Abstract
Introduction
A key part of drug design and development is the optimization of molecular interactions between an engineered drug candidate and its binding target. Thermodynamic characterization provides information about the balance of energetic forces driving binding interactions and is essential for understanding and optimizing molecular interactions.
Areas covered
This review discusses the information that can be obtained from thermodynamic measurements and how this can be applied to the drug development process. Current approaches for the measurement and optimization of thermodynamic parameters are presented, specifically higher throughput and calorimetric methods. Relevant literature for this review was identified in part by bibliographic searches for the period 2004 – 2011 using the Science Citation Index and PUBMED and the keywords listed below.
Expert opinion
The most effective drug design and development platform comes from an integrated process utilizing all available information from structural, thermodynamic and biological studies. Continuing evolution in our understanding of the energetic basis of molecular interactions and advances in thermodynamic methods for widespread application are essential to realize the goal of thermodynamically-driven drug design. Comprehensive thermodynamic evaluation is vital early in the drug development process to speed drug development towards an optimal energetic interaction profile while retaining good pharmacological properties. Practical thermodynamic approaches, such as enthalpic optimization, thermodynamic optimization plots and the enthalpic efficiency index, have now matured to provide proven utility in design process. Improved throughput in calorimetric methods remains essential for even greater integration of thermodynamics into drug design.
Keywords: Affinity, binding, calorimetry, drug design and development, DSC, energetics, enthalpy, entropy, free energy, ITC, ligand, molecular interactions, screening, thermal shift assay, thermodynamics
1. Introduction
Thermodynamics has found increasing adoption in the drug design and development process in both academic and commercial endeavors and is increasingly prevalent alongside longer-standing structure- and molecular modeling-based approaches. The integration of thermodynamic measurements has grown with a better understanding of energetic data, the increasing demonstration of the utility and application of these measurements, and the availability of ever-improving instrumentation. However, as will be discussed in this article, there is still a long way to go. Although the understanding and application of thermodynamic data is growing, there is still much that is not understood about the basis of binding interactions and how these can be interpreted from thermodynamic data. Advances in instrumentation have increased throughput and reduced sample demands, but still only offer moderate throughput for a drug discovery effort that demands much higher. Despite these limitations, useful practical approaches have been developed and advances are being made that, when realized, present a bright future for thermodynamics in drug design and development.
Historically, rational drug design has been based upon seeking structural complementarity and optimizing binding contacts between an engineered drug and a target binding site to generate lead compounds [1]. Of course, drug design is part of a bigger picture involving consideration and optimization of solubility, selectivity, ADMET (absorption, distribution, metabolism, excretion and toxicology) and pharmacokinetic/pharmacodynamic properties, but rational design and engineering of ligands for molecular recognition of a given target is the core of the process and forms the focus of this article. Historical drug design utilized structural information of the target site from X-ray crystallography and NMR alongside molecular modeling of drug-target interactions. Drug development was driven by the goal of optimizing molecular recognition, seeking high affinity compounds which were considered to possess optimal binding interactions. However, a purely structure-based approach is incomplete and it is essential to incorporate complementary approaches to understand the driving forces underlying the molecular interactions of the binding process. Approaches based solely on structural data or often sought binding affinity optimization provide an oversimplified picture of molecular interactions, with isostructural complexes and similar binding affinities potentially hiding disparate binding thermodynamics and providing only one part of the binding picture.
1. Thermodynamics of molecular interactions
2.1 Thermodynamic profile of molecular interactions
Parameters describing a complete thermodynamic profile of molecular interactions are shown in Table 1. The crucial parameter describing the interaction of binding partners is the free energy (ΔG) where both the magnitude and sign describe the likelihood of biomolecular events occurring. A binding event described by a negative free energy (an exergonic process) will occur spontaneously to an extent governed by the magnitude of ΔG. Conversely, positive ΔG values (an endergonic process) indicate that binding will not occur spontaneously but will require energy to drive the interaction, for example, by coupling to another exergonic event, such as ATP hydrolysis, which results in an overall negative ΔG for the entire process. The equilibrium binding constant, or binding affinity, (Ka) provides access to the key parameter of ΔG through equations 1.1 and 1.2 where Ka can be used to calculate the standard Gibbs free energy (ΔGo) which in turn is related to ΔG. ΔGo refers to a standard state of 1 M reactant concentrations at pH 7 and 25 °C where the ratio of actual non-equilibrium concentrations of reactant and product for a particular binding process, defined by the reaction quotient (Q), can be used to calculate ΔG.
Table 1.
Basic thermodynamics of molecular interactions
| ΔG = ΔGo + RT ln Q | Equation 1.1 | |
| ΔGo =−RT ln Ka | Equation 1.2 | |
| ΔG =ΔH − TΔS | Equation 1.3 | |
|
|
Equation 1.4 | |
|
|
Equation 1.5 | |
| ΔH (T) =ΔH (TR) + ΔCp (T − TR) | Equation 1.6 | |
|
|
Equation 1.7 | |
|
|
Equation 1.8 |
ΔGo, standard Gibbs free energy change; Ka, equilibrium binding constant (or binding affinity); ΔG, Gibbs free energy change; Q, reaction quotient; ΔH, enthalpy change; ΔS, entropy change; ΔHvH, van’t Hoff enthalpy change; ΔCp, heat capacity change; R, gas constant equal to 8.31451 J/K.mol; T, temperature of interest; TR, arbitrary reference temperature. Description of the thermodynamic parameters and their interrelationship is provided in the text.
ΔG provides only part of the picture being composed of enthalpic (ΔH) and entropic (ΔS) components. ΔH can be readily measured: most accurately and directly through measurement of heat changes accompanying binding events using calorimetry but also indirectly as the van’t Hoff enthalpy (ΔHvH) through the temperature dependence of Ka as defined by the van’t Hoff relationship (equation 1.4). ΔS is calculated through equation 1.3 with knowledge of ΔH and ΔG to complete the thermodynamic profile. It is important to note that calorimetry measures the global properties of a system reflecting the sum of all coupled processes accompanying binding, such as solvent reorganization and protonation events, and these processes must be deconvoluted from the observed heat changes in order to extract binding energetics. Despite having to deconvolute binding energetics, direct measurement of ΔH via calorimetry is significantly advantageous over van’t Hoff enthalpy determination. The temperature dependency of ΔH can be measured calorimetrically and signifies a non-zero heat capacity change, ΔCp (equation 1.5). A negative ΔCp indicates that the binding complex has a lower heat capacity than the free binding partners and, along with a positive entropy, is typically associated with hydrophobic interactions and conformational changes upon binding [2,3]. The presence of curvature in van’t Hoff plots resulting from non-zero heat capacity changes is often neglected and is a source of discrepancy between ΔHvH and ΔH values [3–6]. The determination of non-zero ΔCp values requires the application of more complex expressions for ΔH, ΔS and ΔG (equations 1.6–1.8).
Enthalpy reflects heat differences between reactants and products of a binding reaction as a result of net bond formation or breakage, with negative values indicating a net release of heat energy with the resulting products at a lower energy level than the reactants. Entropy reveals the ease of distribution of binding energy among molecular energy levels with positive values associated with an increase in disorder, and vice versa. The release of structured water molecules surrounding binding surfaces is commonly considered to be a source of positive entropy as a result of an increase in the disorder of the system but depends on the precise balance of solvent interactions with the free binding partners and within the binding complex. Conversely, an increase in the order of the system through, for example, the introduction of conformational restrictions in the binding complex is reflected by negative entropy values.
The importance of separating ΔG into component ΔH and ΔS terms is well accepted. A consideration of Ka and ΔG alone is insufficient because similar values can mask radically different ΔH and ΔS contributions describing entirely different binding modes, information which is of obvious importance in the rational drug design process. The phenomenon of entropy-enthalpy compensation has received much discussion in the literature and been frequently reported in drug development studies [7–10]. Designed modifications of drug candidates often result in the desired effect on ΔH but with a concomitant undesired effect on ΔS, or vice-versa, yielding little or no net effect on ΔG or Ka that was originally sought. For example, a compound modification resulting in increased bonding will yield a more negative enthalpy but could lead to increased ordering in the binding complex which would be associated with a more negative entropy. Moreover, high affinity, as is often sought in drug design, is not a requirement for high selectivity and specificity [6].
2.2 Thermodynamic optimization of molecular interactions
In terms of rational drug design, there are binding interactions which are significantly easier to engineer and optimize and some that are extremely difficult. Traditionally, synthetic drugs have been based upon achieving selectivity through shape complementary of the binding ligand to the target site and affinity through hydrophobicity [11]. Shape complementary was engineered through pre-shaping and conformationally constraining an initial drug scaffold, followed by affinity optimization through entropy optimization. Favorable binding entropy often arises from the partial or complete release upon binding of structured water molecules surrounding both the drug and binding site. Desolvation entropy is a non-specific force proportional to the hydrophobicity of the drug; hydrophobic effects are by far the most favorable contributor to free energy, estimated at 80 % [12]. The relative ease of increasing binding entropy through the decoration of drug candidates with hydrophobic groups have led to the increasing hydrophobic character of drug candidates as they move through the drug design process. Synthetic, rationally designed drugs have a proportionately greater favorable entropy contribution to binding free energy than natural, biological ligands [13]; however, there is a limit to hydrophobic optimization, reaching a solubility limit where candidates become useless as drugs [11].
