Summary
It is not understood how Hsp104, a hexameric AAA+ ATPase from yeast, disaggregates diverse structures including stress-induced aggregates, prions, and α-synuclein conformers connected to Parkinson disease. Here, we establish that Hsp104 hexamers adapt different mechanisms of intersubunit collaboration to disaggregate stress-induced aggregates versus amyloid. To resolve disordered aggregates, Hsp104 subunits collaborate non-co-operatively via probabilistic substrate binding and ATP hydrolysis. To disaggregate amyloid, several subunits co-operatively engage substrate and hydrolyze ATP. Importantly, Hsp104 variants with impaired intersubunit communication dissolve disordered aggregates but not amyloid. Unexpectedly, prokaryotic ClpB subunits collaborate differently than Hsp104 and couple probabilistic substrate binding to cooperative ATP hydrolysis, which enhances disordered aggregate dissolution but sensitizes ClpB to inhibition and diminishes amyloid disaggregation. Finally, we establish that Hsp104 hexamers deploy more subunits to disaggregate Sup35 prion strains with more stable ‘cross-β’ cores. Thus, operational plasticity enables Hsp104 to robustly dissolve amyloid and non-amyloid clients, which impose distinct mechanical demands.
Introduction
Several fatal neurodegenerative disorders, including Parkinson disease (PD), are connected with the misfolding of specific proteins into soluble toxic oligomers and stable cross-β fibers, termed amyloid (Cushman et al., 2010). Amyloidogenesis is also a severe problem in recombinant protein purification from diverse systems ranging from bacteria to animal cells. Here, overexpressed proteins form inclusions and adopt the amyloid form (Wang et al., 2008). Thus, amyloid frustrates basic structural and functional studies, and limits production of valuable therapeutic proteins in the pharmaceutical sector. The dearth of solutions to these problems reflects a profound gap in our understanding of how cells safely reverse amyloid formation.
Amyloid disaggregation is coupled to degradation in animal cell extracts, but the identity of the disaggregase is unknown (Cohen et al., 2006). Moreover, Hsp110, Hsp70 and Hsp40, the metazoan protein-disaggregase system, cannot rapidly disaggregate amyloid (Shorter, 2011). Perplexingly, animals lack Hsp104 orthologs, which are found in bacteria, fungi, protozoa, chromista and plants. Hsp104 is a hexameric, ring-shaped translocase with 2 AAA+ nucleotide-binding domains (NBDs) per subunit that couple ATP hydrolysis to protein disaggregation (Vashist et al., 2010). In yeast, Hsp104 promotes survival of protein-folding stress by collaborating with Hsp70 and Hsp40 to renature the entire aggregated proteome (Parsell et al., 1994; Vashist et al., 2010). Thioflavin-T (ThT) fluorescence, Congo Red binding, sedimentation, electron microscopy and SDS-resistance have been used to establish that Hsp104 rapidly remodels various amyloid forms, including Sup35 and Ure2 prions. Hsp104 also rapidly eliminates preamyloid oligomers that accumulate prior to fibers (Shorter and Lindquist, 2004, 2006). Thus, Hsp104 enables yeast to harness infectious amyloids, termed prions, for beneficial purposes (Halfmann et al., 2012; but see also Wickner et al., 2011). How Hsp104 disaggregates such a diverse repertoire of structures, ranging from stable amyloid to less stable disordered aggregates (Knowles et al., 2007; Wang et al., 2010) is not understood. This immense substrate diversity imposes extreme mechanical demands on Hsp104. The loss of Hsp104 from metazoa is baffling. Transgenic mice expressing Hsp104 are normal and Hsp104 increases stress tolerance of animal cells (Dandoy-Dron et al., 2006). Moreover, Hsp104 directly remodels PD-associated oligomers and amyloids formed by α-synuclein (α-syn) and rescues rodent models of PD and Huntington disease (HD) (Lo Bianco et al., 2008; Vacher et al., 2005). Thus, Hsp104 could be developed as a therapeutic disaggregase for neurodegenerative disorders (Vashist et al., 2010).
Ideally, to optimize therapy and minimize side effects, Hsp104 would be engineered and potentiated to dissolve specific aggregates central to the disease in question (Vashist et al., 2010). Indeed, Hsp104’s disaggregase activity could be enhanced and tailored for any protein. Thus, substrate-optimized Hsp104 variants could increase protein solubility and enable facile purification of recalcitrant proteins in diverse settings. However, limited structural and mechanistic understanding of Hsp104 hexamers frustrates such endeavors. It is not understood how individual subunits of the Hsp104 hexamer co-ordinate substrate translocation and ATP hydrolysis to solubilize unrelated proteins trapped in energetically and structurally distinct aggregates (Doyle et al., 2007b; Lee et al., 2010; Tessarz et al., 2008; Wendler et al., 2007; Wendler et al., 2009).
Do Hsp104 hexamers use the same mechanism to disaggregate amyloid and non-amyloid clients? Specific mutations in Hsp104 differentially affect its activity against prions and disordered aggregates, as does ATPγS, a slowly hydrolyzable ATP analog, suggesting a mechanistic dichotomy or plasticity (Doyle et al., 2007b; Kurahashi and Nakamura, 2007). This dichotomy might reflect an ability of Hsp104 subunits to collaborate differently to promote dissolution of diverse aggregated structures.
How individual subunits collaborate to promote substrate remodeling is a key question, not only for Hsp104, but for all NTP-fueled, hexameric ring-translocases. Several different intersubunit collaboration models have been proposed including: (a) probabilistic models in which individual subunits function non-co-operatively and independently (Martin et al., 2005); (b) models of subglobal co-operativity where a subset of subunits co-operate (Moreau et al., 2007); and (c) models of global cooperativity where all subunits co-operate in sequence or in concert (Lyubimov et al., 2011). Typically, these models focus on co-ordination of NTPase events. Less attention has been given to how individual subunits within the hexamer contribute to substrate binding and translocation. For example, it is not clear whether globally co-operative ATPase activity must invariably be coupled to globally co-operative substrate handling. A key unresolved issue is whether a single ring-translocase can exploit different modes of intersubunit collaboration to remodel substrates that impose disparate mechanical demands.
