Abstract
MicroRNAs (miRs) are evolutionarily conserved, noncoding RNA molecules of ∼21 nt that regulate the expression of genes that are involved in various biological processes, such as cell proliferation and differentiation. Previously, we reported the presence of a heterogeneous population of mRNAs present in the axons and nerve terminals of primary sympathetic neurons to include the nuclear-encoded mitochondrial mRNA coding for COXIV. Sequence analysis of the 3′UTR of this mRNA revealed the presence of a putative binding site for miR-338, a brain-specific microRNA. Transfection of precursor miR-338 into the axons of primary sympathetic neurons decreases COXIV mRNA and protein levels and results in a decrease in mitochondrial activity, as measured by the reduction of ATP levels. Conversely, the transfection of synthetic anti-miR oligonucleotides that inhibit miR-338 increases COXIV levels, and results in a significant increase in oxidative phosphorylation and also norepinephrine uptake in the axons. Our results point to a molecular mechanism by which this microRNA participates in the regulation of axonal respiration and function by modulating the levels of COXIV, a protein which plays a key role in the assembly of the mitochondrial cytochrome c oxidase complex IV.
Keywords: mitochondria, ATP synthesis, RNA localization, inhibitory RNA, oxidative phosphorylation, local translation, norepinephrine uptake
Introduction
Over the past few years, it has become widely accepted that a distinct subset of neuronal mRNAs are selectively transported to the distal structural/functional domains of the neuron, including the axon and presynaptic nerve terminal. Local proteins synthesized from these mRNAs play a key role in the development of the neuron and the function of the axon and nerve terminal (Campbell et al., 2001; Wu et al., 2005; Poon et al., 2006; Hillefors et al., 2007; Cox et al., 2008). The importance of local protein synthesis for mitochondrial function and viability of distal axons was demonstrated in previous studies (Hillefors et al., 2007). Mitochondria are thought to be closely associated with synapses and tethered to vesicle release sites (Zenisek and Matthews, 2000). Synaptic transmission requires mitochondrial ATP generation and control of local [Ca2+]i for neurotransmitter exocytosis, vesicle recruitment, and potentiation of neurotransmitter release (Chang et al., 2006). Results derived from an invertebrate model system revealed that ∼25% of the total protein synthesized locally in the nerve terminal were destined for the mitochondria (Gioio et al., 2004). Other studies demonstrated that either the inhibition of local protein synthesis or the blockade of local protein transport into the organelle significantly reduced mitochondrial membrane potential and inhibited the mitochondria's ability to restore axonal levels of ATP after KCl-induced depolarization (Gioio et al., 2001, 2004; Hillefors et al., 2007).
Novel molecular mechanisms involving noncoding RNAs have recently been shown to spatially regulate mRNA translation in axons and dendrites. Ashraf et al. (2006) demonstrated that memory-specific patterns of synaptic protein synthesis occur with the induction of a long-term memory in Drosophila, and that these patterns appear to be controlled by the proteasome-mediated degradation of a RISC pathway component. Other studies identified a dendritically localized miR that regulates the expression of the synaptic Limk1 protein, thereby controlling dendritic spine size (Schratt et al., 2006). Importantly, Hengst et al. (2006) have shown that key proteins involved in the RNAi/miR pathway, i.e., RISC complexes can assemble and function in developing axons.
Previously, we reported that several nuclear-encoded mitochondrial mRNAs, such as the mRNA encoding COXIV, were present in the distal axons of rat sympathetic neurons (Hillefors et al., 2007). COXIV has been demonstrated to have an essential role in the assembly of the cytochrome c oxidase complex, suggesting a tight coupling of the local synthesis of cytochrome c oxidase and oxidative phosphorylation (Li et al., 2006). To assess the potential involvement of miRs in the control of the local synthesis of nuclear-encoded mitochondrial proteins in neurons, we analyzed the interrelationship between COXIV and one of its cognate miRs, miR-338. We found that levels of miR-338 increased during axonal outgrowth and maturation, and further demonstrated that this miR can modulate local COXIV levels and oxidative phosphorylation in the distal axons. Together, these findings identify a novel mechanism for the local regulation of axonal protein synthesis and respiration by miR in sympathetic neurons.
Materials and Methods
Neuronal cell cultures.
SCG were obtained from 3-d-old Harlan Sprague Dawley rats, and dissociated neurons plated in the center compartment of Campenot compartmented culture dishes as previously described (Hillefors et al., 2007). Cells were cultured in serum-free medium containing NGF (50 ng/ml) for 14–21 d before use with media changes every 3–4 d. The complete culture media, including NGF was present in both the central and side compartments throughout the culture period and during all experimental procedures. The side compartments, which contained the distal axons used in these experiments, contained no neuronal soma or non-neuronal cells, as judged by phase-contrast microscopy, as well as ethidium bromide and acridine orange staining.
Bioinformatics and miR target prediction.
The miRanda algorithm (John et al., 2004) was used to investigate the 3′UTR sequence of rat COXIV mRNA for putative binding sites of miRs. MiR-338 was selected for further analyses as judged by the low predicted free energy of hybridization with the COXIV mRNA target (−14.9 kcal/mol), and the secondary structure prediction analysis of the COXIV 3′UTR using Mfold (Zuker, 2003). Constructs and primers used in this report were designed using VectorNTI (Invitrogen).
Luciferase reporter gene constructs and luciferase assay.
The sense and antisense strands of oligonucleotides coding for the full rat COXIV 3′UTR (for sequence, see Fig. 1B), or the 3′UTR minus the putative miR-338 targeting site were synthesized (Invitrogen). Oligonucleotides were annealed and ligated into the HindIII and SpeI sites of pmir-Report luciferase vector (Ambion). Cells were cotransfected with pmir-REPORT β-galactosidase (β-gal), and the luciferase reporter constructs containing either the full COXIV 3′UTR or 3′UTR lacking the miR-338-binding site (ΔMTS), together with inhibitors for miR-338 or a nontargeting control (NT), respectively. SCG neurons were also transfected with a control luciferase reporter vector to determine the level of activity that can be achieved for an unmodified pmir-REPORT luciferase. Twenty-four hours after transfection, cells were assayed for firefly luciferase and β-gal expression, and β-gal was used to normalize for differences in transfection efficiency. The Dual-Light luminescent reporter gene assay (Applied Biosystems) was used for the detection of firefly luciferase and β-gal in the same sample.
Figure 1.