Enthalpy is much harder to optimize compared with entropy because, even alongside structural information concerning the geometry of the binding site, engineering of precise atomic interactions a priori is very difficult. Binding enthalpies are in fact comprised of opposing contributions: favorable enthalpy from non-covalent hydrogen bonding and van der Waals interactions and unfavorable enthalpy from the desolvation of polar groups necessary for their participation in binding interactions. The enthalpic desolvation penalty of polar groups is significant, estimated at ~ 30–40 kJ/mol at 25 °C (compared with a tenth of that for the desolvation of non-polar groups) and dominates enthalpy [7]. A significantly favorable contribution from hydrogen bonding and van der Waals interactions is required to overcome the large desolvation penalty. For every ~6 kJ/mol of unfavorable enthalpy (with no change in entropy), binding affinity drops by one order of magnitude [10]. A strong hydrogen bond can contribute approximately −20 kJ/mol of favorable enthalpy but is often opposed by a large entropic loss resulting from structuring and conformational restriction of both the target site and binding ligand [10]. The engineering of conformational constraints into a ligand can overcome much of the entropic penalty associated with its binding. Minimizing the penalty associated with structuring within target binding regions is more difficult but a number of strategies exist [7]. One strategy is to direct hydrogen bonds at already structured regions within or adjacent to the target binding site detected through structural or computational means. Alternatively, multiple hydrogen bonds can be directed to the same location with an entropic penalty for the first hydrogen bond but subsequent bonding to a then structured region devoid of conformational penalty. Extensive thermodynamic measurements can be correlated with systematic modification of the location of ligand hydrogen bonding groups in an attempt to more precisely identify and optimize target binding interactions.
2. Application of thermodynamics to drug design and development
Measurement of binding thermodynamics was envisioned to provide critical information regarding the nature and balance of drug-target interactions. While thermodynamic studies have undoubtedly provided a wealth of important information for a number of drug design studies, its widespread application across unrelated systems has not been realized because the fundamental problem of delineating and understanding molecular interactions underlying drug-target associations is significant and of exceptional difficulty. A lot is known but significantly more is still not understood or poorly approximated. What has resulted is the application of thermodynamic approaches to a given system or related systems where relative assessments of incremental drug design changes can be determined for a series of drug candidates binding to a single target. These approaches have been very successful and have resulted in useful thermodynamic optimization strategies for drug design and development. Selected examples are discussed below.
The enthalpy optimization approach developed by Freire and co-workers resulted from the realization of an observed trend of increasing enthalpy optimization concomitant with the evolution of drug molecules from first-in-class to best-in-class [7]. Extensive thermodynamic analyses of binding energetics of two classes of drugs, HIV-1 protease inhibitors and statin drugs, revealed that first-in-class drug molecules were thermodynamically unbalanced resulting from entropically optimized drug design strategies. Drug development resulted in the evolution from nanomolar binding affinities with zero or unfavorable binding enthalpies of the first drugs to picomolar affinities and favorable enthalpies dominating binding of the later drug generations. This then presented a more efficient drug design strategy, that of seeking enthalpically-driven leads early in the drug development process. A favorable binding enthalpy implies significant, favorable binding interactions necessary to overcome the enthalpic desolvation penalty. However to achieve optimal binding interactions it is necessary to consider all energetic contributions: favorable binding and desolvation entropies must be maximized and unfavorable conformational entropy minimized. The concept of the enthalpy funnel was developed to assess binding energetics at each stage of the drug development process [11]. Here the enthalpic contribution to binding free energy (ΔH/ΔG) was plotted as a function of the logarithm of the binding affinity (log Ka). Low affinity drug hits appear at the opening of the funnel where a wide range of possible enthalpy/entropy combinations, and hence polar/hydrophobic interactions, exist with subsequently narrowing as affinity increases. Recognition of the energetic nature of binding interactions early in the drug discovery process allows the selection of hits that establish the most favorable enthalpic interactions with the binding target for faster thermodynamic optimization of drug leads. Continuing thermodynamic monitoring as optimization proceeds avoids chemical modification that could result in undesirable binding energetics.
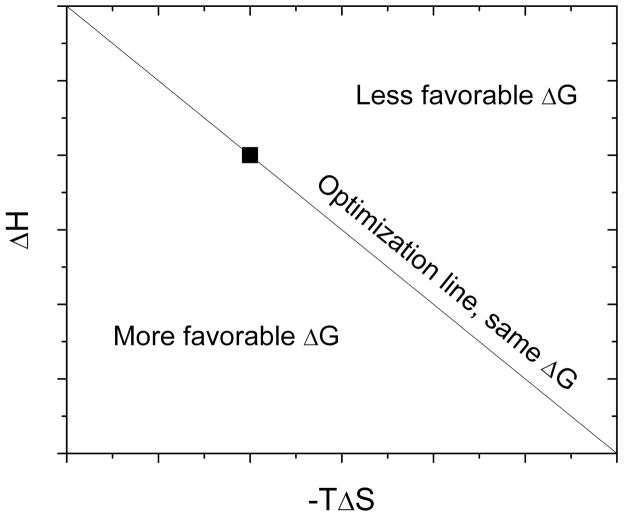
Freire extended the application of a thermodynamic optimization strategy through the construction of a “thermodynamic optimization plot” (TOP) [10]. Here a drug candidate for optimization is characterized calorimetrically and a TOP constructed by plotting binding enthalpy on the y-axis against entropy (in the form of −TΔS) on the x-axis (Figure 1). Chemical modification and subsequent thermodynamic characterization of drug candidates are added to the TOP. Compounds with the same ΔG as the lead compound will fall on the optimization line, those with more positive ΔG (lower affinity) fall above the line and those with more negative ΔG (higher affinity) fall below the line. Six specific regions of binding energetics are defined within the TOP characterized by enthalpy and entropy gains or losses and whether these overcome opposing competing energetic forces. Evaluation of the effect of systematic compound modifications on the binding energetics can then be used as a drug design optimization strategy.
Figure 1.
Example of a Thermodynamic Optimization Plot.
Another experimental strategy incorporating thermodynamic data into drug design is the enthalpic efficiency (EE) parameter which provides an index used for the ranking of compound interactions [13]. This is particularly relevant in fragment-based drug discovery (FBDD). FBDD is used in early drug discovery to seek compound building blocks with high efficiency binding interactions which are used in the construction of larger and more potent drug leads. Screening of compound interactions based on affinity alone would select against lower molecular weight compound fragments binding with low overall affinity and EE is proposed as a more relevant metric. EE relates changes in non-covalent bonding, through measurement of ΔH, to either the size of the molecule (number of non-hydrogen atoms or molecular weight) or the number of polar atoms. Consideration of the number of polar atoms is suggested to yield the most utility as this relates the contribution of hydrogen bond donors and acceptors to the measured ΔH providing a direct reflection of the strength of these bonds [13]. There are only a certain number of possible non-covalent interactions within a given binding site and simply increasing the polar character of a compound can have negative consequences for bioavailability [11,13]. It is therefore important to optimize the quality and not the quantity of interactions, information which is provided by the EE metric, while maintaining the desired thermodynamic binding signature. Other useful metrics have been provided as ligand efficiency measures [14–16]. Of particular relevance is the size-independent enthalpic efficiency (SIHE) and group efficiency (GE). SIHE accounts for the dependence of EE on molecular size and allows comparison of ligands of dissimilar sizes. GE defines the quality of an added structural element and is of utility in lead optimization.
3. Calorimetric measurement of thermodynamics
Calorimetry is widely considered the gold standard for the direct measurement of binding energetics and is considered advantageous over indirect thermodynamic methods that measure Ka values to determine ΔHvH. The direct detection of small heat changes accompanying biological reactions provides a universal detection method for molecular interactions and presents a significant advantage over biochemical assays requiring specific development and optimization for each target studied. Calorimetry is also performed in the solution state and without the need for chemical modification, labeling or immobilization. Isothermal titration calorimetry (ITC) is applied to obtain a binding profile from the titration of aliquots of binding partners at a constant temperature and yields a complete binding profile in ΔG, ΔH and ΔS. Differential scanning calorimetry (DSC) provides a complete thermodynamic profile for the unfolding energetics of a system. The determination of binding energetics is then determined from a consideration of unfolding energetics of a biomolecule in the presence and absence of a binding partner. It is important to distinguish and not confuse the parameters obtained from these methods: the energetic parameters from ITC relate to binding and are denoted with a subscript b; those from DSC relate to thermal unfolding (or melting) of a biomolecule and are denoted with a subscript m. A detailed discussion of these methods and their application is beyond the scope of this article and the reader is referred to a number of reviews available for both methods (for example, [17–19]).
