliferation- or cell cycle-related genes can easily be co-founded as by-stander genes, because the expression of these genes might be altered as an end result of disease progression. With this concern, a previous study has tested the possible effect of those genes by simply subtracting them from embryonic stem (ES) cell signatures [49]. This would be one of solutions for the signature purification to remove unwanted effects of the by-stander genes, although further accurate solutions might be required.
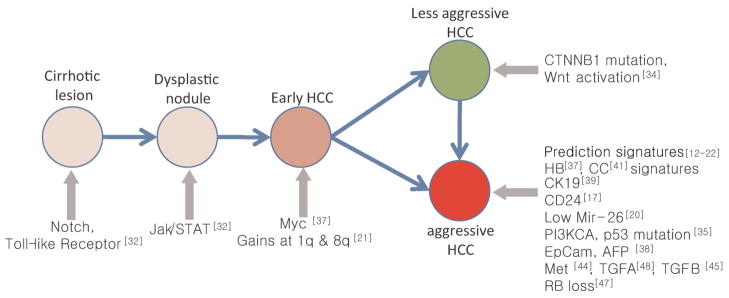
The pathogenesis of HCC is thought to be processed by multi-step pathologic states of chronic cirrhotic lesions, dysplastic nodules, and progression to early and advanced HCC [3,4]. The function signatures as well as phenotype signatures could address key regulators and genetic events during step-by-step progression of tumors (Figure 2). In earlier study, Nam et al. [31] have shown the discrete expression patterns (i.e., phenotype signature) across the multi-step hepatocarcinogenesis from low- to high-grade dysplastic nodules and HCCs. In addition, by performing pathway analysis, Wurmbach et al. [32] have further revealed the dysregulation of the Notch and Toll-like receptor pathways in cirrhosis, followed by dysregulation of Jak/STAT in early carcinogenesis. The application of MYC signature has also revealed its regulatory role during malignant conversion from dysplasia to early HCC [46].
Figure 2.
Genetic aberrations during multi-step hepatocarcinogenesis identified by genome-wide high-throughput profiling studies.
Besides these findings, genetic events such as gene mutation (e.g., TP53, CTNNB1, and KRAS), DNA methylation [50], and other gene expressions (e.g., IGF, VEGFR, CD24) have been noticed to involve the multi-step hepatocarcinogenesis, and their targeted therapies are now under investigation [51]. Signaling pathways such as proliferation (e.g., TGFA, EGF, IGF2, and HGF), differentiation (e.g., Wnt and TGFB), inflammation (e.g., IL6, IFN, and TNFA), and angiogenesis (e.g., VEGF, FGF, PDGF, and angiopoietin) were frequently deregulated resulting in high complexity and heterogeneity of HCC [see details in review [52,53]]. Such multiple and multistep complexity of genetic interaction impedes complete understanding of the genetic pathogenesis of hepatocarcinogenesis. Further extended studies defining phenotype or function signatures with multi-layered platform data including microRNAs, DNA copy numbers, or epigenetic changes are required to further delineate the multi-step hepato-carcinogenesis.
Molecular Targets
In cancer genomics studies, many of researchers are now interested in identifying molecular determinants of disease pathogenesis and novel therapeutic targets. These could be identified from prediction, function, or phenotype signatures. However, since lengthy gene list of those signatures would preclude the further evaluation of target genes, it would be important to select the most promising candidates as minimum as possible. To prioritize the most probable candidate genes, systemic integration of the gene signatures from multiple layers of data sets is thought to be “state-of-art” strategy. Data integration with multiple-layered platforms may have advantage to overcome the limitation of each platform data by reducing biased observation in each data set. For example, recent integrative analysis of microRNA target signatures from different databases could successfully identify novel oncogenic function of mir-101 in prostate cancer [54]. In HCC, cross-species comparative analysis of array-based comparative genomic hybridization (CGH) has revealed novel cancer genes, cIAP1 and Yap [55]. The same group also performed an efficient high-throughput screening of siRNA libraries by analyzing array CGH data, which discovered 13 novel tumor suppressor genes including XPO4 [56]. In addition, we have shown a successful integration of array CGH and gene expression profiling data [57]. Guiding the gene selection strategy by assessing the prognostic impact of the selected genes, we identified 50 potential driver genes including NCSTN and SCRIB. Then, further interrogation of the driver genes by using Connectivity Map database [58] could identify molecular targets (i.e., EGFR, mTOR, and AMPK) of the driver genes suggesting their mechanistic implications. These studies of systematic and integrative analyses of genomic profiles to select candidate target genes can speed up the screening and validation process of cancer-gene discovery and therapeutic targets.