Here, we elucidate that Hsp104 subunits collaborate via radically different mechanisms to disaggregate disordered aggregates versus amyloid. Unexpectedly, the E. coli homolog of Hsp104, ClpB, co-ordinates subunit collaboration differently to Hsp104, even though Hsp104 and ClpB are widely assumed to function by the same mechanism (Doyle and Wickner, 2009). Hsp104 exhibits operational plasticity that confers adaptable disaggregase activity suited for the demands of the yeast proteome, which include prion disaggregation. By contrast, ClpB is finely tuned for optimal disordered aggregate dissolution and has limited ability to dissolve amyloid.
Results
Experimental logic
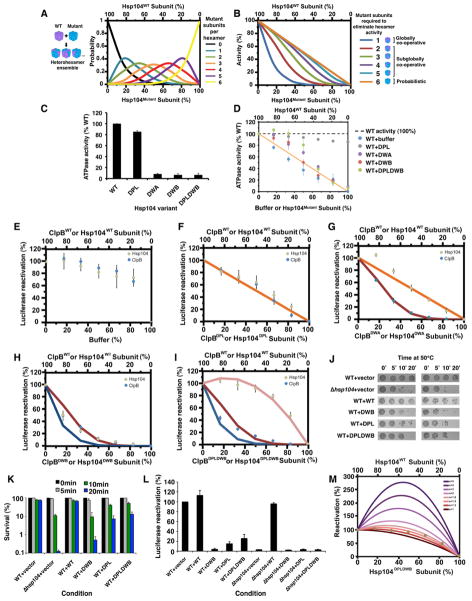
We employed a mutant doping strategy to determine the contribution of individual subunits toward protein disaggregation and thereby define mechanochemical coupling mechanisms of Hsp104 hexamers. Thus, mutant subunits defective in ATP hydrolysis or substrate binding are mixed with wild-type (WT) subunits to yield heterohexamer ensembles with different ratios of WT and mutant protein. This strategy has yielded key insights for other NTP-fueled ring-translocases, but is dependent upon random mixing of mutant and WT subunits at the monomer level (Moreau et al., 2007; Werbeck et al., 2008).
First, we established that Hsp104 forms dynamic hexamers that rapidly exchange subunits on the minute timescale (Figure S1A–J, S2A–J; Supplemental Results) similar to ClpB (Werbeck et al., 2008). This rapid subunit exchange allows generation of heterohexamer ensembles comprised of WT and mutant subunits according to a binomial distribution that varies as a function of the molar ratio of each subunit (Figure 1A, S1D–I; see Extended Experimental Procedures) (Werbeck et al., 2008). Using this distribution, we can predict how disaggregase activity would be inhibited at various mutant:WT ratios if a specified number of mutant subunits inactivate the hexamer (Figure 1B; see Extended Experimental Procedures). Thus, if all 6 subunits must work together, then 1 mutant subunit would abolish hexamer activity (Figure 1B, dark blue curve). If the activity of a single subunit within the hexamer is sufficient, then some activity would still be observed with 5 mutant subunits per hexamer and only 6 mutant subunits would abolish activity (Figure 1B, orange line). By comparing experimental data with theoretical plots, we can determine whether subunit collaboration within Hsp104 hexamers is probabilistic (6 mutant subunits abolish activity), subglobally co-operative (2–5 mutant subunits abolish activity), or globally co-operative (1 mutant subunit abolishes activity).
Figure 1. Hsp104 uses a probabilistic mechanism to dissolve disordered aggregates.
(A) Theoretical Hsp104 hexamer ensembles containing 0 (black), 1 (blue), 2 (green), 3 (orange), 4 (red), 5 (purple) and 6 mutant subunits (yellow) as a function of the fraction of mutant subunit present.
(B) Theoretical activity curves where 1 or more (blue), 2 or more (red), 3 or more (green), 4 or more (purple), 5 or more (light blue) or 6 mutant subunits (orange) are needed to ablate hexamer activity.
(C) WT or mutant Hsp104 ATPase activity. Values represent means±SEM (n=3–5).
(D) Hsp104 was mixed with increasing fractions of mutant Hsp104 proteins or buffer and ATPase activity was assessed. Values represent means±SEM (n=3–5). Orange line: expected ATPase activity if 6 mutant subunits are needed to ablate hexamer activity.
(E–I) Luciferase aggregates were treated with Hsp104 (grey markers), Hsc70 (an Hsp70) and Hdj2 (an Hsp40) plus increasing fractions of buffer (E), Hsp104DPL (F), Hsp104DWA (G), Hsp104DWB (H) or Hsp104DPLDWB (I). Alternatively, luciferase aggregates were treated with ClpB (blue markers), DnaK, DnaJ and GrpE plus increasing fractions of buffer (E), ClpBDPL (F), ClpBDWA (G), ClpBDWB (H) or ClpBDPLDWB (I). Luciferase reactivation (% WT activity) was then assessed. Values represent means±SEM (n=3–4). Theoretical disaggregase activity if 6 (orange line, F, G), 2 or more (red line, G–I) or 1 or more mutant subunits (blue line, H, I) ablate hexamer activity. Pink line (I): simulated activity if a mutant subunit stimulates an adjacent WT subunit 1.4-fold but is inhibitory if adjacent to a mutant subunit.
(J, K) WT yeast carrying the indicated Hsp104 plasmid or Δhsp104 yeast harboring an emptor vector control were treated at 50°C for 0–20min and then spotted (J). The spotting on the right is a 5-fold dilution of the spotting on the left. Alternatively, cells were plated and survival (%) was calculated (K). Values represent means±SEM (n=3).