DSC provides direct and accurate determination of ΔHm,ΔCp,m and melting temperature (Tm) for biomolecule unfolding which can yield a wealth of invaluable information concerning the nature of stabilizing forces present in their active states and the effect of experimental conditions and interacting ligands [17]. The Tm of a biomolecule will increase as a result of preferential ligand binding to its native state and decrease through binding to the unfolded state. An estimate of Ka (and hence ΔGb) can be made from Tm measurements in the absence and presence of binding ligand, with knowledge of the unfolding energetics of the unligated biomolecule. McGhee and Crothers developed statistical mechanical theories to extract binding thermodynamics from the deconvolution of multiphasic DNA unfolding profiles in the presence of less than saturating amounts of ligand or protein [20–23]. Based on Schellman’s early work, analytical treatments have also been developed for protein-ligand systems [17,24–27]. DSC holds a significant advantage in the ability to estimate ultratight binding constants up to 1020 M−1 [27], inaccessible by any other method, although the measurement of ultratight interactions may not be of direct utility in early drug screening and development. A number of assumptions are made during the estimation of binding constants using DSC and should be considered carefully [17]. Treatment of protein-ligand binding assumes a two-state reversible transition with complete saturation of binding sites on the native protein, for which binding saturation must be established through the construction of a plot of Tm as a function of added ligand. Also, both DNA-ligand and protein-ligand approaches assume binding to only the native form and an estimation of the number of binding sites must be determined from unfolding curves at intermediate ligand concentrations or from other methods (e.g. ITC, optical methods). Moreover, determination of Ka is made at Tm which then requires significant and potentially inaccurate extrapolation to a temperature of interest, such as 20 °C. Determination of ΔHb using DSC is calculated from the difference inΔHm in the presence and absence of ligand [17,18], with the thermodynamic binding profile completed through calculation of ΔSb.
ITC is often favored in drug discovery because of its direct measurement of interaction energetics under relevant experimental conditions. The magnitude of the measured heat reflects extent of binding with decreasing heats observed upon saturation of the binding target. Construction of a binding isotherm of interaction heat as a function of titrated ligand yields ΔHb, Ka (hence ΔGb) and binding stoichiometry, with ΔSb calculated from equation 1.3. Measurement of ΔHb as a function of temperature directly yields ΔCp. Access to EE measurements for weakly binding drug fragments requires either the application of a displacement ITC approach for a low affinity ligand [28] or determination of ΔHb alone using the so-called excess sites method [18,29,30]. The excess sites method also permits model-free determination of ΔHb without recourse to the application of a binding model required for fitting of the binding isotherm from a typical saturation ITC approach [18]. Direct determination of binding energetics using ITC is an extremely powerful and useful approach but can be complicated by the observation of the global properties of a system. Observed heats are not solely related to the binding event of interest but reflect the global contribution from all coupled equilibria associated with the binding process [13,31]. These can include solvent interactions and reorganization, conformational changes and protonation events. Observation of this global picture can make it difficult to deconvolute specific interaction energetics. This has led to the utilization of thermodynamic measurements in a focused way to assess the relative binding of a series of compounds within a specific system through an examination of the effect of incremental structural changes or the influence of experimental conditions on changes in observed energetic measurements. In order to realize the full utility of thermodynamic information and its broad application to the characterization of binding interactions it is necessary to significantly advance our understanding of molecular interaction forces.
4. Correlation of thermodynamic data with molecular association
Over the last decades significant experimental advances have occurred directed at achieving the ability to control molecular recognition through rational drug design [12]. The production of proteins and generation of site-specific mutants have allowed critical testing of structure activity relationships. Structural analyses through X-ray crystallography coupled with computational modeling and hydrogen/deuterium exchange experiments provide more relevant dynamic and solvation information. Thermodynamic characterizations of binding interactions are enabling the identification and energetic optimization of lead ligands. These impressive advances have gone a long way towards this goal but there is still a significant task of understanding and designing precise molecular interactions, hampered by a disconnect between structural and thermodynamic data [31]. Apparently isostructural complexes can demonstrate significant differences in thermodynamic profiles [6]. A surprising and disappointing lack of correlation is found between change in polar surface area, reflecting the degree of hydrogen bond formation, and ΔH or between burial of hydrophobic surface area and ΔS [31]. Observations of counterintuitive thermodynamic results have been attributed to an incomplete understanding of molecular association, even in relatively simple systems [12]. Non-covalent interactions, and particularly entropies, are only partially understood, especially as relates to the complexity of the thermodynamics of interactions in aqueous systems. Despite significant and important contributions there is no comprehensive theory of water behavior, nor any molecular level understanding of the hydrophobic effect or of electrostatic interactions in water. A better account of water solvation at interacting surfaces and protein plasticity, particularly within the active site, is essential when attempting to engineer precise biomolecule-ligand interactions, and may go a long way towards a greater understanding of observed entropy-enthalpy compensation. A comprehensive understanding of non-covalent interactions and their deconvolution within energetic measurements is a monumental goal. However, initial attempts have been made in the interpretation of the molecular basis of thermodynamic data and the correlation with structural data [6,31]. Ongoing incremental advances can be made through the synergy of a growing body of data and evolving experimental and theoretical approaches. Very recently, the development of an NMR entropy meter is an encouraging new approach which determines the entropy landscape of a protein on an atomic level with great utility for rational drug design [32].
Bissantz et al. recently compiled and reviewed an extensive collection of publicly available X-ray structure data and experimental and theoretical studies of model systems, providing information for a multitude of molecular interactions and its practical application to molecular design [33]. The authors cautioned against forming causal relationships concerning energetic contributions to molecular interactions based on too few observations for the wider application to other systems. Complexities arise from a number of factors including an inability to deconvolute effects of a multitude of molecular interactions, their highly nonadditive nature, the sensitivity of interaction energetics to structural context, and the difficulties of accurately modeling all factors (e.g. cooperativity, mobility, solvation) associated with a binding event. Valuable insight could be provided by carefully designed studies probing the energetic contributions of specific features of the binding partners. Extensive biophysical characterization of precise and controlled variation in structural ligand features with the use of well-defined, readily available model systems, such as carbonic anhydrase [12,34], can provide crucial thermodynamic data for the dissection of interaction energetics for rational drug design. While such data have undeniable value their extension to other systems is still challenging. A recent analysis of the experimental binding thermodynamics of 100 protein-ligand complexes provided some interesting food for thought [35]. While good correlation between ΔH or −TΔS and ΔG was observed in certain systems, the overall correlation was surprisingly poor. This creates challenges for the predictive power of enthalpies or entropies in assessing overall binding free energies. A further surprising observation came from the correlation of enthalpy and entropy with molecular size which found a primarily enthalpic, not entropic, effect associated with decreasing ligand efficiency of larger ligands. This is counterintuitive to the expectation that larger, floppier ligands would be expected to incur larger conformational entropic penalties and suggests that the conformational penalty is outweighed by other enthalpic energetic factors. Both of these observations emphasize the complexity of molecular interactions and the structural-thermodynamic puzzle which needs to be solved.
5. Higher throughput thermodynamic measurements
6.1 Thermal shift assays
The wider application of thermodynamic approaches and a growing body of high quality data is essential in advancing the utility of the energetic characterization of molecular interactions in the drug development process. The historic locus of calorimetry in specialized research labs and disadvantages of high sample requirements and low throughput has led to the later integration of calorimetry into drug development and has limited its use as a secondary or tertiary screening method, or for validation of other assays. The demand of high throughput drug screening methods has led to the adoption of thermodynamic measurements in another guise, that of high throughput thermal shift assays. The basis of this method has been previously introduced in the description of DSC measurements, that of estimating ligand binding affinity from shifts in the thermal stability of a biomolecule target and for which DSC represents a low throughput example. The most commonly used method is the fluorescence thermal shift assay (FTSA) which involves monitoring the fluorescence properties of an environmentally sensitive hydrophobic dye, commonly sypro orange, which binds specifically to internal, hydrophobic surfaces of a target protein as these become exposed during thermal unfolding [36]. The fluorescence dye signal therefore provides an indirect measurement of protein stability. In some cases, intrinsic protein fluorescence or endogenous fluorescent cofactors or prosthetic groups can serve as monitors of thermal unfolding. FTSA technology was developed to serve as a high throughput universal screening technique for the evaluation of ligand interactions with target proteins from a large compound library.
The original development of FTSA for application to drug discovery was achieved by 3-Dimensional Pharmaceuticals, later acquired by Johnson & Johnson, in the form of ThermoFluor® [37–39]. ThermoFluor was developed as a rapid, miniaturized, universally applicable thermal shift assay in a high-density 384 well microplate format with low volume and sample consumption. High throughput FTSA can also be conducted using commercially available RT-PCR thermocycler instruments [40]. Drug discovery approaches routinely employ functional screening of hit molecules using biological assays such as enzymological and cell assays. A major advantage of FTSA, and other biophysical techniques (ITC, DSC, analytical ultracentrifugation, NMR, etc.) is that it provides a target-independent binding assay. Functional assays can require significant development in terms of assay design and the production of stable, pure and charcterized reagents. They might also detect the desired functional effect of a hit ligand but will not provide any information concerning the mechanism of action or mode of binding. Furthermore, functional assays are useless if protein function is unknown or there is no accessible direct activity readout. FTSA is a broadly applicable screening method independent of any knowledge of the type or function of a biomolecule. Importantly, screening of a functional probe ligand library can reveal important information concerning the unknown function of a protein target [38].