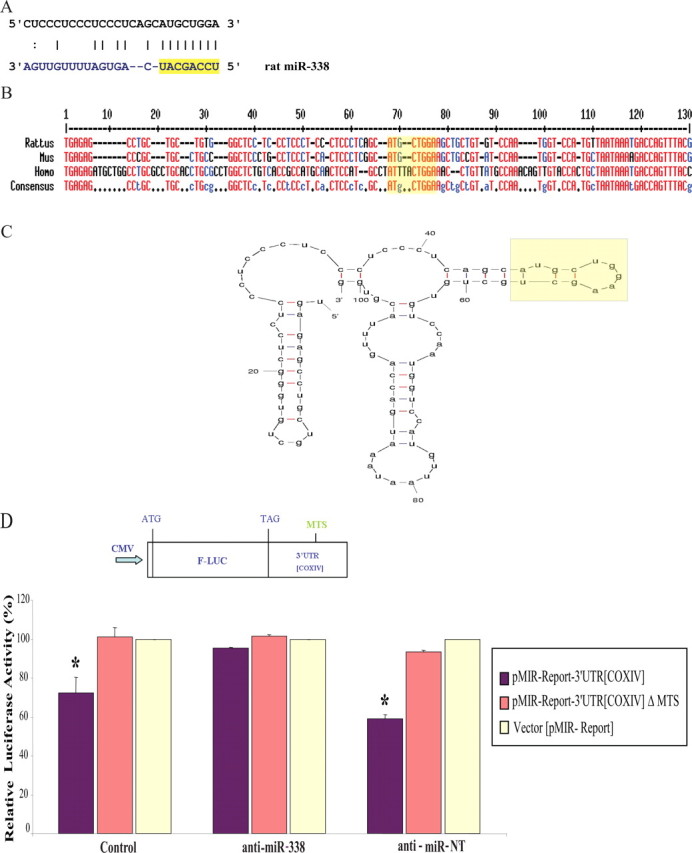
MiR-338 targets rat COXIV. A, Putative miR-338 binding site in the rat COXIV 3′UTR. The 8 nt seed sequence is marked in yellow. B, C, Sequence (B) and secondary structure (C) of the COXIV 3′UTR, as determined by secondary structure prediction analysis (Mfold). The miR-338 target site (MTS) is indicated in yellow. D, The luciferase reporter plasmids carrying the firefly luciferase coding sequence (F-LUC) attached to the entire 3′UTR of rat COXIV (wild-type) or the 3′UTR in which the miR-338 target site was deleted (ΔMTS) are diagrammed. Luciferase expression was driven by the cytomegalovirus promoter (CMV). Luciferase activity of wild-type or ΔMTS COXIV 3′UTR reporter genes in the absence (control) or presence of the anti-miR-338 (25 nm), or a nontargeting (NT) anti-miR oligonucleotide (25 nm), is shown. Luciferase reporter constructs were cotransfected in equal ratios with the control plasmids coding for β-galactosidase (0.5 μg for each plasmid per transfection). The ratio of reporter to control plasmids in relative luminescence units was normalized for each reporter and plotted as a percentage of sham-transfected control value. Anti-miR oligonucleotide transfections were performed within 4 h following plasmid DNA transfection. Error bars represent the SEM for n = 10 cultures. *p < 0.05.
Immunocytochemical analyses.
SCG neurons grown in Campenot chambers for 15 d were first fixed with 4% paraformaldehyde in PBS for 1 h at ambient temperature. After washing in PBS, neurons were subsequently permeabilized with 0.3% Triton X-100 in PBS for 30 min and blocked with 10% normal donkey serum in PBS for 1 h. Rabbit polyclonal antibodies against DICER and eIF2c were kindly provided by Dr. Neil Smalheiser (University of Illinois, Chicago, IL). Incubation with anti-DICER or anti-eIF2c antibodies diluted 1:700 and 1:1000, respectively, in blocking solution (2% normal donkey serum in PBS), was performed overnight at 4°C. Cells were then incubated with donkey Cy3-labeled, affinity-purified anti-rabbit IgG (Jackson ImmunoResearch) diluted 1:200 in blocking solution for 1 h and washed in PBS. Images were captured using a Nikon Eclipse TE 300 fluorescence microscope equipped with a Nikon Digital Sight DS-L1 camera.
Analyses of miR and COXIV mRNA.
For in situ hybridization of miR-338 and the scramble nontargeting miR, digoxigenin-labeled antisense-locked nucleic acid (LNA) oligonucleotides were obtained from Exiqon. Tailed LNA oligonucleotides were purified and used for overnight hybridization at 37°C. The probe concentration in the hybridization mix was 0.3 μm. All other hybridization conditions and procedures were previously described (Hillefors et al., 2007).
For the analysis of miR-338 expression by quantitative RT-PCR, distal axons located in the side chambers and soma and proximal axons in the central chamber were harvested separately, and total RNA prepared using the Cells-to-Signal lysis buffer (Ambion), and used directly for reverse transcription. The TaqMan microRNA Assay (Applied Biosystems) was used to quantify the expression of the mature miR-338 and values expressed relative to β-actin mRNA. TaqMan MicroRNA Assays use stem-looped primers that enable a two-step quantification of miRs present in a sample. In the first step, stem-looped primers anneal to target mature miRs and extend the length of the molecule by reverse transcription PCR. In the second step, a real-time PCR that involves a forward primer, a reverse primer, and a TaqMan probe quantifies the number of mature miR molecules present in a sample based on fluorescent emission of a reporter dye (Chen et al., 2005).
For COXIV mRNA analyses, quantitative RT-PCR was performed on total RNA prepared from SCG axons and cell somas using the Cells-to-Signal lysis buffer (Ambion). RT-PCR was done essentially as previously described (Hillefors et al., 2007). The relative levels of each transcript were normalized to β-actin mRNA to provide an internal control for reverse transcription and axonal density. RNA values are expressed relative to control by the comparative threshold method (CT).
Preparation of siRNA for COXIV silencing.
Two independent siRNAs targeting rat COXIV were tested for silencing in SCG neurons. The siRNA sense and antisense strands were purchased from Dharmacon with the following sequences: siRNA-1, sense 5′-GGAGUGUUGUGAAGAGUGAUU-3′, antisense 5′-UCACUCUUCACAACACUCCUU-3′; siRNA-2, sense 5′-CCUCAUACCUUUGAUCGUGUU-3′, antisense 5′-CACGAUCAAAGGUAUGAGGUU-3′; and scramble control, sense 5′-UAGCGACUAAACACAUCAA-3′, antisense 5′-UAAGGCUAUGAAGAGAUAC-3′. The capacity of each siRNA to reduce the expression of COXIV was determined by transient transfection into the distal axons, or soma and proximal axons as explained below. The expression level of COXIV was determined by qRT-PCR and Western blotting using rabbit monoclonal antibodies against rat COXIV (Cell Signaling Technology).
Transfection of neurons with luciferase reporter plasmids, siRNAs, miR precursors, and anti-miRs.
The transfection of reporter gene plasmids into the neuronal soma located in the central compartment of the Campenot cultures was conducted using NeuroPorter (Genlantis) according to the manufacturer's instructions. The double-stranded RNA that mimics endogenous rat precursor miR-338, and miR-NT, used as a nontargeting precursor control, were obtained from Ambion. In addition, the miR inhibitor, anti-miR-338, as well as nontargeting control anti-miR-NT were obtained from Ambion. The introduction of small RNAs (miRs or siRNAs, each at 25 nm final concentration) into the distal axons located in the side chambers of the culture dishes, or the soma and proximal axons in the center compartment was accomplished by lipofection using siPORT NeoFX (Ambion). The Campenot compartmented culture used in the present studies contained two lateral compartments that harbored the distal axons, both were transfected independently, and the total RNA were tested separately from each other to increase the sample number for the analyses.
Immunoblot assay.
Distal axons in the side chambers or soma and proximal axons in the central chamber were harvested separately and lysed in 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% NP-40, and Complete protease inhibitor mixture (Roche). Equal amounts of each lysate were applied to Hybond ECL nitrocellulose membranes (GE Healthcare). Membranes were blocked using the reagent provided with the ECL plus Western blotting detection kit (GE Healthcare) for 1 h and incubated for 1 h with rabbit monoclonal antibodies against either rat COXIV, or rat β-actin (Sigma). Membranes were washed in 1× TBS-T and incubated with horseradish peroxidase-labeled secondary antibody for 1 h at room temperature. After washing, membranes were developed with the ECL plus Western blotting detection reagents. Dot blots were quantified using the NIH software Image J. The specificity of the antibodies used was assessed by SDS-PAGE Western blot analysis. A single band was detected at the expected molecular weight corresponding to COXIV and β-actin polypeptides, respectively.