(L) WT or Δhsp104 yeast expressing luciferase and the indicated Hsp104 variant were shifted to 44°C, treated with cycloheximide, and allowed to recover at 30°C. Luciferase activity (% of the WT+vector control) was determined. Values represent means±SEM (n=3).
(M) Adjacent pairs of WT-WT or WT-mutant subunits determine hexamer activity, whereas adjacent mutant subunits have no activity. Each adjacent WT-WT pair has an activity of 1/6. By contrast, adjacent WT-mutant pairs have a stimulated activity (s), and the effect of various values of s is depicted. Brown markers indicate experimental luciferase disaggregation data obtained with Hsp104DPLDWB.
See also Supplementary Figure S1, S2 and S3.
Hsp104 uses a probabilistic mechanism to dissolve disordered aggregates
To define how Hsp104 subunits co-ordinate substrate binding, we employed Hsp104DPL, which harbors Y257A and Y662A mutations in the NBD1 and NBD2 channel loops that impair substrate binding (Lum et al., 2008). Importantly, Hsp104DPL has WT ATPase activity (Figure 1C), incorporates into WT hexamers just as well as WT Hsp104 (Figure S1D–F, S2J), and has minimal effect on total ATPase activity when mixed with WT Hsp104 (Figure 1D, grey markers).
We assembled heterohexamer ensembles of WT Hsp104 and Hsp104DPL and assessed disaggregase activity against disordered luciferase aggregates. Dilution of Hsp104 with buffer had little effect, whereas addition of Hsp104DPL caused a roughly linear decline in disaggregase activity (Figure 1E, F). Similar data were obtained with heat-denatured GFP aggregates and heat-denatured citrate synthase (CS) aggregates (Figure S3A, B). This tolerance of Hsp104 hexamers to Hsp104DPL subunits suggests that for disordered aggregates, Hsp104 translocates substrate in a probabilistic manner. Thus, a single WT subunit per hexamer can catalyze disaggregation.
This probabilistic mechanism of substrate handling is conserved over 2 billion years of evolution to E. coli ClpB. ClpB displayed a roughly linear decline in luciferase disaggregation activity in response to a substrate binding defective variant, ClpBDPL (Y251A:Y653A) (Weibezahn et al., 2004), whereas buffer had no effect (Figure 1E, F).
This non-cooperative substrate handling was surprising since Hsp104 co-operatively hydrolyzes ATP (Hattendorf and Lindquist, 2002). To determine the role of individual subunits with respect to ATP hydrolysis, we utilized ATPase-defective Hsp104DWA, which harbors K218T and K620T mutations in the NBD1 and NBD2 Walker A motifs. These mutations severely inhibit ATP hydrolysis (Figure 1C) by reducing affinity for ATP, but do not impair hexamerization at the Hsp104 concentrations employed here (Schirmer et al., 2001). Indeed, Hsp104DWA incorporated into WT hexamers just like WT Hsp104 (Figure S1C–E, S1G, S2J). Doping revealed that Hsp104DWA subunits inhibited total ATPase activity slightly less than predicted by a linear response (Figure 1D, compare purple markers to orange line). Strikingly, Hsp104DWA subunits elicited a roughly linear decline in luciferase, GFP and CS disaggregation by Hsp104 (Figure 1G, S3C, D). Thus, Hsp104 couples probabilistic ATPase activity and substrate handling to disordered aggregate dissolution, indicating that the Hsp104 power stroke can be generated by ATP hydrolysis in a single subunit.
ClpB hexamers are tuned differently to Hsp104 hexamers
These findings were surprising because mutant doping with ClpB indicated highly cooperative ATP hydrolysis powers disordered aggregate dissolution (Hoskins et al., 2009). Indeed, ATPase-defective ClpBDWA (K212T:K611T) caused a sharp non-linear decline in ClpB disaggregase activity, consistent with 2 mutant subunits abolishing hexamer activity (Figure 1B, G). We also assessed ClpBDWB, which bears E279Q and E678Q mutations in the NBD1 and NBD2 Walker B motifs. ClpBDWB forms hexamers that bind but do not hydrolyze ATP (Weibezahn et al., 2003). Doping ClpBDWB elicited a sharp non-linear decline in disaggregase activity such that 1–2 mutant subunits abolish hexamer activity (Figure 1B, H). Thus, unlike Hsp104, ClpB subunits couple highly collaborative ATPase activity to probabilistic substrate handling to dissolve disordered aggregates.
ClpBDWB is a substrate ‘trap’ (Weibezahn et al., 2003), which might poison WT hexamers by not releasing substrate rather than by perturbing intersubunit co-ordination of ATP hydrolysis. To address this issue, we constructed ClpBDPLDWB in which the substrate-binding pore loops and Walker B motifs are mutated (Y251A:E279Q:Y653A:E678Q). Doping ClpBDPLDWB caused a sharp decline in luciferase reactivation such that 1–2 mutant subunits ablated activity (Figure 1I). Thus, substrate binding by ClpBDWB does not poison WT hexamers. Rather, 5–6 ClpB subunits per hexamer must hydrolyze ATP for protein disaggregation. Surprisingly, this highly co-ordinated ATPase pattern of ClpB hexamers is coupled to stochastic substrate binding (Figure 1F–H).