INTEGRATIVE ANALYSIS UNVEILS COMPLEXITY
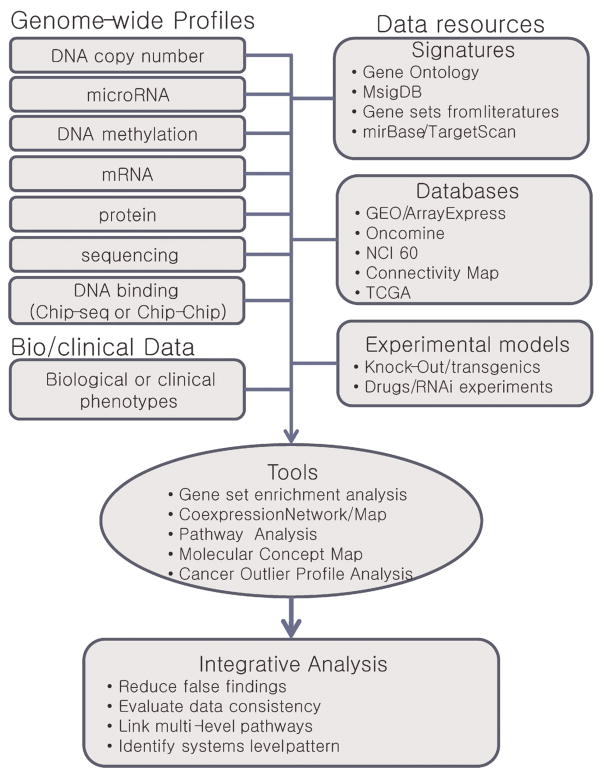
As shown in previous studies, appropriate integration of independent genomic data had a great advantage to identify novel molecular targets. Huge data repositories of microarray data, Gene Expression Omnibus (GEO) [59] and ArrayExpress [60] has enabled the researchers to perform integrative data analysis. In addition, as one of pioneering resources for integrative genomic profiling, National Cancer Institute 60 (NCI60) cell line panel has been established [61]. NCI60 had multi-dimensional data sets for 60 cancer cell lines including profiles of mRNAs, proteins, microRNAs, DNA copy numbers, gene mutations, and drug responses. These multilayered data integration could unveil molecular complexity by linking phenotypes and gene expression signatures [61–63]. Similar approaches for the cell-line based screen of candidate drugs have shown an efficient improvement of the evaluation process for anti-cancer agents [see review in [64]]. In addition, recent developments of huge data repositories such as The Cancer Genome Atlas (TCGA) [65], Connectivity Map, and Oncomine [66] will also be useful resources for integrative analysis of cancer genome, providing more qualified molecular targets [67,68].
Interaction of signatures is one of the challenging issues in the future integrative analyses. Recently, Chinnaiyan’s group has succeeded integrative data analyses using the gene signatures from multidimensional data sets. They developed a newer technique, “molecular concept map” (MCM) to address higher-level patterns across multiple and disparate microarray experiments [69]. Differing from the previous studies linking one-to-one or one-to-many association between signatures and phenotypes, MCM has linked many-to-many association among function and phenotype signatures by integrating different data types of pathways, proteins, and networks in an unbiased approach. The same group also developed an algorithm, that is, Cancer Outlier Profile Analysis (COPA) [70]. By identifying outlier expression across cancer samples, it could successfully reveal novel and uncharacterized oncogenic alterations. In another study, network analysis by constructing a BRCA-centered network (BCN) could identify novel susceptibility genes in breast cancer [71]. Such attempts on higher-level systematic approaches by integrating multiple and multi-level data resources would be promising to delineate genomic determinants in cancer development and progression (summarized in Figure 3); however, only a few of such integrative approaches have been applied to liver cancers.
Figure 3.
Strategies and resources for integrative analysis of genomic profiling data. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
CONCLUSION
We have reviewed the current status of the HCC gene expression profiling studies particularly the signature outcomes. Our classification of gene expression signatures as prediction, phenotype, function, and molecular targets would be helpful to address the distinction and the current trends of the application of genomic data to cancer research.
The accumulation of genome-wide “omics” data has provided us new opportunities to understand disease mechanisms at systems level. Moreover, emerging new technologies such as massively parallel sequencing and their applications (e.g., RNA-seq and ChiP-Seq) enabled researchers to investigate new parameters, such as mutations, alternative splicing, epigenetic silencing, and DNA-protein interactions which have not been resolved by current microarray technologies [72–74]. However, as data become complex and the study purposes are diverse, more careful attention on appropriate study design, reporting, and analytical strategies become essential in genomics studies. We believe that the systems view of cancers by constructing integrative atlas of genomic profiles with appropriate study design and analysis strategies would provide great potential for improving cancer management as well as understanding the cancer pathobiology.
Acknowledgments
This project was supported by a grant from the National R&D Program for Cancer Control, Ministry for Health and Welfare, Republic of Korea (0920280), and the Basic Research Laboratory (BRL) program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2010-0001200).