Mitochondrial functional assays.
The Alamar Blue (AB) reduction assay was used to examine the metabolic activity of mitochondria in SCG neuronal cultures transfected with anti-miR-338, miR-338 precursor, or nontargeting short oligonucleotides in the axon and soma compartments. AB is an oxidation-reduction indicating dye that produces a colorimetric change in response to metabolic activity. The AB assay evaluates the metabolic activity of cells based on the reduction of resazurin (blue and nonfluorescent) to resorufin (pink and highly fluorescent) in the presence of metabolically active cells (Nakayama et al., 1997). This indicator is set up to detect oxidation by the whole of the electron transport chain. With AB the cells remain fully functional and healthy, unaffected by the presence of the indicator. Ten percent AB (AbD Serotec) was added to either axonal side chambers or center chambers immediately after lipofection, and the cultures were incubated for 24–48 h as indicated. Absorbance was quantified by the measurements at 570 and 595 nm, using a microplate reader (UVSpectramax, Molecular Devices), and triplicate samples run from each side compartment sample. The relative levels of AB absorbance from anti-miR-338 or miR precursor lipofected axons were compared with nontargeted miR and sham-transfected controls, and expressed as percentage of control.
ATP levels were assessed using CellTiter-Glo luminescent cell viability assay from Promega using the manufacturer's instruction. Briefly, SCG neurons were transfected with anti-miR-338 or the nontargeting control oligonucleotide (anti-miR-NT). After transfection (24 h), the distal axons and soma were independently harvested, and CellTiter-Glo reagent (100 μl) was added to lyse the soma and distal axons, and the luminescence was then recorded in a luminometer with an integration time of 1 s per well. The luminescent signals for the anti-miR-338- and anti-miR-NT-treated cells were normalized to the relative luminescent signal of mock-treated cells.
Statistical analysis.
Quantitative data are presented as the mean ± SEM. Student's t test was used to determine significant differences between two groups. One-way ANOVA was used to analyze significant differences among multiple groups; p ≤ 0.05 was considered significant.
Results
MiR-338 responsive element in rat COXIV mRNA
The relatively short rat COXIV 3′UTR contains a specific 8-mer which has the potential to function as a putative hybridization site for miR-338 (Fig. 1A). A RNA secondary structure prediction analysis using Mfold (Zuker, 2003) revealed that the apparent miR-338 target site (MTS) was positioned on a hairpin-loop structure, in an exposed position, that might facilitate miR accessibility (Fig. 1C). The predicted MTS in COXIV 3′UTR had a low predicted free energy of hybridization with the cognate miR (−14.9 kcal/mol), suggesting a stable miR:MTS duplex within the 8 nt seed region at the 5′ end of the miR. This seed sequence is an important determinant of miR-induced repression of gene expression (Doench and Sharp, 2004).
To assess whether miR-338 can specifically target COXIV mRNAs, dissociated superior cervical ganglia (SCG) neurons were isolated from 3-d-old rats and cultured in Campenot multicompartment chambers for 14–21 d and subsequently transfected with either a luciferase reporter plasmid containing the COXIV 3′UTR, or a control luciferase plasmid (Fig. 1D). When compared with the control condition, the presence of the COXIV 3′UTR reduced luciferase activity ∼25–50%. Luciferase levels did not change when the anti-miR-338 was cotransfected with this reporter plasmid, indicating that COXIV is specifically targeted by miR-338. Results of a deletion experiment confirmed that the putative MTS was specifically targeted by miR-338. For example, if the 8-mer seed sequence was removed from the 3′UTR construct, luciferase levels did not vary from those of the control reporter (Fig. 1D). Identical results were obtained using the rat neuroblastoma B35 cell line (data not shown).
MiR-338 abundance during axonal outgrowth and maturation
Although miR-338 is known to be specifically expressed in neuronal tissue (Kim et al., 2004; Wienholds et al., 2005), little is known about its abundance and function during neuronal maturation and axonal outgrowth. Using in situ hybridization (ISH), the subcellular localization of miR-338 RNA within SCG neurons cultured in Campenot multicompartment chambers (Campenot, 1977) was examined. Unlike the scrambled miR probe, used as a negative control, hybridization with the miR-338-specific probe revealed the presence of miR-338 in the distal axons (Fig. 2A), as well as the soma and proximal axons (Fig. 2B). In addition, the miR-338 expression during axonal outgrowth was assessed using a TaqMan microRNA assay. MiR-338 expression levels were normalized to β-actin mRNA, which is relatively abundant in the axons of these neurons (Eng et al., 1999). After 3 d in culture, miR-338 was expressed at low levels throughout the neuron, but after 21 d in culture, its relative abundance had increased approximately threefold to fourfold in the soma, and sixfold in distal axons (Fig. 2E,F), suggesting that this miR may play a role in neuronal maturation, particularly in the elaboration of the structure and function of SCG axons.
Figure 2.
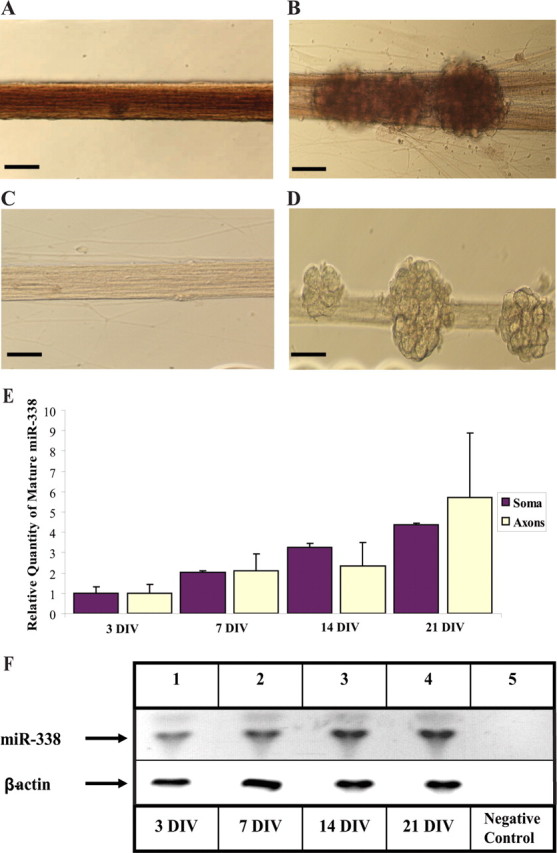
Mature miR-338 is expressed in SCG neurons and is present in proximal and distal axons. A–D, Visualization of the presence of miR-338 at 21 d in culture by ISH in SCG axons (A), or soma and proximal axons (B); the scramble control probes show no significant staining in the soma or axons (C, D); scale bars, 10 μm. E, miR-338 levels increase during axonal outgrowth. The TaqMan miR Assay was used to quantify mature miR-338 isolated from either the soma and proximal axons (Soma), or distal axons (Axons) of SCG neurons at various DIV. Levels of miR-338 are expressed relative to β-actin mRNA which was used as an internal control. Error bars represent the SEM for n = 6 independent experiments. F, Samples obtained from the PCRs to quantify miR-338 in the distal axons (E) were size-fractionated on 4% agarose gels containing ethidium bromide and visualized using UV absorption (254 nm wavelength). Levels of β-actin mRNA served as an internal control. Negative control, PCR minus cDNA.