Hsp104 hexamers tolerate multiple subunits defective in ATP hydrolysis and substrate binding
We obtained dissimilar results with Hsp104. Hsp104DWB (E285Q:E687Q) and Hsp104DPLDWB (Y257A:E285Q:Y662A:E687Q) have little ATPase activity and incorporate into WT hexamers as predicted (Figure 1C, S1D, S1E, S1H, S1I, S2J). Like Hsp104DWA, Hsp104DWB subunits caused a roughly linear decline in total ATPase activity (Figure 1D, compare red markers to orange line). By contrast, Hsp104DPLDWB had little effect on total ATPase activity unless the fraction of mutant subunit exceeded 50%, in which case inhibition was similar to Hsp104DWB (Figure 1D, compare green to red markers). Similar to ClpB, 1–2 Hsp104DWB subunits per hexamer abolished disaggregase activity (Figure 1H, S3E, F). Unlike ClpB, inhibition was partially rescued by the substrate-binding loop mutations in Hsp104DPLDWB (Figure 1I, S3G, H). Thus, it is not the ATPase defect, but ‘substrate trapping’ by a single Hsp104DWB subunit that poisons a hexamer with 5 WT subunits. Indeed, Hsp104DWA confers a similar ATPase defect to Hsp104DWB (Figure 1D), but cannot interact with substrate (Bosl et al., 2005) and does not elicit a sharp decline in disaggregase activity (Figure 1G, H). Consistent with these in vitro findings, Hsp104DWB has a more severe dominant negative effect than Hsp104DPL or Hsp104DPLDWB on Hsp104 function in thermotolerance and luciferase disaggregation in vivo (Figure 1J–L).
The response to Hsp104DPLDWB subunits was unusual. Rather than a linear decline, we observed little effect at low fractions of Hsp104DPLDWB and a sharp decline when the fraction of Hsp104DPLDWB subunit exceeded 66.7% (Figure 1I). We could model this behavior if we imposed rules whereby a mutant subunit stimulates the activity of an adjacent WT subunit by ~1.4-fold, but exerts an inhibitory effect if it is adjacent to a mutant subunit (Figure 1I, S3G, H, compare pink line to grey markers; See Extended Experimental Procedures and Figure 1M). Thus, Hsp104 hexamers operate via principles distinct to those of ClpB hexamers. The Hsp104 hexamer displays greater plasticity. It tolerates a wider variety of subunit inactivating events without gross perturbations in disaggregase activity. For example, an Hsp104 subunit that: (a) binds but cannot hydrolyze ATP, and, (b) is unable to engage substrate, can stimulate the disaggregase activity of an adjacent subunit. In ClpB, a single subunit with these properties inactivates the entire hexamer.
Hsp104 remodels diverse amyloids, whereas ClpB has limited activity
Hsp104 plasticity might ensure inheritance of numerous beneficial prions (Alberti et al., 2009). By contrast, although E. coli exploits functional amyloid on the cell surface, it is not known to harbor cytoplasmic prions and barely supports Sup35 prion formation (Barnhart and Chapman, 2006; Garrity et al., 2010). Indeed, ClpB has limited ability to remodel Sup35 prions (Reidy et al., 2012; Shorter and Lindquist, 2004). However, it is unknown whether this limitation extends to other amyloids.
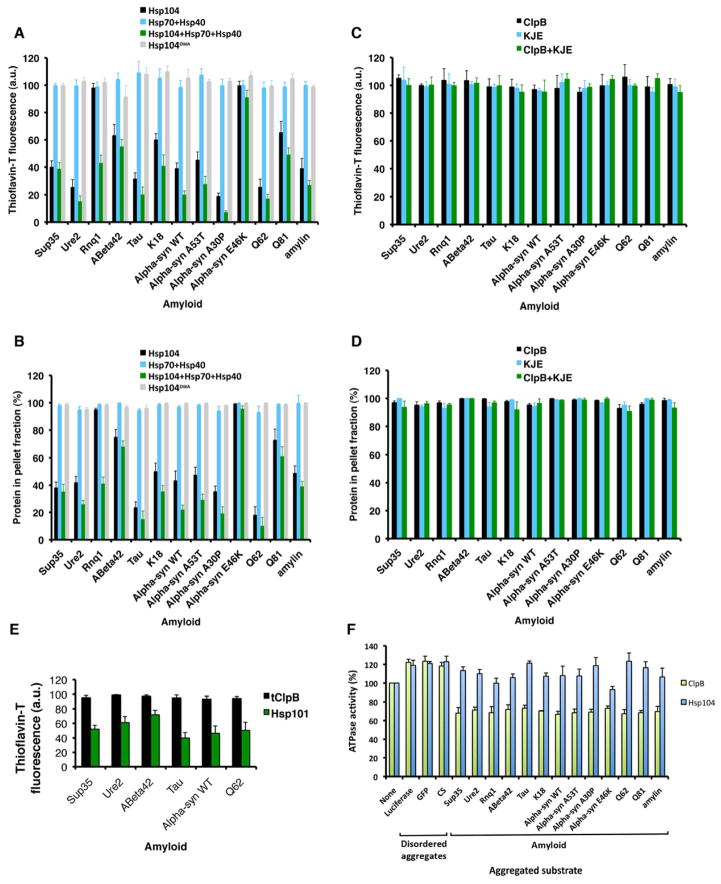
We tested whether Hsp104 and ClpB disaggregated various amyloids formed by proteins with diverse primary sequences, including yeast prion proteins: Sup35, Ure2 and Rnq1; proteins linked to Alzheimer Disease, PD, HD or type 2 diabetes: Aβ42, tau (and K18, a tau fragment), α-syn (WT, and PD-linked variants: A53T, A30P and E46K), polyglutamine (Q62 and Q81) and amylin (Cushman et al., 2010). Hsp104DWA was inactive, but Hsp104 remodeled the majority of these amyloids in a manner that was slightly enhanced by Hsp70 (Ssa1) and Hsp40 (Sis1), which were inactive alone (Figure 2A, B). Rnq1 prions were an exception that necessitated Hsp70 and Hsp40, whereas α-synE46K, Aβ42 and Q81 amyloids were generally more refractory (Figure 2A, B). Thus, Hsp70 and Hsp40 are not always essential for Hsp104 to disaggregate diverse cross-β structures. We suggest that a generic feature of amyloid unleashes Hsp104 disaggregase activity in the absence of Hsp70 and Hsp40.