FTSA approaches utilize the thermodynamic coupling between ligand binding and biomolecule unfolding applying established thermodynamic relationships to describe the unfolding and binding properties of the biomolecule-ligand system under study. Experimental and analysis protocols are described elsewhere [36–40]. Initial analyses involve an exploration of biomolecule unfolding properties and optimization of assay conditions, followed by binding studies against test ligands to obtain relative binding affinities. The influence of binding conditions (relative concentrations of binding partners, pH, detergent, substrates, cofactors, competing ligands, etc.) can reveal important information about binding mechanism, cooperativity and stoichiometry. Reported results have demonstrated high precision and good agreement with biological assays; observed discrepancies with biological assays can be attributed to differences in mechanisms of action or assay conditions [39]. Significant disadvantages of funtional assays, such as interference from absorbing compounds or loss of protein through binding to plastic assay hardware or to precipitated compounds, often have little effect on Tm values under the typical conditions of FTSA [39]. A much wider dynamic range of affinity measurements is possible with FTSA, compared with functional assays, with no real upper limit of affinity [38]. However, there are a number of limitations of FTSA; an extensive examination of these is provided in a number of recent articles [2,36,40,41]. Technical limitations include high background fluorescence for proteins with significant hydrophobic surfaces as a result of the binding of reporter dye before biomolecule unfolding, significant binding of the reporter dye to the protein target or competition between the dye and the test compound [36]. These difficulties arise from the use of a reporter dye in a non-direct binding assay and necessitates conducting careful control experiments. Scenarios resulting in false positive and false negative observations are also described [36]; it is essential to correlate binding results from FTSA with other binding or functional assays as part of a comprehensive drug discovery strategy.
More serious drawbacks of FTSA, and thermal shift assays in general, arise from assumptions and approaches implemented during data analysis [2,36,40,41]. For FTSA, the thermal unfolding process in the presence of the reporter dye is irreversible, yet is often described by a model for a two-state, cooperative, reversible process. This is justified by the demonstration of systems with little or no reversibility conforming to equilibirum thermodynamics and reports of general agreement of binding data obtained from thermal shift assays and ITC [40]. However, binding of the dye to the unfolded form of the protein necessarily perturbs the binding equilibrium according to the Le Chatelier-Braun principle and may introduce errors into the determination of binding energetics [2]. Moreover, multiphasic transitions may occur as a result of a number of factors and would require additional analysis [36]. The typical and most relevant interaction scheme for drug discovery screening is consideration of binding to the native state of the target biomolecule. However, binding to the unfolded state or both the native and unfolded states are possible and will have significant effects on observed shifts in thermal shift assay approaches; moreover, competitive, noncompetitive, uncompetitive binding or differences in interaction stoichiometry need to be considered [36,40,41]. Assaying a single ligand concentration during a compound screening study could provide misleading information and it is important to explore a range of ligand and biomolecule concentrations during the study of any binding system.
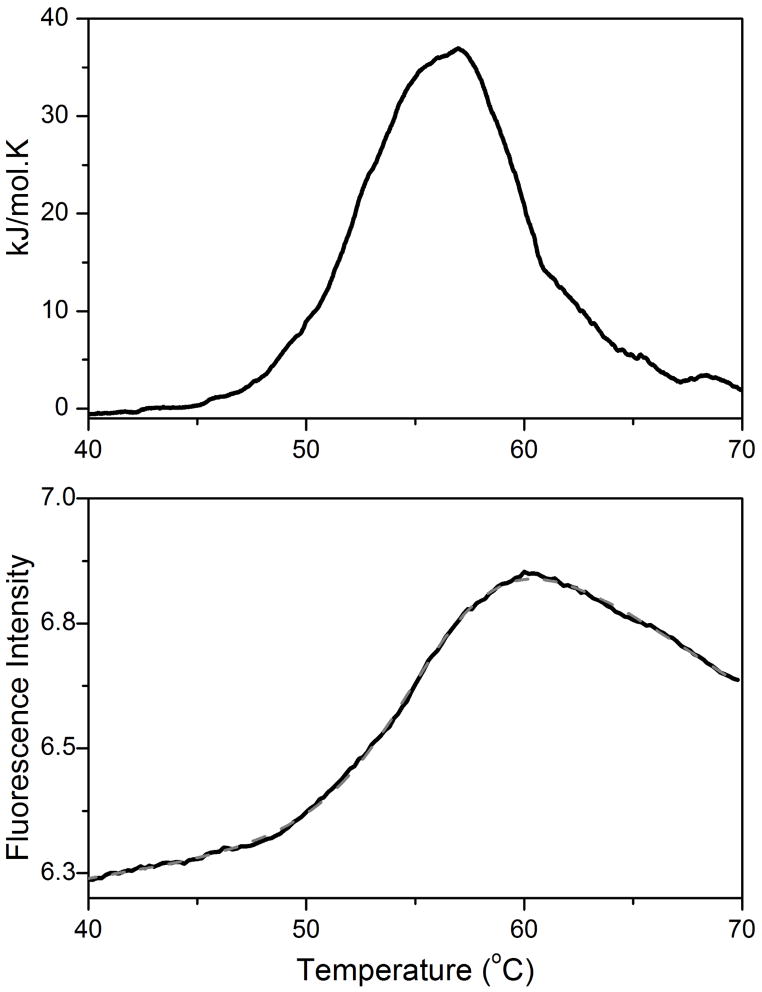
Despite the above caveats, the FTSA does seem to provide apparently accurate thermodynamic data for at least some systems. Figure 2 shows a comparison of lysozyme unfolding measured by DSC and by FTSA. The FTSA melting curve was fit to a two-state unfolding model including linear pre- and post-transition baselines. The van’t Hoff enthalpy estimate of 397±17 kJ/mol is in good agreement with the calorimetric enthalpy of 387±12 kJ/mol.
Figure 2.
Representative DSC data (top panel) and FTSA data (bottom panel) for lysozyme unfolding at 0.06630 g/L.
A number of important considerations should be made when interpreting observations from thermal shift assays. Larger thermal shifts are observed for entropically-driven binding compared with enthalpically-driven when the binding constants are the same; these observations are true for binding to both the native (positive shifts) and unfolded (negative shifts) states [41]. Binding affinities are therefore underestimated for ligands with very high binding enthalpies and overestimated for high binding entropies. Tm is dependent not only on the binding affinity but also on the enthalpy of binding such that a range of binding affinities and enthalpies can result in the same observed Tm shifts [41]. These competing effects must be considered carefully when interpreting thermal shift data. Tight, enthalpically-driven binding to the native state coupled with weaker, entropically-driven binding to the unfolded state can nullify any thermal shift or result in a negative thermal shift, masking significant interaction with the native state and potentially eliminating the test ligand from the drug discovery screen. The issue is further exacerbated by the need to extrapolate thermal shifts to a common reference temperature for comparison. Small extrapolation to the Tm value of the ligand exhibiting the least thermal shift will minimize extrapolation errors but typically extrapolation is made to a physiological temperature of interest which can represent a significant extrapolation. Waldron and Murphy have shown that such extrapolation can alter the relative interaction affinities of test ligands such that consideration of affinities directly from Tm shifts could led to erroneous conclusions concerning the relative potency of drug candidates [41].
It is clear then that Tm shifts cannot be directly translated into physiological relevant binding affinities but are dependent upon the accurate estimation of enthalpies and heat capacities of unfolding and binding and the unbound ligand concentration. Waldron and Murphy provide an account of the potential errors associated with the determination of accurate ΔCp values including the role of pKa shifts and buffer and pH choices [41]. It is critical to fully characterize binding energetics under a number of biomolecule and ligand concentrations and as a function of solution conditions to explore the effect of factors such as pH and pKa. The most robust approach for the determination of unfolding and binding energetics lies in a combined DSC and ITC approach where direct measurement of enthalpy and heat capacity values of unfolding and binding can be made. Other methods, including FTSA, while providing good monitoring of thermal unfolding do not provide good sensitivity to binding energetics [36,41]. This is partly because of the fact that the melting transition is dominated by unfolding energetics which are typically disproportionately large compared to binding energetics and consequently makes the determination of binding energetics difficult. Moreover, indirect measurement of energetics using methods such as FTSA provide only van’t Hoff estimates and do not represent true energetic determinations using calorimetric techniques. The assumption of negligible heat capacity changes and use of a general binding enthalpy for standalone FTSA approaches is inadequate. Reports exist interpreting thermal shift data with the assumption of a general binding enthalpy for a series of test ligands [37,42]. It is unwise to assume a general enthalpy to describe the undefined binding behaviour of ligands; for some compounds the assumed value may be reasonable but for others it will not. A number of studies have shown that similar ligands although binding with a similar overall binding free energy have significantly different binding enthalpies [41]. Binding enthalpy values can have a substantial effect on the calculation of binding affinities and use of an incorrect enthalpy can have a significantly adverse effect [40,41].