Regulation of axonal levels of COXIV mRNA and protein by miR-338
To investigate the possibility that mature miRs might function in the axons, the expression and localization of Dicer and the RNA-induced silencing complex (RISC) component eIF2c in the distal axons, as well as proximal axons and soma of sympathetic neurons was studied by immunocytochemistry. A recent report has shown that DRG axons in culture are capable of autonomously silencing a gene without the contribution of the cell body, thereby spatially regulating gene expression (Hengst et al., 2006). Antibodies against rat eIF2c and DICER were used to visualize RISC complexes in SCG neurons by fluorescence microscopy. DICER and eIF2c antibodies revealed the presence of granule-like structures in the cell bodies and along the entire length of the axon (Fig. 3A,B). These granular structures are believed to represent complexes of RNA and proteins (RNP particles). The demonstration that the proteins DICER and eIF2c are localized in the distal axons supports the hypothesis that microRNA plays a role in the regulation of mRNA levels in the axons.
Figure 3.
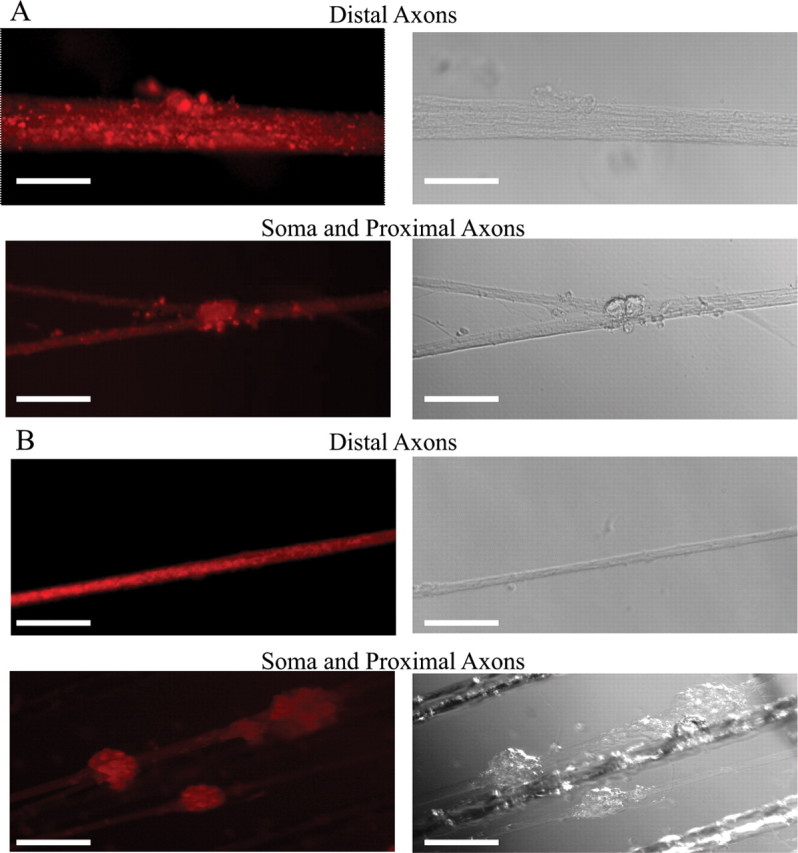
Localization of Dicer and eIF2c in the soma and axons of sympathetic neurons. A, Immunohistochemical examination of Dicer revealed granule-like structures visible in the soma and along the entire length of the axons of rat SCG neuronal cultures (15 DIV). B, Immunohistochemical analyses further visualized eIf2c in the axons and soma of SCG neurons using antibodies against this protein. Reactions conducted with Cy3-labeled secondary antibody alone produced only faint fluorescence. Scale bars: A, distal axons: 10 μm; soma and proximal axons: 50 μm; B, 50 μm.
To assess pre-miR processing in the distal axons, SCG axons were transfected with the precursor miR-338. Subsequently, the steady-state levels of mature miR-338 was determined using specific stem-loop primers for reverse transcription (RT) of mature miR-338 followed by real-time TaqMan within a two-step RT-PCR assay. Transfection with precursor miR-338 resulted in an ∼10- and 42-fold increase in mature miR-338 levels compared with the endogenous miR-338 levels in sham-transfected axons 1 and 4 h after transfection, respectively. After 24 h, we observed a 42,000-fold increase in mature miR-338 levels in the axons, compared with miR-338 levels in sham-transfected control axons (data not shown). These results demonstrated that the distal axons of SCG neurons have the capability of processing microRNA precursors to the mature form of the molecule.
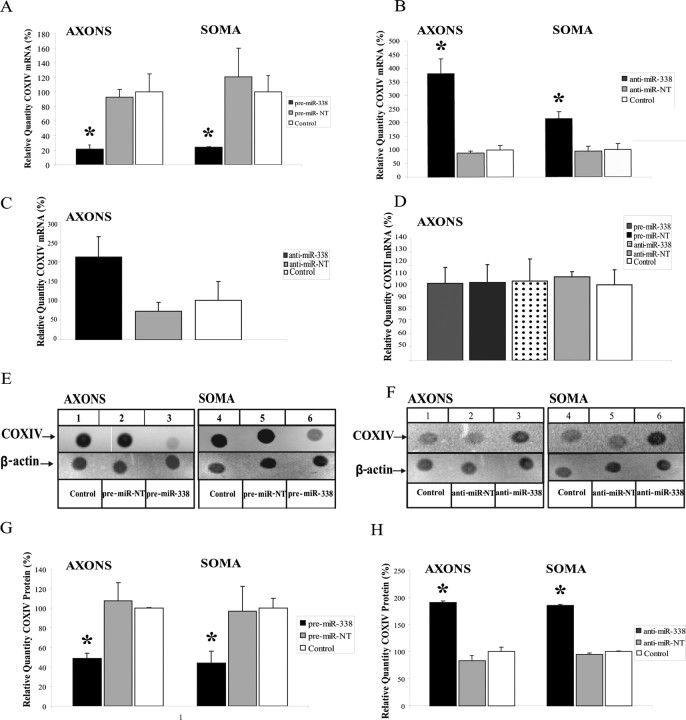
To explore whether mature miR-338 regulates COXIV mRNA levels in the distal axons of SCG neurons, we monitored COXIV mRNA levels after transfecting the soma or the distal axons with the miR-338 precursor (pre-miR-338). COXIV mRNA levels decreased ∼80% in both neuronal compartments when compared with the nontargeting pre-miR-NT (Fig. 4A). Conversely, lipofection of anti-miR-338 into the distal axons resulted in a twofold increase in COXIV mRNA as early as 4 h after axonal transfection (Fig. 4C), and in a 3.5-fold increase within 24 h (Fig. 4B), when compared with the nontargeting anti-miR control. The rapid nature of the response argues against the possibility that increases in axonal COXIV mRNA levels derive from the transport of mitochondria located in the cell soma. To evaluate this postulate, the levels of COXII mRNA, which codes for one of the three subunits of the COX complex IV encoded by the mitochondrial genome, was assessed by qRT-PCR. In contrast to the alterations observed in COXIV mRNA levels after miR-338 inhibition, no difference in COXII mRNA levels was observed after transfection (Fig. 4D). Together, these findings demonstrate that anti-miR-338-induced elevation of COXIV mRNA levels is not mediated by significant increases in the number of mitochondria present in the distal axons.