Figure 2. Hsp104 disaggregates diverse amyloids, whereas ClpB does not.
(A–D) Sup35, Ure2, Rnq1 or Aβ42, tau, K18, α-synWT, α-synA53T, α-synA30P, α-synE46K, Q62, Q81 and amylin amyloids were treated with the indicated combination of Hsp104, Ssa1, Sis1, or Hsp104DWA (A, B) or ClpB, DnaK, DnaJ, and GrpE (C, D). Fiber integrity was assessed by ThT fluorescence (A, C) or sedimentation (B, D). Values represent means±SEM (n=3).
(E) Sup35, Ure2, Aβ42, tau, α-synWT and Q62 amyloids were treated with tClpB or Hsp101. Fiber integrity was assessed by ThT fluorescence. Values represent means±SEM (n=3).
(F) ATPase activity of ClpB or Hsp104 in the presence of the indicated aggregated substrate. Values represent means±SEM (n=3). See also Supplementary Figure S4 and Table S1.
ClpB had limited ability to disaggregate amyloid with or without Hsp70 (DnaK) and Hsp40 (DnaJ) (Figure 2C, D). Indeed, we varied ClpB, DnaK, DnaJ and GrpE concentration (0–50μM), incubation time (0–96h), and ATP concentration (0–25mM), but could not establish conditions where ClpB disaggregated amyloid. Similarly, ClpB from T. thermophilus was unable to disaggregate amyloid, whereas the A. thaliana homolog, Hsp101, remodeled various amyloids (Figure 2E).
The low ClpB activity might reflect a lack of unknown co-factors that enable amyloid disaggregation. However, E. coli cytosol had limited ability to disaggregate amyloid and did not stimulate ClpB (Figure S4A). By contrast, yeast cytosol remodeled diverse amyloids, whereas Δhsp104 yeast cytosol did not unless supplemented with Hsp104 (Figure S4B). Thus, the failure of E. coli cytosol to stimulate amyloid disaggregation by ClpB indicated that co-factors were not missing and that ClpB has limited amyloid-disaggregase activity.
The inability of ClpB to disaggregate amyloid (Figure 2C, D) might reflect a reduced binding affinity for amyloid. Yet, the Kd of ClpB and Hsp104 for each amyloid and disordered aggregate used here was similar and ranged from ~30–100nM (Table S1). Thus, some aspect of amyloid antagonizes ClpB but not Hsp104 after initial engagement.
ClpB is more sensitive than Hsp104 to ATPase-defective subunits (Figure 1G, I). Thus, amyloid might inhibit the ATPase activity of sufficient ClpB subunits per hexamer to ablate activity. Indeed, amyloids inhibited ClpB ATPase activity by ~30%, whereas disordered aggregates stimulated by ~20% (Figure 2F). Hsp104 ATPase activity was stimulated by disordered aggregates and several amyloids, but some amyloids had no effect (Figure 2F). Thus, amyloid specifically inhibits ClpB ATPase activity, which might explain ClpB’s limited amyloid-disaggregase activity.
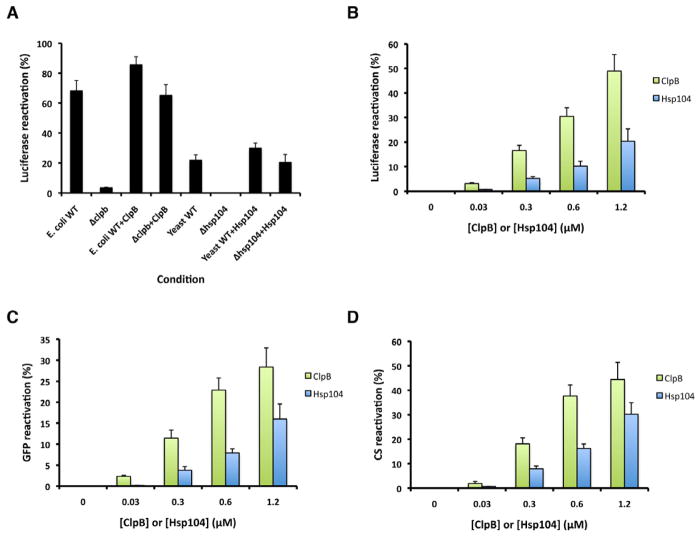
ClpB reactivates disordered aggregates more effectively than Hsp104
E. coli cytosol was more active than yeast cytosol in reactivating aggregated luciferase, whereas Δclpb E cytosol was inactive, but could be rescued by pure ClpB (Figure. coli 3A). Accordingly, ClpB was more effective than Hsp104 in disordered aggregate dissolution (Figure 3B–D). Thus, ClpB appears more adapted to resolve disordered aggregates that accrue upon protein-folding stress, but is ineffective against amyloid.
Figure 3. ClpB reactivates disordered aggregates more effectively than Hsp104.
(A) Luciferase aggregates were treated with the indicated combination of E. coli WT cytosol, E. coli Δclpb c ytosol, ClpB, yeast WT cytosol, yeast Δhsp104 cytosol or Hsp104. Luciferase reactivation was assessed (% of total recoverable activity). Values represent means±SEM (n=3).
(B–D) Disordered luciferase aggregates (B), disordered GFP aggregates (C) or disordered CS aggregates (D) were treated with ClpB, DnaK, DnaJ and GrpE or Hsp104, Ssa1 and Sis1. Reactivation was then assessed (% of total recoverable activity). Values represent means±SEM (n=3).
See also Table S1.