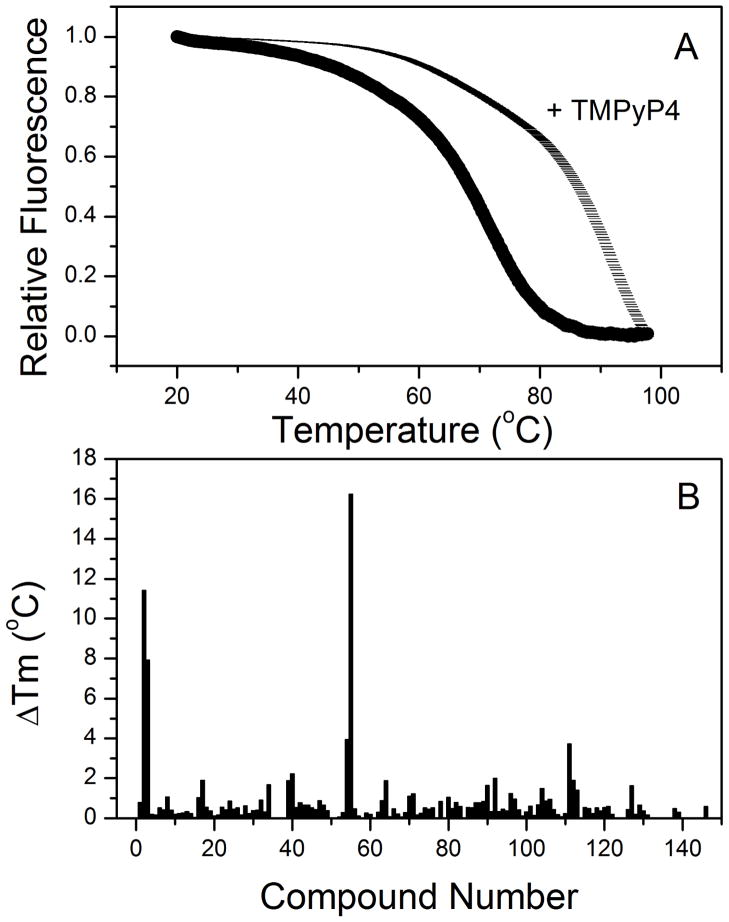
The simplicity and speed of FTSA make it an attractive technique for the primary screening of compound binding provided sufficient caution is exercised in data interpretation with essential input of energetic information using calorimetric methods. A demonstration of the synergy of multiple analysis methods is provided by the following example. FTSA was implemented using an Applied Biosystems StepOnePlus™ Real-Time PCR system to test the 160 best hits from an in silico screen of ligands targeted to the human telomere quadruplex DNA structure [43]. A FRET pair was incorporated into an antiparallel human telomeric quadruplex to provide direct fluorescence monitoring of the denturation event and circumvented issues described earlier from the use of an external fluorescence reporter dye. Figure 3A shows representative melting curves for the FRET-labeled quadruplex in the presence and absence of a known quadruplex binding agent, the cationic porphyrin meso-5,10,15,20-Tetrakis-(N-methyl-4-pyridyl)porphine (TMPyP4). Figure 3B shows the shifts in melting temperatures for 150 compounds selected by virtual screening of the Zinc Database for binding to a quadruplex receptor site. These FTSA results validate the actual binding to a quadruplex of some of the compounds selected by the virtual screen. As part of the drug discovery platform FTSA was utilized as a primary screen of ligand binding with comprehensive secondary validation employing binding studies (calorimetry and spectroscopy), structural selectivity studies (competition dialysis), structural simulations (molecular dynamics) and functional assays (biological assays and tumor inhibition studies).
Figure 3.
Example of a FTSA. Thermal unfolding of the DNA quadruplex structure labeled with a FRET pair (5′FAM-AGGG(TTAGGG)3-TAMR) was used to select for ligand binding. (A) Representative data for the melting of the quadruplex structure alone or in the presence of a cationic porphyrin meso-5,10,15,20-Tetrakis-(N-methyl-4-pyridyl)porphine (TMPyP4). (B) Measured Tm shifts for 150 compounds screened in the assay. Data taken from Holt, 2011 [43].
6.2 Higher throughput calorimetry
Although FTSA has provided much utility in the drug discovery process through its versatility and high throughput screening capacity, energetics determined directly using calorimetry still represent the gold standard. To address the primary issues of calorimetric techniques, those of low throughput and high sample consumption, significant effort has been invested by both researchers and instrument manufacturers. Almost a decade ago, an autosampling DSC was introduced by MicroCal (now part of GE Healthcare) which reported continuous operation with smaller sample volumes and high scanning rates of up to 250 °C/h with rapid cooling and equilibration between scans which facilitated sample throughput of up to 50 samples in a 24 hour period [42]. The instrument design was aimed at the drug discovery industry in order to provide higher throughput for screening of binding constants or testing of solution conditions for formulation stability studies. Design efforts were also focused on data analysis software with the incorporation of functionality which enabled rapid execution of reference scan correction and normalization of large numbers of data sets. TA Instruments have also recently launched a higher throughput automated DSC instrument with arguably better sensitivity and performance for continuous automated unattended operation, with ongoing development of data analysis software for batch processing of data sets. Both instruments provide much increased throughput and improved sample requirements from the previous generation of manual load instruments. Similar improvements have been made in the design of ITC instruments by both manufacturers with automated higher throughput and lower volume models now commercially available.
Despite these significant advances there is a limit to the sample requirements and throughput that can be achieved by the current design of instruments. To achieve truly low sample, high throughput calorimetric instruments it is essential to utilize microscale technology and move away from the current design of serial measurements to the capability of making parallel measurements. Important steps have been taken in both of these areas. Torres et al. provide a good review of the progress that has been made and a summary of developed technologies [44]. The application of advancing microscale technology has explored different chamber designs (either closed chamber [flow] or open chamber [batch]) and thermal detectors (either thermopiles or thermistors). The advantages and disadvantages of these designs are discussed including the effect on sample volumes and evaporation, measurement sensitivity and fabrication challenges.
Two closed-chamber, thermopile, microelectromechanical system (MEMS) flow devices have been reported with encouraging results. A device developed by Wang et al. [45] consisted of microfluidic and thermal chips; the thermal chips incorporated polyimide diaphragms on which polydimethylsiloxane sample chambers were mounted and a pair of thin film thermopile detectors. The polymeric materials afforded a low cost device with good thermal isolation and achieved results for the unfolding of lysozyme and RNase A reference materials in good agreement with accepted values using just 1 μL of sample [45]. Initial results conducted at relatively fast scan rates (5 °C/min) and high sample concentrations (~20 g/L) demonstrated low thermal noise and fast thermal response and the device sensitivity was subsequently improved to allow analysis at more relevant biomolecule concentrations of ~1 g/L [46]. Lercher and co-workers developed a thermopile chip calorimeter with four independently readable thermopile sections deposited onto a silicon nitride membrane with a micro-machined flow channel of 10–26 μL [47–49]. The device demonstrated a heat power detection limit of about 10 nW and was applied to measure the metabolic heat production of microorganisms and enzymatic reactions. Most recently, biotin binding and DNA hybridization binding reactions have been measured [50], although this involved the immobilization of one interacting partner on streptavidin-coated beads and was a move away from the measurement of free, non-immobilized binding partners in calorimetry. These devices represent significant advances in detection and sample requirements but, although with potential for fabrication into calorimeter arrays, are, at present, still only single monitoring units.
Actual prototype calorimeter arrays have been demonstrated for two open chamber calorimeter designs. Although no longer trading, Vivactis developed the Microplate Differential Calorimetry (MiDiCal) technology [51,52] which incorporated a 96-well microtitre-plate device consisting of 48 monitoring units comprising a sample and a reference well located at the hot and cold junction of a thermopile. The fully automated system also comprised an integrated eight channel nanodispenser robotic sampler and software for data acquisition and analysis. When applied to the quantification of ascorbic acid in food and pharmaceutical products, slow kinetics were observed for the monitored reaction of ascorbic acid and ascorbate oxidase resulting from limited diffusion of oxygen cosubstrate from the surrounding air toward the center of the wells [51]. However, with the use of a mathematical model describing the heat generation, fluxes and reactant diffusion in the microplate wells, results were obtained in good agreement with HPLC reference measurements achieving a maximal measurement time of ~60 s per sample in a reaction volume of 12.5 μL with a dynamic range of 0.8–350 mM.
A second device, the enthalpy array, incorporated a detector cell consisting of two adjacent detector regions each containing two thermistors combined in an interconnected Wheatstone bridge [53,54]. Arrays of detector cells were fabricated on a thin polyimide membrane which provided thermal isolation and allows for low cost large-scale fabrication. In each detector cell, a liquid handling system was used to deposit two 250 nL drops of reactants on one of the detector regions and two drops of reference material on the other. After thermal equilibration the drops were mixed in both regions at the same time and the differential temperature measurement between the two regions monitored. The device incorporated a polymer cap to minimize drop evaporation and was housed in a temperature-controlled measurement chamber. An initial design using an electrostatic merging and mixing method revealed limited sensitivity related to poor mixing of sample volumes but this was subsequently improved by incorporating individually addressable magnetic stirring mechanisms for rapid sample mixing in each region. Device performance was validated through the measurement of a number of well characterized enzymatic and binding reactions with results in good accord with the accepted values [54]. The sensitivity of each detector was found to be ~1000 nJ, corresponding with the ability to monitor a ligand-binding reaction of approximately −40 kJ/mol binding enthalpy at a concentration of 50 μM in a sub-microliter sample volume [53].