Figure 4.
miRNA-338 reduces COXIV expression in SCG neurons. A, B, Quantification of COXIV mRNA levels in the soma and distal axons of SCG neurons transfected with 25 nm pre-miR-338 (A), or with 25 nm anti-miR-338 (B), as determined by qRT-PCR 24 h after oligonucleotide transfection. COXIV mRNA levels are expressed relative to β-actin mRNA. Error bars represent the SEM for n = 3 samples. *p < 0.05. qRT-PCR revealed a doubling in COXIV mRNA levels 4 h after anti-miR-338 transfection into the distal axons of SCG neurons (C). Transfection of SCG axons with precursor or anti-miR-338 oligonucleotides did not affect the relative abundance of COXII mRNA in the distal axons as shown by qRT-PCR using primers to amplify a 100 bp segment of COXII mRNA coding region (D). E, F, Immunoblots of axonal or somal protein lysates of SCG neurons transfected with miR-338 precursor (E), anti-miR-338 (F), or nontargeting short oligonucleotides (NT). β-Actin was used as a loading control. G, H, Immunoblots of axonal or somal protein lysates of SCG neurons transfected with either miR-338 precursor (G) or anti-miR-338 (H) were quantified using Image J. Quantification showed a significant change in COXIV protein levels under the influence of miR-338 (p < 0.05, t test in all comparisons).
To assess whether miR-338-mediated modulation of mRNA also correlated with an alteration in axonal COXIV protein levels, miR-338 was over-expressed in SCG neurons by transfection of miR-338 precursor RNA, and COXIV protein levels were evaluated by immunoblot analysis. Overexpression of miR-338 lead to a ∼40% decrease in COXIV protein levels in both the soma and distal axons of SCG neurons (Fig. 4E,G). Inhibition of endogenous miR-338 by its specific anti-miR in the soma and distal axons led to a twofold to fourfold increase in the expression of COXIV protein, while having no effect on β-actin protein levels (Fig. 4F,H).
MiR-338-mediated COXIV control of axonal mitochondrial function and neurotransmitter uptake
To determine whether miR-338-dependent COXIV downregulation affects mitochondrial function in the distal axons, axons in the side compartments of the Campenot cultures were transfected with either pre-miR-338, or nontargeting pre-miR-NT. Alamar Blue (AB), a redox dye used to assess metabolic activity of mitochondria was added to the culture media and the samples were incubated for 24 h before the measurement of the relative levels of the reduced and oxidized forms of the dye. The overexpression of miR-338 significantly reduced mitochondrial oxygen consumption in the axons as determined by the reduced form of AB (Fig. 5A). Consistent with the decrease in the COXIV levels and a subsequent decrease in mitochondrial metabolic activity upon overexpression of miR-338, axons transfected with anti-miR-338 displayed a ∼50% increase in metabolic oxygen consumption compared with the nontargeting miR (Fig. 5B). In addition, an anti-miR-338-mediated increase of mitochondrial activity could also be observed in the soma and proximal axons, suggesting a cellular role of miR-338 in the regulation of COXIV levels. Further assessment of the change in mitochondrial activity mediated by miR-338 was observed by the measurement of axonal ATP levels 48 h after transfection of distal axons with anti-miR-338. Using the luminometric ATP assay, a marked increase of axonal ATP levels upon local inhibition of endogenous miR-338 was observed, further underlining the importance of the miR-338 in the regulation of axonal respiration (Fig. 5C).
Figure 5.
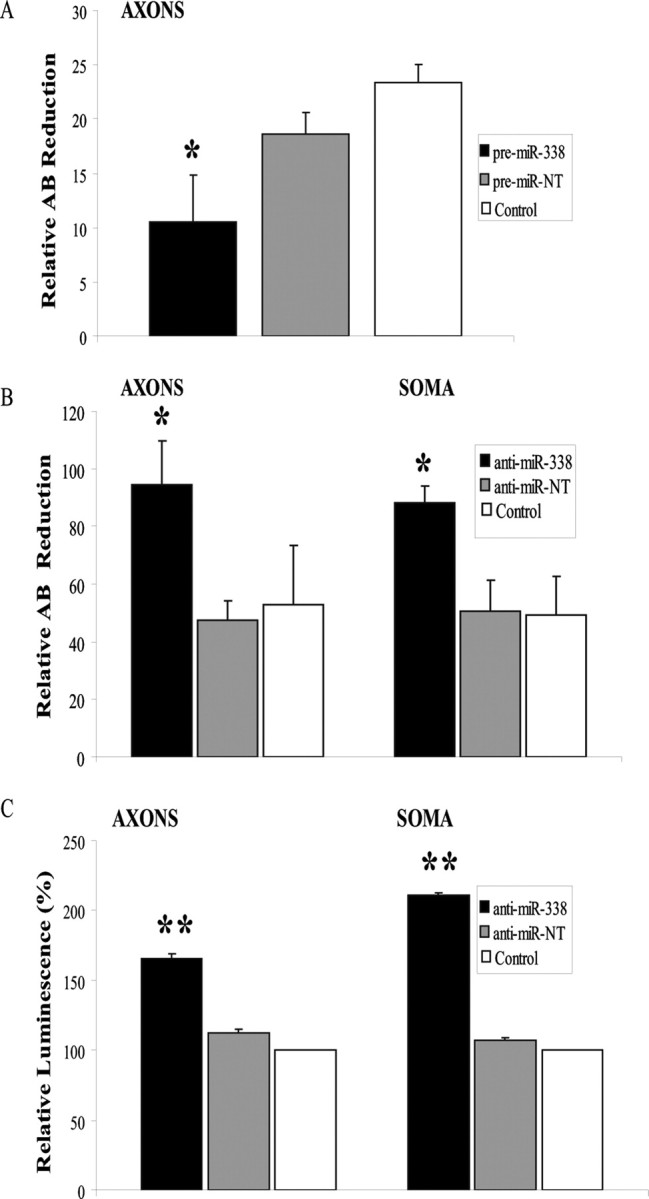
MiR-338 mediated alteration in COXIV levels modulates metabolic activity of mitochondria in the soma and distal axons of sympathetic neurons. A, B, SCG neurons were transfected with precursor miR-338 (A), anti-miR-338 (B), or nontargeting short oligonucleotides in the axon or soma compartments, respectively. AB (10%) was added to the culture media and cells were incubated for 24 h. AB data represent means ± SEM for three independent measures; *p < 0.05. ATP levels were measured in anti-miR-338 transfected axons or soma (C). Values were plotted in arbitrary luminescence units, using the luciferase cell viability assay. Data represent mean ± SEM; one-way ANOVA, **p < 0.0001.