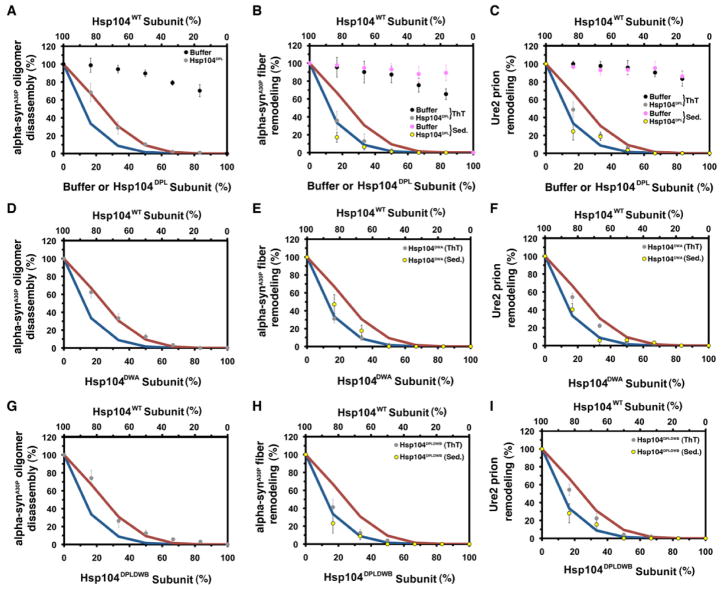
Hsp104 uses a distinct mechanism to resolve toxic oligomers and amyloids
Next, we analyzed Hsp104-catalyzed disassembly of toxic preamyloid oligomers and amyloid formed by the PD-linked α-synA30P and Ure2 prions. Disassembly of α-synA30P oligomers, α-synA30P amyloid and Ure2 prions by Hsp104 was very sensitive to Hsp104DPL (Figure 4A–C), Hsp104DWA (Figure 4D–F) and Hsp104DPLDWB (Figure 4G–I). Hsp104’s ability to disassemble α-SynA30P oligomers was abolished by ~2 mutant subunits per hexamer (Figure 4A, D, G), whereas α-SynA30P amyloid and Ure2 prion disassembly was ablated by 1 mutant subunit per hexamer (Figure 4B, C, E, F, H, I). Thus, more subunits must work together to disaggregate amyloid compared to disordered aggregates. These data suggest that Hsp104 hexamers switch to a highly co-operative mode of ATP hydrolysis and substrate handling to disassemble preamyloid oligomers and amyloids.
Figure 4. Hsp104 exploits co-operative mechanisms to remodel preamyloid α-synA30P oligomers, α-synA30P amyloids and Ure2 prions.
(A–I) α-synA30P oligomers (A, D, G), α-synA30P amyloid (B, E, H) or Ure2 prions (C, F, I) were treated with Hsp104, Ssa1 and Sis1 plus increasing fractions of buffer (A, B, C), Hsp104DPL (A, B, C), Hsp104DWA (D, E, F) or Hsp104DPLDWB (G, H, I). Oligomer remodeling was assessed by filter trap and amyloid remodeling was assessed by ThT fluorescence (grey or black markers) or sedimentation (purple or yellow markers). Activity was converted to % WT activity. Values represent means±SEM (n=2–4). Expected activity if 1 or more (blue line, A–I), or 2 or more (red line, A–I) mutant subunits ablate hexamer activity.
See also Supplementary Figure S4.
The response to mutant subunits (Hsp104DPL, Hsp104DWA and Hsp104DPLDWB) was invariant for amyloid remodeling, whereas the same mutant subunits elicit diverse responses in disordered aggregate dissolution (e.g. compare Figure 4B, E, H to Figure 1F, G, I). Thus, amyloids make more stringent demands on how Hsp104 subunits must collaborate to promote disaggregation.
Missing co-factors might enable Hsp104 to disaggregate amyloid using a probabilistic mechanism as for disordered aggregates. For example, Hsp26 can assist Hsp104 in protein disaggregation (Duennwald et al., 2012). However, neither Hsp26 nor Δhsp104 yeast cytosol (to provide the entire cohort of molecular chaperones) altered the response of Hsp104 to Hsp104DPL subunits in luciferase or Ure2 prion disaggregation (Figure S4C, D). Thus, missing co-factors are unlikely to alter the mechanism by which Hsp104 subunits collaborate to disaggregate disordered aggregates versus amyloid.