Abbreviations
- HCC
hepatocellular carcinoma
- CC
cholangio-carcinoma
- CGH
comparative genomic hybridization
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. Hepatocellular carcinoma: Recent trends in the United States. Gastroenterology. 2004;127:S27–S34. doi: 10.1053/j.gastro.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 4.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: From genes to environment. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 5.Villanueva A, Toffanin S, Llovet JM. Linking molecular classification of hepatocellular carcinoma and personalized medicine: Preliminary steps. Curr Opin Oncol. 2008;20:444–453. doi: 10.1097/CCO.0b013e328302c9e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoshida Y, Toffanin S, Lachenmayer A, Villanueva A, Minguez B, Llovet JM. Molecular classification and novel targets in hepatocellular carcinoma: Recent advancements. Semin Liver Dis. 2010;30:35–51. doi: 10.1055/s-0030-1247131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 8.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radmacher MD, McShane LM, Simon R. A paradigm for class prediction using gene expression profiles. J Comput Biol. 2002;9:505–511. doi: 10.1089/106652702760138592. [DOI] [PubMed] [Google Scholar]
- 10.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhodes DR, Chinnaiyan AM. Integrative analysis of the cancer transcriptome. Nat Genet. 2005;37:S31–S37. doi: 10.1038/ng1570. [DOI] [PubMed] [Google Scholar]
- 12.Iizuka N, Oka M, Yamada-Okabe H, et al. Oligonucleotide microarray for prediction of early intrahepatic recurrence of hepatocellular carcinoma after curative resection. Lancet. 2003;361:923–929. doi: 10.1016/S0140-6736(03)12775-4. [DOI] [PubMed] [Google Scholar]
- 13.Ye QH, Qin LX, Forgues M, et al. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med. 2003;9:416–423. doi: 10.1038/nm843. [DOI] [PubMed] [Google Scholar]
- 14.Lee JS, Chu IS, Heo J, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 15.Budhu A, Forgues M, Ye QH, et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10:99–111. doi: 10.1016/j.ccr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 16.Wang SM, Ooi LL, Hui KM. Identification and validation of a novel gene signature associated with the recurrence of human hepatocellular carcinoma. Clin Cancer Res. 2007;13:6275–6283. doi: 10.1158/1078-0432.CCR-06-2236. [DOI] [PubMed] [Google Scholar]
- 17.Woo HG, Park ES, Cheon JH, et al. Gene expression-based recurrence prediction of hepatitis B virus-related human hepatocellular carcinoma. Clin Cancer Res. 2008;14:2056–2064. doi: 10.1158/1078-0432.CCR-07-1473. [DOI] [PubMed] [Google Scholar]
- 18.Hoshida Y, Villanueva A, Kobayashi M, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 20.Ji J, Shi J, Budhu A, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poon TC, Wong N, Lai PB, Rattray M, Johnson PJ, Sung JJ. A tumor progression model for hepatocellular carcinoma: Bioinformatic analysis of genomic data. Gastroenterology. 2006;131:1262–1270. doi: 10.1053/j.gastro.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 22.Hernandez-Vargas H, Lambert MP, Le Calvez-Kelm F, et al. Hepatocellular carcinoma displays distinct DNA methylation signatures with potential as clinical predictors. PLoS ONE. 2010;5:e9749. doi: 10.1371/journal.pone.0009749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: Ready for clinical use? J Natl Cancer Inst. 2010;102:464–474. doi: 10.1093/jnci/djq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dupuy A, Simon RM. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J Natl Cancer Inst. 2007;99:147–157. doi: 10.1093/jnci/djk018. [DOI] [PubMed] [Google Scholar]
- 25.Quackenbush J. Microarray analysis and tumor classification. N Engl J Med. 2006;354:2463–2472. doi: 10.1056/NEJMra042342. [DOI] [PubMed] [Google Scholar]
- 26.Shi L, Reid LH, Jones WD, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan C, Oh DS, Wessels L, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 28.Sotiriou C, Piccart MJ. Taking gene-expression profiling to the clinic: When will molecular signatures become relevant to patient care? Nat Rev Cancer. 2007;7:545–553. doi: 10.1038/nrc2173. [DOI] [PubMed] [Google Scholar]
- 29.Sherman M. Recurrence of hepatocellular carcinoma. N Engl J Med. 2008;359:2045–2047. doi: 10.1056/NEJMe0807581. [DOI] [PubMed] [Google Scholar]
- 30.Wang XW, Thorgeirsson SS. Transcriptome analysis of liver cancer: Ready for the clinic? J Hepatol. 2009;50:1062–1064. doi: 10.1016/j.jhep.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nam SW, Park JY, Ramasamy A, et al. Molecular changes from dysplastic nodule to hepatocellular carcinoma through gene expression profiling. Hepatology. 2005;42:809–818. doi: 10.1002/hep.20878. [DOI] [PubMed] [Google Scholar]
- 32.Wurmbach E, Chen YB, Khitrov G, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 33.Ura S, Honda M, Yamashita T, et al. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098–1112. doi: 10.1002/hep.22749. [DOI] [PubMed] [Google Scholar]
- 34.Chiang DY, Villanueva A, Hoshida Y, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–6788. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katoh H, Ojima H, Kokubu A, et al. Genetically distinct and clinically relevant classification of hepatocellular carcinoma: Putative therapeutic targets. Gastroenterology. 2007;133:1475–1486. doi: 10.1053/j.gastro.2007.08.038. [DOI] [PubMed] [Google Scholar]