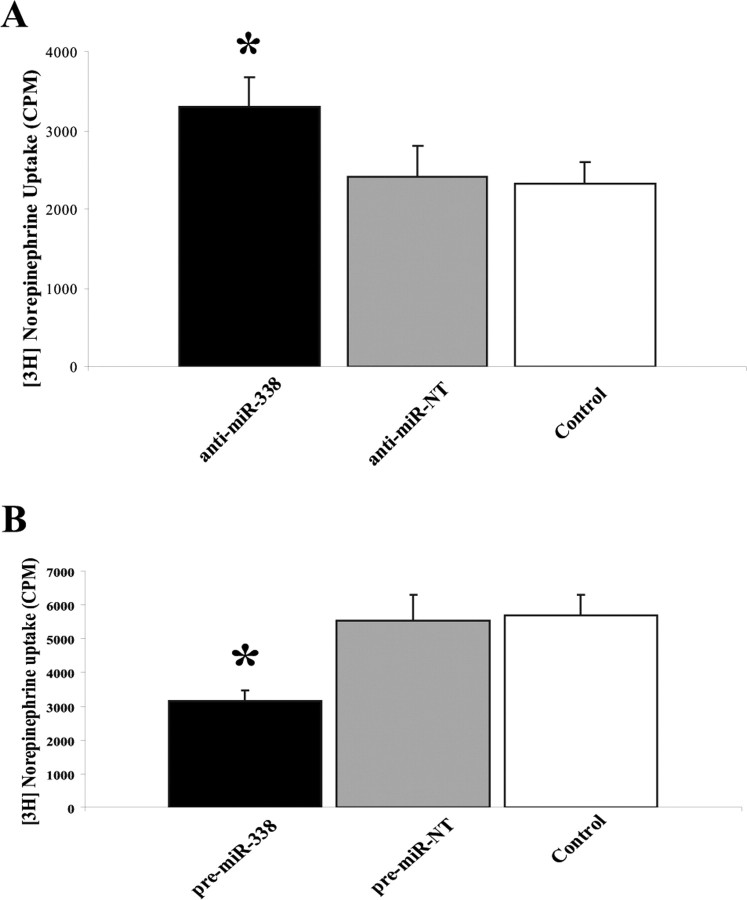
To investigate the impact of miR-338-mediated changes in mitochondrial ATP synthesis and respiration on axonal function, distal axons cultured in compartmented culture dishes were transfected with the precursor miR-338 or anti-miR-338, incubated in medium containing [3H] NE (2 μCi/ml), and NE uptake was than measured by liquid scintillation spectrometry. Under these experimental conditions, preincubation of control axons with desipramine (1 μm), a NE uptake inhibitor, reduced the content of [3H] NE in distal axons by ∼85% (mean CPM in control axons: 4465.8 ± 580 SEM; mean CPM in desipramine-treated axons: 708 ± 266.48 SEM, independent t test, two-tailed p value = 0.0002, n = 12 measurements). As shown in Figure 6, A and B, transfection of distal axons with anti-miR-338 or precursor miR-338 resulted in a 50% increase, or 50% decrease in norepinephrine uptake, respectively. Together, these results establish that modulation of miR-38 levels alters ATP synthesis and metabolic rates, as well as NE uptake in the distal SCG axons.
Figure 6.
MiR-338 modulates norepinephrine uptake in the axons of sympathetic neurons. A, B, Distal axons were transfected with anti-miR-338 (A), precursor miR-338 (B), or nontargeting short oligonucleotides (NT). After transfection (24 h), axons were incubated for 120 min in the presence of [3H]NE (2 μCi/ml). NE uptake into Triton X-100 treated axons was measured by liquid scintillation spectrometry. *p < 0.0002. Norepinephrine uptake was subsequently assessed as outlined in A. Uptake data represent mean CPM ±SEM for six independent measures; *p < 0.002.
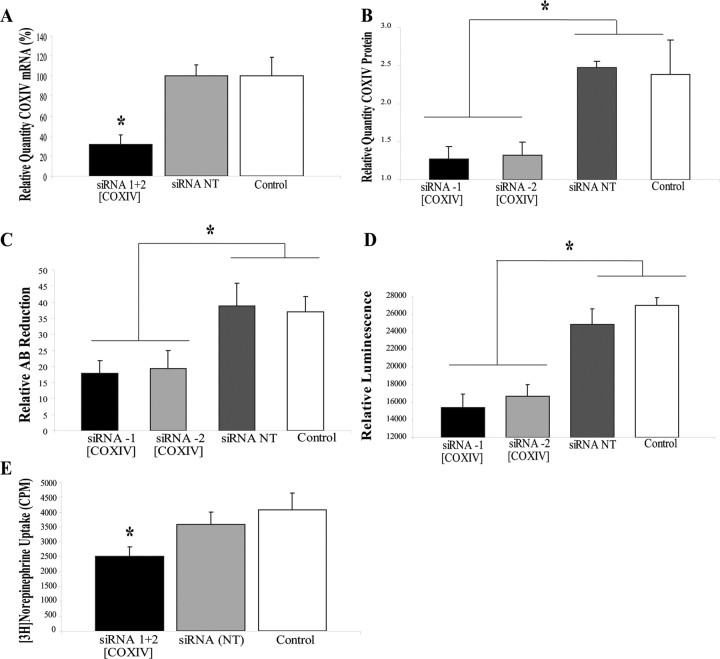
To further evaluate the postulate that the axonal effects of miR-338 are mediated, at least in part, by the local synthesis of COXIV, siRNAs targeted against COXIV mRNA were lipofected into the distal axons. After 24 h, COXIV mRNA and protein levels were reduced by 75 and 50%, respectively (Fig. 7A,B). In contrast to these findings, the introduction of siRNA into the axons had no effect on somal levels of COXIV mRNA and protein 24 h after transfection. Similar to the findings obtained with miR-338, reduction in COXIV expression by RNA interference resulted in a significant decrease in basal oxygen consumption (Fig. 7C), and axonal ATP levels (Fig. 7D) compared with sham controls. Transfection of distal axons with nontargeted siRNA (NT) had no effect on these experimental variables. Additionally, knockdown of the COXIV levels resulted in a 25–30% reduction in the uptake of [3H]NE into the distal axons (Fig. 7E). These data provide evidence that the local translation of COXIV mRNA plays a key role in regulating the oxidative capacity of the axon.
Figure 7.
SiRNA-mediated knockdown of axonal COXIV levels decreases axonal respiration and ATP levels and diminishes NE uptake in distal axons. Two independent siRNA oligonucleotides (25 nm) targeted against COXIV mRNA were introduced in distal axons of SCG neurons by lipofection and COXIV mRNA (A) and protein levels (B) quantitated 24 h later by qRT-PCR and immunoblot analyses, respectively. Knockdown of axonal COXIV expression reduced axonal oxygen consumption (C), ATP levels (D), and [3H]NE uptake (E). Values are expressed as mean ± SEM and statistical significance evaluated by one-way ANOVA. *p < 0.03.
Discussion
The regulation of gene expression by miRs represents a remarkable mechanism of posttranscriptional regulation, widely used in plants and animals. MiRs are single-stranded RNA molecules of ∼21–23 nt in length, first discovered in Caenorhabditis elegans as regulators of genes involved in developmental timing, and are now believed to modulate the expression of a myriad of genes in animals and plants (Krichevsky et al., 2003; Ambros, 2004; Kosik and Krichevsky, 2005; Stark et al., 2005; Kosik, 2006; Ambros and Chen, 2007; Foshay and Gallicano, 2007). They are highly conserved and involved in the regulation of a subset of biological processes such as cell proliferation, apoptosis, metabolism, cell differentiation, and morphogenesis (Alvarez-Garcia and Miska, 2005; Ambros and Chen, 2007; Fiore et al., 2008; Ivanovska et al., 2008; Nelson et al., 2008; Scalbert and Bril, 2008). The present studies identify an axonally localized miR that regulates the expression of COXIV, a key protein within the electron transfer chain in mitochondria, thereby controlling local levels of ATP production in the axons of sympathetic neurons. We hypothesized that the noncoding RNA miR-338 acts as a local regulator of COXIV by binding to the 3′UTR of its mRNA, thereby modulating mitochondrial oxidative phosphorylation. To investigate the mechanism of miR-338 repression, luciferase gene reporter constructs that contained the COXIV 3′UTR were used and their expression studied in transfected SCG cells. We found that the presence of the 3′UTR of COXIV significantly repressed luciferase activity, while cotransfection of the specific inhibitor anti-miR-338, but not the nontarget anti-miR, increased the activity of luciferase in SCG neurons. In addition, we found that overexpression or inhibition of miR-338 in the soma or axons altered COXIV mRNA and protein levels, as determined by qRT-PCR and dot-blot analysis.