Hsp104 switches mechanism to disaggregate distinct Sup35 prion strains
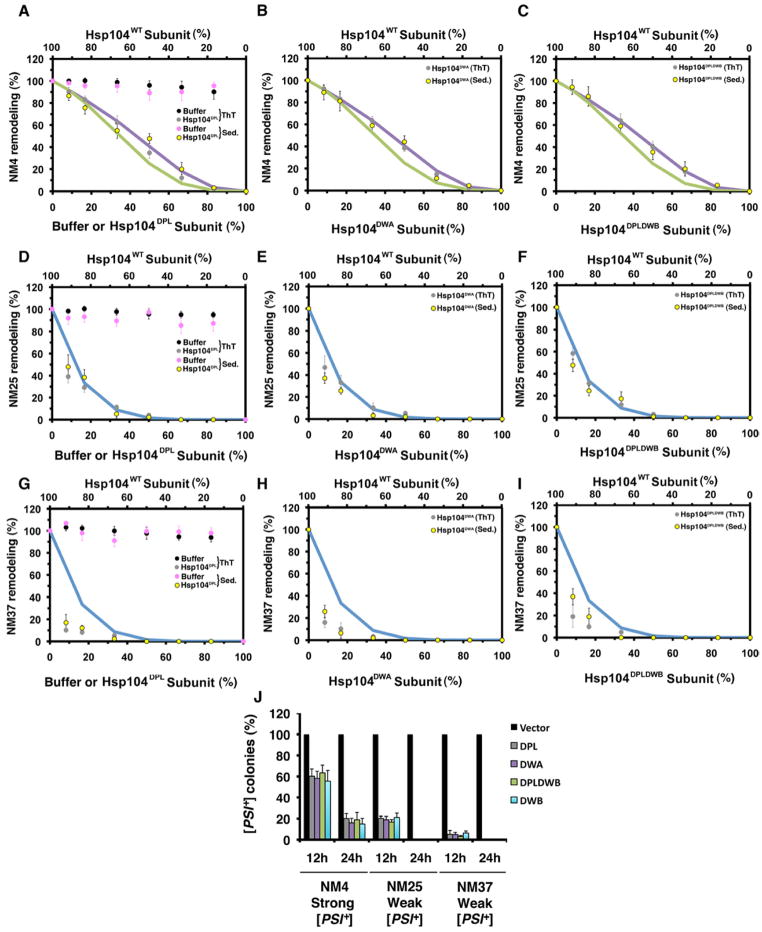
Amyloidogenic proteins form structurally distinct amyloid ‘strains’, which can vary in stability and confer distinct phenotypes (Cushman et al., 2010). Hsp104 subunits might collaborate differently to disaggregate distinct amyloid strains formed by the same protein. To examine this possibility, we exploited Sup35’s prion domain, termed NM, which spontaneously forms different prion strains at different temperatures. NM prions formed at 4°C, termed NM4, possess a shorter, less stable amyloid core (Tm~54°C) with distinctive intermolecular contacts and give rise to ‘strong’ [PSI+] variants in vivo (Krishnan and Lindquist, 2005) (Figure S5). Here, ‘strength’ refers to the nonsense suppression phenotype caused by prion-mediated depletion of soluble Sup35 (Shorter and Lindquist, 2005). NM prions formed at 25°C or 37°C, termed NM25 and NM37, harbor longer, more stable amyloid cores (Tm~81°C for NM25 and Tm~86°C for NM37) with intermolecular contacts distinct from NM4, and give rise to ‘weak’ [PSI+] variants in vivo (Krishnan and Lindquist, 2005) (Figure S5). NM4, NM25 and NM37 provide an opportunity to assess Hsp104 activity against alternative prion structures formed by the same primary sequence.
Remodeling each NM prion strain required a different mode of intersubunit collaboration by Hsp104. Thus, NM4 remodeling was less sensitive than NM25 or NM37 to Hsp104DPL (Figure 5A, D, G), Hsp104DWA (Figure 5B, E, H), Hsp104DPLDWB (Figure 5C, F, I) or Hsp104DWB (data not shown). NM4 remodeling was ablated by ~3–4 mutant subunits per hexamer, whereas NM25 remodeling was ablated by 1 mutant subunit per hexamer (see Figure 1B). NM37 remodeling was unusually sensitive to mutant subunits (Figure 5G–I) suggesting that more than 1 hexamer is needed to remodel this strain. Thus, as the length of the cross-β core of the NM prion increases and encroaches further into C-terminal sequence (Figure S5), the mechanism by which Hsp104 subunits collaborate switches to become more co-operative. For NM4, a subglobal co-operative mechanism will suffice, whereas NM25 requires global co-operativity.
Figure 5. Hsp104 switches mechanism to remodel distinct Sup35 prion strains.
(A–I) NM4, NM25 or NM37 prions were treated with Hsp104, Ssa1 and Sis1 plus increasing fractions of buffer (A, D, G), Hsp104DPL (A, D, G), Hsp104DWA (B, E, H) or Hsp104DPLDWB (C, F, I). Remodeling was monitored by ThT fluorescence (grey or black markers) or sedimentation (purple or yellow markers). Activity was converted to % WT activity. Values represent means±SEM (n=2–4). Expected activity if 4 or more (purple line, A–C), 3 or more (green line, A–C), or 1 or more (blue line, D–I) mutant subunits ablate hexamer activity.
(J) Strong and weak [PSI+] curing by Hsp104DPL, Hsp104DWA, Hsp104DPLDWB or Hsp104DWB overexpression. Values represent means±SEM (n=3).
See also Supplementary Figure S5.
Next, we tested the efficacy by which mutant Hsp104 subunits disrupt propagation of different [PSI+] variants by Hsp104 in vivo (Chernoff et al., 1995). In accord with our in vitro data, Hsp104DPL, Hsp104DWA, Hsp104DWB and Hsp104DPLDWB more readily disrupted propagation of weak [PSI+] encoded by NM37 or NM25 than propagation of strong [PSI+] encoded by NM4 (Figure 5J). Thus, Hsp104-driven remodeling of weak [PSI+] prions (NM25 and NM37) is more sensitive to Hsp104DPL, Hsp104DWA, Hsp104DWB and Hsp104DPLDWB subunits than Hsp104-driven remodeling of strong [PSI+] prions (NM4) in vitro and in vivo (Figure 5A–J). Importantly, these Hsp104 variants were equally effective in disrupting the Hsp104-catalyzed remodeling of a given [PSI+] variant in vitro and in vivo (Figure 5A–J). Unlike their effects on thermotolerance or in vivo luciferase reactivation, Hsp104DWB was not a more effective dominant negative than Hsp104DPL or Hsp104DPLDWB (Figure 1J–L, 5J). Thus, the mechanism by which Hsp104 remodels prions versus disordered aggregates differs in vivo.