- 36.Boyault S, Rickman DS, de Reynies A, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 37.Lee JS, Heo J, Libbrecht L, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 38.Yamashita T, Forgues M, Wang W, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 39.Andersen JB, Loi R, Perra A, et al. Progenitor-derived hepatocellular carcinoma model in the rat. Hepatology. 2010;51:1401–1409. doi: 10.1002/hep.23488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cairo S, Armengol C, De Reynies A, et al. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell. 2008;14:471–484. doi: 10.1016/j.ccr.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 41.Woo HG, Lee JH, Yoon JH, et al. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res. 2010;70:3034–3041. doi: 10.1158/0008-5472.CAN-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mishra L, Banker T, Murray J, et al. Liver stem cells and hepatocellular carcinoma. Hepatology. 2009;49:318–329. doi: 10.1002/hep.22704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van’t Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452:564–570. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- 44.Kaposi-Novak P, Lee JS, Gomez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest. 2006;116:1582–1595. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coulouarn C, Factor VM, Thorgeirsson SS. Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology. 2008;47:2059–2067. doi: 10.1002/hep.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaposi-Novak P, Libbrecht L, Woo HG, et al. Central role of c-Myc during malignant conversion in human hepatocarcinogenesis. Cancer Res. 2009;69:2775–2782. doi: 10.1158/0008-5472.CAN-08-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mayhew CN, Carter SL, Fox SR, et al. RB loss abrogates cell cycle control and genome integrity to promote liver tumorigenesis. Gastroenterology. 2007;133:976–984. doi: 10.1053/j.gastro.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 48.Lee JS, Chu IS, Mikaelyan A, et al. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat Genet. 2004;36:1306–1311. doi: 10.1038/ng1481. [DOI] [PubMed] [Google Scholar]
- 49.Ben-Porath I, Thomson MW, Carey VJ, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee S, Lee HJ, Kim JH, Lee HS, Jang JJ, Kang GH. Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am J Pathol. 2003;163:1371–1378. doi: 10.1016/S0002-9440(10)63495-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minguez B, Tovar V, Chiang D, Villanueva A, Llovet JM. Pathogenesis of hepatocellular carcinoma and molecular therapies. Curr Opin Gastroenterol. 2009;25:186–194. doi: 10.1097/MOG.0b013e32832962a1. [DOI] [PubMed] [Google Scholar]
- 52.Zender L, Villanueva A, Tovar V, Sia D, Chiang DY, Llovet JM. Cancer gene discovery in hepatocellular carcinoma. J Hepatol. 2010;52:921–929. doi: 10.1016/j.jhep.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008;48:1312–1327. doi: 10.1002/hep.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varambally S, Cao Q, Mani RS, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyl-transferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zender L, Spector MS, Xue W, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zender L, Xue W, Zuber J, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woo HG, Park ES, Lee JS, et al. Identification of potential driver genes in human liver carcinoma by genomewide screening. Cancer Res. 2009;69:4059–4066. doi: 10.1158/0008-5472.CAN-09-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 59.Barrett T, Suzek TO, Troup DB, et al. NCBI GEO: Mining millions of expression profiles—Database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parkinson H, Sarkans U, Shojatalab M, et al. ArrayExpress—A public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2005;33:D553–D555. doi: 10.1093/nar/gki056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scherf U, Ross DT, Waltham M, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 62.Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 63.Park ES, Rabinovsky R, Carey M, et al. Integrative analysis of proteomic signatures, mutations, and drug responsiveness in the NCI 60 cancer cell line set. Mol Cancer Ther. 2010;9:257–267. doi: 10.1158/1535-7163.MCT-09-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 2010;10:241–253. doi: 10.1038/nrc2820. [DOI] [PubMed] [Google Scholar]
- 65.Sharma SV, Haber DA, Settleman J, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 68.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009 doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tomlins SA, Mehra R, Rhodes DR, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 70.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 71.Pujana MA, Han JD, Starita LM, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet. 2007;39:1338–1349. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- 72.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 74.Maher CA, Kumar-Sinha C, Cao X, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]