The local regulation of gene expression by noncoding RNA is facilitated at different levels in the distal compartments of neurons (Kosik, 2006). In axons, the RNA-induced silencing complex (RISC) may modulate the expression of genes by one of two mechanisms: direct cleavage (in the case of perfect complementarities between the miR guide strand and the mRNA target) or translational attenuation (in cases of partial complementarities between the miR guide strand and the 3′ UTR of target genes) (Doench et al., 2003; Doench and Sharp, 2004; Anderson et al., 2008). The first mechanism is likely to apply to miR-338-mediated modulation of COXIV levels, even though miRBase tool data suggested an imperfect hybridization between miR-338 and the COXIV 3′UTR. This does not preclude the possibility that translation attenuation is an early event in lowering of COXIV protein levels in the distal axons followed by mRNA degradation (Mathonnet et al., 2007).
Our studies demonstrate that regulation of local translation of nuclear-encoded mitochondrial genes is an important contributor for the maintenance of mitochondrial function in the axon. In previous studies, we have shown that several nuclear-encoded mitochondrial mRNAs were present in distal axons and that the local translation of mRNAs is crucial for mitochondrial function. For example, the inhibition of local protein synthesis for 4 h resulted in a lowering of mitochondrial membrane potential, oxygen consumption and ATP synthetic capacity (Hillefors et al., 2007). These findings suggest that key proteins regulating mitochondrial activity are rapidly turned over in the distal axons. Interestingly, Li et al. (2006) showed that COXIV plays a rate-limiting role in the assembly of enzyme complex IV and that dysfunctional cytochrome c oxidase resulted in a comprised mitochondrial membrane potential, as well as decreased respiration and ATP levels. These deficits could ultimately have marked effects on axonal growth and function.
To assess the potential regulatory impact of miR-338, we searched the miRBase for nuclear-encoded mitochondrial mRNAs that contained putative miR-338 target sites. This search revealed 18 nuclear-encoded mRNAs (Table 1), ∼1% of the total mRNAs which encode mitochondrial proteins. Subsequently, we selected a subset of these mRNAs (n = 8), representing a diversity of mitochondrial functions, and tested for their presence in the distal axons of SCG neurons using qRT-PCR. This experiment identified 4 mRNAs that were present in the axon (data not shown). This finding suggests that miR-338 might regulate multiple, diverse mitochondrial functions by modulating the expression of specific nuclear-encoded mitochondrial genes in the soma or distal parts of SCG neurons.
Table 1.
List of nuclear-encoded mitochondrial target genes of miR-338
| Target genes | Name | Energy (kcal/mol) | p value (×10−02) | RefSeq (transcript ID) |
|---|---|---|---|---|
| Mecr | Trans-2-enoyl-CoA reductase | −25.34 | 0.00006 | NM_017209 |
| Atp5g1* | ATP synthase lipid-binding protein | −24.82 | 1.296 | NM_017311 |
| Hspd1* | 60 kDa heat shock protein (Hsp60) | −23.86 | 0.979 | NM_022229 |
| Mrps18a | mitochondrial ribosomal protein S18A | −23.12 | 0.207 | NM_198756 |
| Sars2 | Seryl-tRNA synthetase, mitochondrial precursor | −21.96 | 0.325 | NM_001008859 |
| Sardh | Sarcosine dehydrogenase(SarDH) | −21.68 | 0.237 | NM_053664 |
| Fdxr | NADPH:adrenodoxin oxidoreductase (ferredoxin reductase) | −21.6 | 0.011 | NM_024153 |
| Clybl+ | Citrate lyase β subunit-like protein | −21.28 | 4.057 | XM_240311 |
| Dlst* | Dihydrolipoamide succinyltransferase component of 2-oxoglutarate dehydrogenase | −20.94 | 0.015 | NM_001006981 |
| Hspa9a_predicted | Heat shock 70 kDa protein 9 | −20.1 | 0.054 | NM_001100658 |
| Iars2 | Isoleucine-tRNA synthetase 2, mitochondrial | −18.28 | 4.446 | XM_344185.3 |
| Ucp1 | Mitochondrial brown fat uncoupling protein 1 (UCP 1) | −16.92 | 0.724 | NM_012682 |
| Slc25a20 | Carnitine/acylcarnitine translocase | −16.81 | 0.293 | NM_053965 |
| Mrps10 | Mitochondrial 28S ribosomal protein S10 | −16.42 | 4.704 | NM_001008859 |
| Cox4i1* | Cytochrome c oxidase polypeptide IV | −14.87 | 0.332 | NM_017202 |
| Timm10+ | Mitochondrial import inner membrane translocase subunit Tim10. | −14.67 | 2.402 | NM_172074 |
| Top1mt+ | Mitochondrial topoisomerase I | −14.48 | 0.247 | NM_001002798 |
| Cyp11a1+ | Cytochrome P450 11A1, mitochondrial precursor | −12.33 | 2.168 | NM_017286 |
Computational prediction analysis (miRBase) identified 1009 putative miR-338 target genes, of which 18 were nuclear-encoded mitochondrial genes (miRBase, http://microrna.sanger.ac.uk).
*Gene products detected in both distal axons and cell soma by RT-qPCR. +Gene products detected in cell soma only by RT-qPCR.
Our studies further provide evidence that the “overexpression” of miR-338 in the axons diminished COXIV levels and subsequently axonal oxidative phosphorylation. This reduction in respiration had minimal effects on neuronal morphology or viability. The lack of morphological abnormalities in the neuron expressing reduced oxidative phosphorylation actively has been demonstrated by other investigations into axonal transport of mitochondria (Stowers et al., 2002; Verstreken et al., 2005). One possibility for the stability of the morphology is that intracellular transport of ATP alleviates the global effects caused by an increase in miR-338. It is also possible that only minimal respiration and ATP production are required to maintain neuronal morphology and viability under basal conditions. However, a reduction in ATP production and reduced oxidative phosphorylation may compromise additional axonal functions under prolonged activity or stress that are not observed through morphological alteration. Previous studies demonstrated that ATP and Ca2+ are needed locally at the synapses and mitochondria are highly abundant in axon terminals and are essential for vesicle cycling and neurotransmitter release and uptake (Shepherd and Harris, 1998; Rowland et al., 2000). In the Drosophila mutant, Milton, a defect in synaptic transmission is associated with the loss of mitochondria from the axon terminal (Stowers et al., 2002). Mitochondria are important regulators of cell survival and death and a dysfunction of mitochondrial energy metabolism leads to reduced ATP production, impaired calcium buffering, and increased generation of reactive oxygen species (ROS) (Beal, 2007; Petrozzi et al., 2007) that are implicated in a subset of neurodegenerative diseases, such as Alzheimer's disease (Stokin et al., 2005; Stokin and Goldstein, 2006), and Parkinson's disease (Murdock et al., 2000). In addition, the generation of ROS contributes to aging since overexpression of mitochondrially localized antioxidant enzymes lengthens life span in Drosophila (Ruan et al., 2002; Calingasan et al., 2008).
In conclusion, we have provided evidence that the noncoding miR-338 is a novel regulator of mitochondrial oxidative phosphorylation and axonal function (e.g., NE uptake) in the distal axons through local modulation of COXIV levels. These findings point to a novel mechanism for a soma-independent regulation of respiration in distal axons through neuronal microRNA. In future studies, we will use the elegant Campenot culture system to further characterize the subset of miRs that regulates a distinct set of target genes involved in axonal maintenance and function.