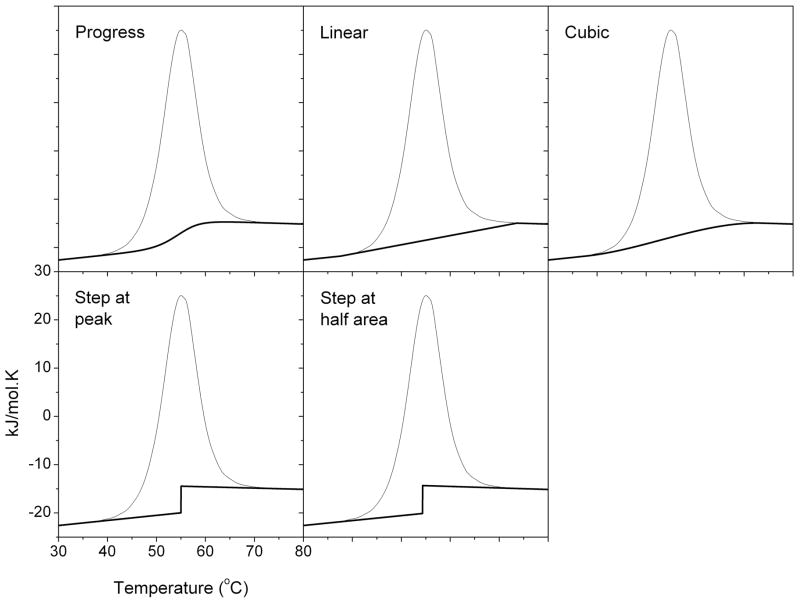
Impressive steps have been taken towards the realization of true high throughput calorimeters; however, to effectively handle the volume of data that will be generated in high throughput applications it is also essential to have efficient analysis approaches. Batch data handling processes developed alongside higher throughput DSC instrument designs are a good start for data management but a pressing issue lies in the high throughput interpretation and analysis of DSC sample baselines. Functionality for the rapid analysis of Tm shifts was incorporated into data analysis software for the higher throughput instrument developed by MicroCal. Analysis of Tm shifts avoids the need for the selection of a sample baseline and simply measures the difference in peak maxima relative to a defined reference scan. While the analysis of Tm shifts has some utility, the ability to rapidly analyze the complete calorimetric profile for full thermodynamic characterization is needed. Cooper et al. have previously discussed the issue of sample baseline selection and showed that this can have a significant effect on determined parameters [26]. Baseline interpretation remains very subjective and currently requires the manual execution of a baseline selection and subtraction procedure. Examples of some of the available baseline subtraction options are shown in Figure 4 with the effects of baseline selection on the analysis of DSC data summarized in Table 2. Data shown are for the unfolding of lysozyme, a well-studied reference material [55]. Table 1 here and analyses by Cooper et al. using modeled data show that variation as a result of baseline selection is not insignificant, with 5–8 % variation in enthalpy and ~0.5 °C in unfolding temperature. Such variations for a well-behaved reference system and modeled data would be expected to increase for the large number of possibly ill-defined systems tested during drug development activities. The manual evaluation and selection of sample baselines for vast data sets would also present an unfeasible bottleneck in data processing. To coordinate with the progress in high throughput data acquisition it is therefore essential to begin to explore the development of automated baseline analyses as part of the better alignment of calorimetric analyses with drug development efforts.
Figure 4.
Examples of sample baseline selection options for the analysis of DSC data. Experimental data for unfolding of lysozyme are shown.
Table 2.
Effects of the selection of different sample baselines on the analysis of DSC data
| Baseline | Tmax (°C) | ΔH (kJ/mol) |
|---|---|---|
| Progress | 54.85 | 385 |
| Linear | 55.02 | 414 |
| Cubic | 54.92 | 399 |
| Step at peak | 55.01 | 389 |
| Step at half area | 54.32 | 385 |
6. Conclusions
Thermodynamic characterization of binding interactions provide invaluable information concerning the molecular forces underlying binding events. Understanding and optimizing these forces in the context of structural information and measured functional effects is key to driving successful drug development efforts. Disparate binding modes with radically different component enthalpic and entropic contributions can be masked by similar affinities and free energies, with structural modification of candidate ligands often resulting in entropy-enthalpy compensation with little change in overall affinity. Measurement of affinities alone are therefore an inadequate assessment of binding interactions and detailed thermodynamic measurements are essential. The implementation of thermodynamic approaches to the measurement of binding interactions has resulted in the development of a number of useful strategies for the thermodynamic optimization of drug-target interactions. Freire and co-workers have successfully applied an enthalpic optimization approach and use of a thermodynamic optimization plot for the evaluation of compound modifications with optimal binding energetics. The use of an enthalpic efficiency index has also been suggested for the identification of compound fragment scaffolds with high efficiency binding interactions. These and other studies have highlighted the value of thermodynamic characterization and demonstrated practical approaches to their measurement. The delineation of thermodynamics into molecular interaction forces is complex and only partially described. However, the utilization of thermodynamic approaches in a focused manner to assess the effect of small and incremental changes in compound structure within a given binding system presents a very viable and practical strategy which will continue to add to our understanding of molecular interactions. The direct determination of binding energetics in the solution state on unmodified, unlabeled, non-immobilized samples using calorimetry represents the best method for thermodynamic characterization of binding interactions. However, high sample consumption and low throughput of traditional calorimetric methods has limited its application to a secondary validation role in drug development. Other surrogate methods namely FTSA has seen broader, earlier application for rapid screening of drug candidates. The utility of thermal shift data as a standalone method can be limited by the difficulties of uncoupling competing effects of enthalpically- and entropically-driven binding to both native and unfolded states and the assumption of generalized binding energetics during data analysis. The most valuable approach at present seems to lie in the utilization of rapid screening methods such as FTSA alongside calorimetric measurements as part of a comprehensive thermodynamic characterization. The development of the next generation calorimetric instruments have yielded lower sample requirements and automated, higher throughput measurements; the beginnings of progress has also been made in efficient data handling and analysis. Further advances have been made in the fabrication of microscale devices and calorimeter arrays that allow parallel measurement of microliter or sub-microliter sample volumes and could realize true high throughput application of comprehensive thermodynamic analyses for the drug development field.
7. Expert Opinion
The most effective drug design and development platform comes from a comprehensive integrated process utilizing all available information from complementary techniques including structural, thermodynamic and biological assays. Drug candidates must ultimately demonstrate the required functional properties assessed with biological assays but the process first involves engineering of desired drug-target interactions. Importantly, rational ligand design has evolved from primarily structural input to a more complete approach which appreciates the balance of energetic forces driving molecular association. A significant body of work exists comprising structural and thermodynamic data, theoretical studies of model systems, and the interpretation and correlation of these data for application to molecular design. However, to realize the goal of thermodynamically-driven drug design significant evolution must occur in two areas: (1) our understanding of the energetic basis of molecular interactions and (2) advances in thermodynamic approaches for widespread application. Currently, much work needs to be done to bridge the connection between structural and thermodynamic information which is of significant complexity because of the enormity of interactions occuring during a binding process and our limited appreciation of these events at the molecular level. Our understanding of biomolecule dynamics and plasticity, the complexity of water behavior and molecular interactions in this environment, observed compensation of binding energetics and the deconvolution of non-covalent interactions is still being developed. By continuing to add to the existing body of structural and thermodynamic data and its application to the study of molecular interactions we will advance our understanding and use of these data for drug development. Crucial to this process is the ability to incorporate broadly applicable, flexible and rapid approaches for thermodynamic characterization. Calorimetry is a highly versatile universal detector of binding reactions performing measurments in solution on unlabeled, unmodified and unimmobilized binding partners. These features afford significant advantages over other methods such as biochemical assays requiring target specific development and optimization. Also, the ability to directly measure component energetic parameters of enthalpy and entropy via calorimetry is essential in defining molecular interactions that is not obtained from measurement of binding affinity or free energy alone. Comprehensive thermodynamic evaluation is vital early in the drug development process to speed drug development towards an optimal energetic interaction profile. Practical approaches, such as enthalpic optimization, thermodynamic optimization plots and the enthalpic efficiency index, have provided more utility in the earlier optimization of binding energetics. However, in its current form calorimetry has significant drawbacks in high sample consumption and low throughput which has significantly restricted its use to secondary validation of drug interactions. Higher throughput thermodynamic methods in the guise of the thermal shift assay were developed to provide utility in early, rapid screening of compound libraries but significant limitations in data interpretation and analysis highlight the necessity for comprehensive calorimetric characterization. The urgent need is the development of high throughput calorimetric approaches which can be used as a universal, rapid, primary screening tool for the characterization of molecular interactions. So far, important advances have been made in the development of current instrumentation to provide reduced sample requirements and automated, higher throughput. However, more exciting and relevant advances have appeared in the fabrication of microscale devices and calorimeter arrays. Significant encouraging results have been obtained but further development in the expansion of the sample array and measurement sensitivity and reproducibility is required before these devices can receive mainstream adoption. The successful development of micro calorimeter arrays are vital in extending the utility of calorimetric approaches, enabling the crucial jump from automated serial measurements to the ability to perform rapid, parallel measurements with only micro- or nanoliter sample requirements. As important as progress in instrumentation is the development of efficient processes for data handling and analysis. The incorporation of some batch processing procedures have appeared with the development of the current generation of commercially available automated single cell calorimeters. However, an essential development that is yet to be realized is the incorporation of automated routines for baseline procedures. This is essential for the efficient and accurate handling of larger volumes of data that will inevitably appear with the incorporation of high throughput calorimeter arrays. Current baseline routines are manually implemented and as such are highly subjective and slow; automated analysis approaches could reduce variability and increase speed. It is important to ensure that high quality goes hand-in-hand with high quantity to provide the full utility of thermodynamic data in the drug development process.