Footnotes
This work was supported by the Division of Intramural Research Programs of the National Institute of Mental Health. We thank Dr. Neil Smalheiser (University of Illinois) for the kind gift of the DICER and eIF2c antibodies. We thank Elena Perry for assistance with the miR maturation studies. We express our gratitude to Dr. Howard Nash (National Institute of Mental Health) for a critical evaluation of this manuscript.
References
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Ambros V, Chen X. The regulation of genes and genomes by small RNAs. Development. 2007;134:1635–1641. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- Anderson EM, Birmingham A, Baskerville S, Reynolds A, Maksimova E, Leake D, Fedorov Y, Karpilow J, Khvorova A. Experimental validation of the importance of seed complement frequency to siRNA specificity. RNA. 2008;14:853–861. doi: 10.1261/rna.704708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Beal MF. Mitochondria and neurodegeneration. Novartis Found Symp. 2007;287:183–192. doi: 10.1002/9780470725207.ch13. discussion 192–196. [DOI] [PubMed] [Google Scholar]
- Calingasan NY, Ho DJ, Wille EJ, Campagna MV, Ruan J, Dumont M, Yang L, Shi Q, Gibson GE, Beal MF. Influence of mitochondrial enzyme deficiency on adult neurogenesis in mouse models of neurodegenerative diseases. Neuroscience. 2008;153:986–996. doi: 10.1016/j.neuroscience.2008.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DS, Regan AG, Lopez JS, Tannahill D, Harris WA, Holt CE. Semaphorin 3A elicits stage-dependent collapse, turning, and branching in Xenopus retinal growth cones. J Neurosci. 2001;21:8538–8547. doi: 10.1523/JNEUROSCI.21-21-08538.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campenot RB. Local control of neurite development by nerve growth factor. Proc Natl Acad Sci U S A. 1977;74:4516–4519. doi: 10.1073/pnas.74.10.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DT, Honick AS, Reynolds IJ. Mitochondrial trafficking to synapses in cultured primary cortical neurons. J Neurosci. 2006;26:7035–7045. doi: 10.1523/JNEUROSCI.1012-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol. 2008;10:149–159. doi: 10.1038/ncb1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng H, Lund K, Campenot RB. Synthesis of β-tubulin, actin, and other proteins in axons of sympathetic neurons in compartmented cultures. J Neurosci. 1999;19:1–9. doi: 10.1523/JNEUROSCI.19-01-00001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore R, Siegel G, Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochim Biophys Acta. 2008;1779:471–478. doi: 10.1016/j.bbagrm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Foshay KM, Gallicano GI. Small RNAs, big potential: the role of microRNAs in stem cell function. Curr Stem Cell Res Ther. 2007;2:264–271. doi: 10.2174/157488807782793781. [DOI] [PubMed] [Google Scholar]
- Gioio AE, Eyman M, Zhang H, Lavina ZS, Giuditta A, Kaplan BB. Local synthesis of nuclear-encoded mitochondrial proteins in the presynaptic nerve terminal. J Neurosci Res. 2001;64:447–453. doi: 10.1002/jnr.1096. [DOI] [PubMed] [Google Scholar]
- Gioio AE, Lavina ZS, Jurkovicova D, Zhang H, Eyman M, Giuditta A, Kaplan BB. Nerve terminals of squid photoreceptor neurons contain a heterogeneous population of mRNAs and translate a transfected reporter mRNA. Eur J Neurosci. 2004;20:865–872. doi: 10.1111/j.1460-9568.2004.03538.x. [DOI] [PubMed] [Google Scholar]
- Hengst U, Cox LJ, Macosko EZ, Jaffrey SR. Functional and selective RNA interference in developing axons and growth cones. J Neurosci. 2006;26:5727–5732. doi: 10.1523/JNEUROSCI.5229-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillefors M, Gioio AE, Mameza MG, Kaplan BB. Axon viability and mitochondrial function are dependent on local protein synthesis in sympathetic neurons. Cell Mol Neurobiol. 2007;27:701–716. doi: 10.1007/s10571-007-9148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson AL, Linsley PS, Cleary MA. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28:2167–2174. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci U S A. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Kosik KS, Krichevsky AM. The elegance of the MicroRNAs: a neuronal perspective. Neuron. 2005;47:779–782. doi: 10.1016/j.neuron.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Park JS, Deng JH, Bai Y. Cytochrome c oxidase subunit IV is essential for assembly and respiratory function of the enzyme complex. J Bioenerg Biomembr. 2006;38:283–291. doi: 10.1007/s10863-006-9052-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, Filipowicz W, Duchaine TF, Sonenberg N. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- Murdock DG, Christacos NC, Wallace DC. The age-related accumulation of a mitochondrial DNA control region mutation in muscle, but not brain, detected by a sensitive PNA-directed PCR clamping based method. Nucleic Acids Res. 2000;28:4350–4355. doi: 10.1093/nar/28.21.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama GR, Caton MC, Nova MP, Parandoosh Z. Assessment of the Alamar Blue assay for cellular growth and viability in vitro. J Immunol Methods. 1997;204:205–208. doi: 10.1016/s0022-1759(97)00043-4. [DOI] [PubMed] [Google Scholar]
- Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrozzi L, Ricci G, Giglioli NJ, Siciliano G, Mancuso M. Mitochondria and neurodegeneration. Biosci Rep. 2007;27:87–104. doi: 10.1007/s10540-007-9038-z. [DOI] [PubMed] [Google Scholar]
- Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci. 2006;26:13390–13399. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland KC, Irby NK, Spirou GA. Specialized synapse-associated structures within the calyx of Held. J Neurosci. 2000;20:9135–9144. doi: 10.1523/JNEUROSCI.20-24-09135.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H, Tang XD, Chen ML, Joiner ML, Sun G, Brot N, Weissbach H, Heinemann SH, Iverson L, Wu CF, Hoshi T. High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc Natl Acad Sci U S A. 2002;99:2748–2753. doi: 10.1073/pnas.032671199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalbert E, Bril A. Implication of microRNAs in the cardiovascular system. Curr Opin Pharmacol. 2008;8:181–188. doi: 10.1016/j.coph.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Shepherd GM, Harris KM. Three-dimensional structure and composition of CA3→CA1 axons in rat hippocampal slices: implications for presynaptic connectivity and compartmentalization. J Neurosci. 1998;18:8300–8310. doi: 10.1523/JNEUROSCI.18-20-08300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Stokin GB, Goldstein LS. Axonal transport and Alzheimer's disease. Annu Rev Biochem. 2006;75:607–627. doi: 10.1146/annurev.biochem.75.103004.142637. [DOI] [PubMed] [Google Scholar]
- Stokin GB, Lillo C, Falzone TL, Brusch RG, Rockenstein E, Mount SL, Raman R, Davies P, Masliah E, Williams DS, Goldstein LS. Axonopathy and transport deficits early in the pathogenesis of Alzheimer's disease. Science. 2005;307:1282–1288. doi: 10.1126/science.1105681. [DOI] [PubMed] [Google Scholar]
- Stowers RS, Megeath LJ, Górska-Andrzejak J, Meinertzhagen IA, Schwarz TL. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron. 2002;36:1063–1077. doi: 10.1016/s0896-6273(02)01094-2. [DOI] [PubMed] [Google Scholar]
- Verstreken P, Ly CV, Venken KJ, Koh TW, Zhou Y, Bellen HJ. Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron. 2005;47:365–378. doi: 10.1016/j.neuron.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenisek D, Matthews G. The role of mitochondria in presynaptic calcium handling at a ribbon synapse. Neuron. 2000;25:229–237. doi: 10.1016/s0896-6273(00)80885-5. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]