Article highlights.
Thermodynamic characterization of binding interactions is essential for understanding molecular forces driving binding events. A comprehensive integrated approach involving thermodynamic, structural and functional data provides a fuller understanding for the optimization of molecular interactions during drug development.
Key thermodynamic parameters are the binding free energy (ΔG), enthalpy (ΔH), entropy (ΔS) and heat capacity change (ΔCp). ΔG indicates the likelihood and extent of biomolecular interactions and is composed of ΔH and ΔS components, which define the molecular forces governing the binding process. It is the balance of these forces that determines the overall thermodynamics of molecular interactions.
Molecular interactions and the relationship between structural and thermodynamic data are complex and only partially understood. Similar binding affinities and free energies can be observed for disparate binding modes with radically different ΔH and ΔS contributions. Structural modification of candidate ligands can result in compensating changes in ΔH and ΔS with little overall impact on interaction affinities. Detailed thermodynamic measurements are therefore essential and are most useful when applied in a focused way to measure the effect of small and incremental changes in ligand design within a well-characterized binding system.
Useful practical approaches to the optimization of binding energetics have been developed and include an enthalpic optimization approach, use of a thermodynamic optimization plot (TOP) and an enthalpic efficiency (EE) index.
The direct measurement of binding energetics using calorimetry represents the gold standard for the thermodynamic characterization of molecular interactions but requires advances in true high throughput and microscale approaches to realize comprehensive thermodynamic measurements as a primary tool in drug design.
Abbreviations (in order of first appearance)
- ADMET
absorption, distribution, metabolism, excretion and toxicology
- TOP
thermodynamic optimization plot
- EE
enthalpic efficiency
- FBDD
fragment-based drug discovery
- ITC
isothermal titration calorimetry
- DSC
differential scanning calorimetry
- FTSA
fluorescence thermal shift assay
- FRET
fluorescence resonance energy transfer
- MEMS
microelectromechanical system
- MiDiCal
microplate Differential Calorimetry
References
- 1.Henry CM. Structure-based drug design. Chem Eng News. 2001;79(23):69–74. [Google Scholar]
- 2**.Holdgate GA. Thermodynamics of binding interactions in the rational drug design process. Expert Opin Drug Discov. 2007;2(8):1103–1114. doi: 10.1517/17460441.2.8.1103. Very comprehensive and useful discussion of the thermodynamic characterization of binding interactions with a critical discussion of different experimental approaches and the application of thermodynamic measurements to the drug design process. [DOI] [PubMed] [Google Scholar]
- 3.Holdgate GA, Ward WHJ. Measurements of binding thermodynamics in drug discovery. Drug Discov Today. 2005;10(22):1543–1550. doi: 10.1016/S1359-6446(05)03610-X. [DOI] [PubMed] [Google Scholar]
- 4.Chaires JB. Possible origin of differences between van’t hoff and calorimetric enthalpy estimates. Biophys Chem. 1997;64(1–3):15–23. doi: 10.1016/s0301-4622(96)02205-3. [DOI] [PubMed] [Google Scholar]
- 5.Naghibi H, Tamura A, Sturtevant JM. Significant discrepancies between van’t hoff and calorimetric enthalpies. Proc Natl Acad Sci USA. 1995;92(12):5597–5599. doi: 10.1073/pnas.92.12.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6**.Chaires JB. Calorimetry and thermodynamics in drug design. Ann Rev Biophys. 2008;37:135–151. doi: 10.1146/annurev.biophys.36.040306.132812. Excellent review illustrating, with several pertinent examples, the value of obtaining complete thermodynamic profiles in drug design. [DOI] [PubMed] [Google Scholar]
- 7**.Freire E. Do enthalpy and entropy distinguish first in class from best in class? Drug Discov Today. 2008;13(19–20):869–874. doi: 10.1016/j.drudis2008.07.005. Excellent discussion of the use of thermodynamics to guide drug development with particular focus on the enthalpic optimization approach. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8*.Cooper A. Heat does not come in different colours: Entropy-enthalpy compensation, free energy windows, quantum confinement, pressure perturbation calorimetry, solvation and the multiple causes of heat capacity effects in biomolecular interactions. Biophys Chem. 2001;93(2–3):215–230. doi: 10.1016/s0301-4622(01)00222-8. Useful discussion of entropy-enthalpy compensation and its relevance to biological systems. [DOI] [PubMed] [Google Scholar]
- 9.Sharp K. Entropy enthalpy compensation: Fact or artifact? Protein Sci. 2001;10(3):661–667. doi: 10.1110/ps.37801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10*.Freire E. A thermodynamic approach to the affinity optimization of drug candidates. Chem Biol Drug Des. 2009;74(5):468–472. doi: 10.1111/j.1747-0285.2009.00880.x. Description of the thermodynamic optimization plot and its application to drug development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11*.Ruben AJ, Kiso Y, Freire E. Overcoming roadblocks in lead optimization: A thermodynamic perspective. Chem Biol Drug Des. 2006;67(1):2–4. doi: 10.1111/j.1747-0285.2005.00314.x. Introduction of the enthalpy funnel approach and its use for faster thermodynamic optimization of drug leads during drug development. [DOI] [PubMed] [Google Scholar]
- 12*.Whitesides GM, Krishnamurthy VM. Designing ligands to bind proteins. Q Rev Biophys. 2005;38(4):385–395. doi: 10.1017/S0033583506004240. Interesting perspective on the progress towards the goal of true rational drug design highlighting important technological developments alongside limitations in our current understanding of molecular interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13**.Ladbury JE, Klebe G, Freire E. Adding calorimetric data to decision making in lead discovery: A hot tip. Nat Rev Drug Discov. 2010;9(1):23–27. doi: 10.1038/nrd3054. Helpful overview describing the utility and use of thermodynamic data in lead discovery and optimization with specific application of ITC. Discusses the use of an enthalpic efficiency metric to identify optimal drug interactions and provide a valuable complementary approach for the discovery and optimization of drug leads. [DOI] [PubMed] [Google Scholar]
- 14.Ferenczy GG, Keser GM. Thermodynamics guided lead discovery and optimization. Drug Discov Today. 2010;15(21–22):919–932. doi: 10.1016/j.drudis.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 15.Ferenczy GG, Keser GM. Enthalpic efficiency of ligand binding. J Chem Inf Model. 2010;50(9):1536–1541. doi: 10.1021/ci100125a. [DOI] [PubMed] [Google Scholar]
- 16.Meanwell NA. Improving drug candidates by design: A focus on physicochemical properties as a means of improving compound disposition and safety. Chem Res Toxicol. 2011;24(9):1420–1456. doi: 10.1021/tx200211v. [DOI] [PubMed] [Google Scholar]
- 17**.Bruylants G, Wouters J, Michaux C. Differential scanning calorimetry in life science: Thermodynamics, stability, molecular recognition and application in drug design. Curr Med Chem. 2005;12(17):2011–2020. doi: 10.2174/0929867054546564. Detailed review of the use of DSC to assess the stability and ligand interactions of biomolecules with discussion of practical approaches and examples with a focus on application to formulation and drug discovery studies. [DOI] [PubMed] [Google Scholar]
- 18.Garbett NC. The use of calorimetry to study ligand-DNA interactions. In. In: Aldrich-Wright J, editor. Metallointercalators. Springer-Verlag/Wien; Heidelberg, Germany: 2011. pp. 299–324. [Google Scholar]
- 19.Pierce MM, Raman CS, Nall BT. Isothermal titration calorimetry of protein-protein interactions. Methods. 1999;19(2):213–221. doi: 10.1006/meth.1999.0852. [DOI] [PubMed] [Google Scholar]
- 20.Spink CH, Wellman SE. Thermal denaturation as a tool to study DNA-ligand interactions. Methods Enzymol. 2001;340:193–211. doi: 10.1016/s0076-6879(01)40423-x. [DOI] [PubMed] [Google Scholar]
- 21.Chaires JB, Shi X. Thermal denaturation of drug-DNA complexes: Tools and tricks. In: Waring MJ, editor. Sequence-specific DNA binding agents. Royal Society of Chemistry; Cambridge: 2006. pp. 130–151. [Google Scholar]
- 22.Crothers DM. Statistical thermodynamics of nucleic acid melting transitions with coupled binding equilibria. Biopolymers. 1971;10(11):2147–2160. doi: 10.1002/bip.360101110. [DOI] [PubMed] [Google Scholar]
- 23.McGhee JD. Theoretical calculations of the helix-coil transition of DNA in the presence of large, cooperatively binding ligands. Biopolymers. 1976;15 (7):1345–1375. doi: 10.1002/bip.1976.360150710. [DOI] [PubMed] [Google Scholar]
- 24.Schellman JA. The factors affecting the stability of hydrogen-bonded polypeptide structures in solution. J Phys Chem. 1958;62(12):1485–1494. [Google Scholar]
- 25.Jelesarov I, Bosshard HR. Isothermal titration calorimetry and differential scanning calorimetry as complementary tools to investigate the energetics of biomolecular recognition. J Mol Recognit. 1999;12:3–18. doi: 10.1002/(SICI)1099-1352(199901/02)12:1<3::AID-JMR441>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 26**.Cooper A, Nutley MA, Wadood A. Differential scanning microcalorimetry. In: Chowdhry BZ, Harding SE, editors. Protein-ligand interactions: Hydrodynamics and calorimetry: A practical approach. Oxford University Press; Oxford, UK: 2001. pp. 287–318. Excellent review of DSC with comprehensive description of experimental procedures and analysis considerations and approaches for the characterization of biomolecule unfolding and ligand binding. Discussion of the DSC sample baseline issue. [Google Scholar]
- 27.Brandts JF, Lin L-N. Study of strong to ultratight protein interactions using differential scanning calorimetry. Biochemistry. 1990;29(29):6927–6940. doi: 10.1021/bi00481a024. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y-L, Zhang Z-Y. Low-affinity binding determined by titration calorimetry using a high-affinity coupling ligand: A thermodynamic study of ligand binding to protein tyrosine phosphatase 1b. Anal Biochem. 1998;261(2):139–148. doi: 10.1006/abio.1998.2738. [DOI] [PubMed] [Google Scholar]
- 29.Ren J, Jenkins TC, Chaires JB. Energetics of intercalation reactions. Biochemistry. 2000;39(29):8439–8447. doi: 10.1021/bi000474a. [DOI] [PubMed] [Google Scholar]
- 30.Bishop GR, Ren J, Polander BC, Jeanfreau BD, Trent JO, Chaires JB. Energetic basis of molecular recognition in a DNA aptamer. Biophys Chem. 2007;126(1–3):165–175. doi: 10.1016/j.bpc.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Ladbury JE. Calorimetry as a tool for understanding biomolecular interactions and an aid to drug design. Biochem Soc Trans. 2010;38(4):888–893. doi: 10.1042/BST0380888. [DOI] [PubMed] [Google Scholar]
- 32.Schwalbe H, Rinnenthal J. Thermodynamics: The world is flat. Nat Chem Biol. 2010;6(5):312–313. doi: 10.1038/nchembio.357. [DOI] [PubMed] [Google Scholar]
- 33.Bissantz C, Kuhn B, Stahl M. A medicinal chemist’s guide to molecular interactions. J Med Chem. 2010;53(14):5061–5084. doi: 10.1021/jm100112j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott AD, Phillips C, Alex A, Flocco M, Bent A, Randall A, O’Brien R, Damian L, Jones LH. Thermodynamic optimization in drug discovery: A case study using carbonic anhydrase inhibitors. Chem Med Chem. 2009;4:1985–1989. doi: 10.1002/cmdc.200900386. [DOI] [PubMed] [Google Scholar]
- 35.Reynolds CH, Holloway MK. Thermodynamics of ligand binding and efficiency. ACS Med Chem Lett. 2011;2(6):433–437. doi: 10.1021/ml200010k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36**.Zhang R, Monsma F. Fluorescence-based thermal shift assays. Curr Opin Drug Discov Develop. 2010;13(4):389–402. Critical discussion of fluorescence-based thermal shift assays including applications, advantages and disadvantages, experimental protocols and data analysis and interpretation. [PubMed] [Google Scholar]
- 37*.Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, Carver T, Asel E, Springer BA, Lane P, Salemme FR. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screen. 2001;6(6):429–440. doi: 10.1177/108705710100600609. Original description of the ThermoFluor technology and its application to drug discovery. [DOI] [PubMed] [Google Scholar]
- 38.Cummings MD, Farnum MA, Nelen MI. Universal screening methods and applications of thermofluorR. J Biomol Screen. 2006;11(7):854–863. doi: 10.1177/1087057106292746. [DOI] [PubMed] [Google Scholar]
- 39.Todd MJ, Salemme FR. Direct binding assays for pharma screening. Genetic Engineering News. 2003;23(3) [Google Scholar]
- 40*.Lo M-C, Aulabaugh A, Jin G, Cowling R, Bard J, Malamas M, Ellestad G. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal Biochem. 2004;332(1):153–159. doi: 10.1016/j.ab.2004.04.031. Report of the use of an inexpensive, commercially available RT-PCR thermocycler for protein thermal shift assays. Comparison of results from ITC and enzymatic assays and discussion of data analysis approaches and pitfalls. [DOI] [PubMed] [Google Scholar]
- 41**.Waldron TT, Murphy KP. Stabilization of proteins by ligand binding: Application to drug screening and determination of unfolding energetics. Biochemistry. 2003;42(17):5058–5064. doi: 10.1021/bi034212v. Use of simulation to demonstrate the complexity of biomolecule stability and ligand interactions underlying thermal shift observations with emphasis on drug screening applications. Discussion of appropriate caution and approaches necessary for the interpretation of thermal shift data and determination of reliable binding energetics. [DOI] [PubMed] [Google Scholar]
- 42.Plotnikov V, Rochalski A, Brandts M, Brandts JF, Williston S, Frasca V, Lin L-N. An autosampling differential scanning calorimeter instrument for studying molecular interactions. Assay Drug Dev Technol. 2002;1(1–1):83–90. doi: 10.1089/154065802761001338. [DOI] [PubMed] [Google Scholar]
- 43.Holt PA, Buscaglia R, Trent JO, Chaires JB. A discovery funnel for nucleic acid binding drug candidates. Drug Dev Res. 2011;72(2):178–186. doi: 10.1002/ddr.20414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44**.Torres FE, Recht MI, Coyle JE, Bruce RH, Williams G. Higher throughput calorimetry: Opportunities, approaches and challenges. Curr Opin Struct Biol. 2010;20:598–605. doi: 10.1016/j.sbi.2010.09.001. the progress in higher throughput calorimetry, with particular discussion of the development and limitations of higher throughput instrumentation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Wang B, Lin Q. Demonstration of mems-based differential scanning calorimetry for determining thermodynamic properties of biomolecules. Sensors and Actuators B. 2008;134:953–958. [Google Scholar]
- 46.Wang B, Lin Q. A mems differential scanning calorimeter for thermodynamic characterization of biomolecules. 2011 IEEE 24th International Conference on Micro Electro Mechanical Systems (MEMS); Cancun, Mexico. 2011; Abs 821–824. [Google Scholar]
- 47.Lerchner J, Wolf A, Wolf G, Baier V, Kessler E, Nietzsch M, Krugel M. A new micro-fluid chip calorimeter for biochemical applications. Thermochim Acta. 2006;445(2):144–150. [Google Scholar]
- 48.Lerchner J, Wolf A, Schneider H-J, Mertens F, Kessler E, Baier V, Funfak A, Nietzsch M, Krugel M. Nano-calorimetry of small-sized biological samples. Thermochim Acta. 2008;477(1–2):48–53. [Google Scholar]
- 49.Lerchner J, Maskow T, Wolf G. Chip calorimetry and its use for biochemical and cell biological investigations. Chem Eng Process. 2008;47(6):991–999. [Google Scholar]
- 50.Ahmad LM, Towe B, Wolf A, Mertens F, Lerchner J. Binding event measurement using a chip calorimeter coupled to magnetic beads. Sens Actuators B Chem. 2010;145(1):239–245. [Google Scholar]
- 51.Vermeir S, Nicolai BM, Verboven P, Van Gerwen P, Baeten B, Hoflack L, Vulsteke V, Lammertyn J. Microplate differential calorimetric biosensor for ascorbic acid analysis in food and pharmaceuticals. Anal Chem. 2007;79 (16):6119–6127. doi: 10.1021/ac070325z. [DOI] [PubMed] [Google Scholar]
- 52.Verhaegen K, Baert K, Simaels J, Van Driessche W. A high-throughput silicon microphysiometer. Sens Actuators A Phys. 2000;82(1–3):186–190. [Google Scholar]
- 53*.Torres FE, Kuhn P, De Bruyker D, Bell AG, Wolkin MV, Peeters E, Williamson JR, Anderson GB, Schmitz GP, Recht MI, Schweizer S, et al. Enthalpy arrays. Proc Natl Acad Sci U S A. 2004;101(26):9517–9522. doi: 10.1073/pnas.0403573101. Description of enthalpy arrays and their application to the measurement of biomolecular interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Recht MI, Torres FE, De Bruyker D, Bell AG, Klumpp M, Bruce RH. Measurement of enzyme kinetics and inhibitor constants using enthalpy arrays. Anal Biochem. 2009;388(2):204–212. doi: 10.1016/j.ab.2009.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hinz H-J, Schwarz FP. Measurement and analysis of results obtained on biological substances with differential scanning calorimetry. Pure Appl Chem. 2001;73(4):745–759. [Google Scholar]