Hsp104 switches mechanism to disaggregate disordered aggregates versus prions
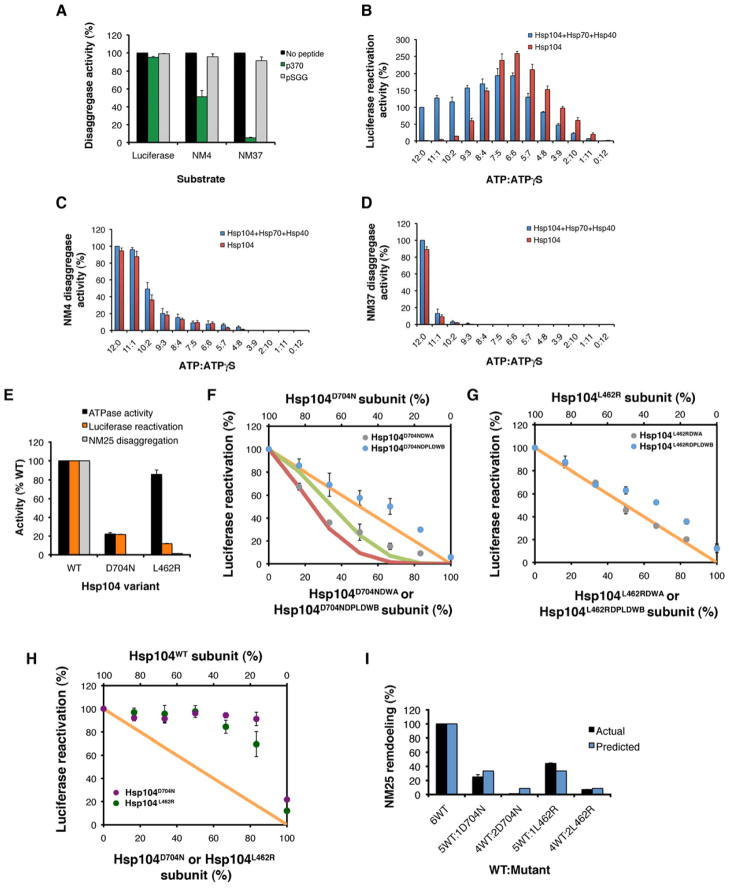
We confirmed that Hsp104 switches mechanism to resolve disordered aggregates versus prions using 2 strategies that do not employ mutant subunits. First, we used p370, a short peptide that competitively inhibits Hsp104-substrate binding (Lum et al., 2008). Importantly, Hsp104-catalyzed luciferase reactivation was insensitive to a 20-fold excess of p370, whereas NM4 remodeling was inhibited and NM37 remodeling was abolished (Figure 6A). A negative control peptide, pSGG, had no effect (Figure 6A). Thus, in accord with Hsp104DPL doping (Figure 1F, 5B, H), amyloid disaggregation by Hsp104 is more sensitive to inhibition of substrate binding than disordered aggregate dissolution.
Figure 6. Selective ablation of amyloid disaggregase activity by p370, ATPγS, Hsp104D704N or Hsp104L462R subunits.
(A) Luciferase aggregates, NM4 prions or NM37 prions were treated with Hsp104, Ssa1 and Sis1 plus buffer, p370 or pSGG. Disaggregase activity was converted to % activity in the absence of peptide. Values represent means±SEM (n=2).
(B) Luciferase aggregates were treated with Hsp104, Hsc70 and Hdj2 or Hsp104 alone plus various ATP:ATPγS ratios. Disaggregase activity was converted to % activity of Hsp104, Hsc70 and Hdj2 plus ATP. Values represent means±SEM (n=3).
(C, D) NM4 prions (C) or NM37 prions (D) were treated with Hsp104, Ssa1 and Sis1 or Hsp104 alone plus various ATP:ATPγS ratios. Disaggregase activity was converted to % activity of Hsp104, Ssa1 and Sis1 plus ATP. Values represent means±SEM (n=3).
(E) Comparison of ATPase activity, luciferase reactivation activity, and NM25 disaggregase activity of WT Hsp104, Hsp104D704N, and Hsp104L462R. Observed activity was converted to % WT activity. Values represent means±SEM (n=2–3).
(F) Luciferase aggregates were treated with Hsp104D704N, Hsc70 and Hdj2 plus increasing fractions of Hsp104D704NDWA (grey markers) or Hsp104D704NDPLDWB (blue markers). Luciferase reactivation was converted to % Hsp104D704N activity. Expected activity if 6 (orange line), 3 or more (green line), or 2 or more (red line) mutant subunits ablate hexamer activity. Values represent means±SEM (n=3).
(G) Luciferase aggregates were treated with Hsp104L462R, Hsc70 and Hdj2 plus increasing fractions of Hsp104L462RDWA (grey markers) or Hsp104L462RDPLDWB (blue markers). Luciferase reactivation was converted to % Hsp104L462R activity. Orange line: expected activity if 6 mutant subunits are needed to ablate hexamer activity. Values represent means±SEM (n=3).
(H) Luciferase aggregates were treated with Hsp104, Hsc70 and Hdj2 plus increasing fractions of Hsp104D704N (purple markers) or Hsp104L462R (green markers). Luciferase reactivation was converted to % WT Hsp104 activity. Orange line: expected activity if 6 mutant subunits are needed to ablate hexamer activity. Values represent means±SEM (n=2).
(I) NM25 was treated with Hsp104, Ssa1 and Sis1 plus increasing fractions of Hsp104D704N or Hsp104L462R. Remodeling was monitored by ThT fluorescence. Activity was converted to % WT Hsp104 activity. Predicted activity (blue bars) if 1 mutant subunit ablates hexamer activity. Values represent means±SEM (n=2).
Next, we examined the effect of various ratios of ATP and ATPγS, a slowly hydrolyzable ATP analog. We kept the total nucleotide concentration constant, but varied the ATP:ATPγS ratio from 12:0 to 0:12. Luciferase reactivation by Hsp104, Hsp70 and Hsp40 was largely unaffected by increasing fractions of ATPγS. Optimal activity was observed at 7:5 or 6:6 ATP:ATPγS, and a ratio of 4:8 ATP:ATPγS supported activity similar to reactions with just ATP (Figure 6B). Activity was even detected at 1:11 ATP:ATPγS (Figure 6B). Hsp104 alone was inactive with ATP, but addition of ATPγS unleashed activity, and a 6:6 ATP:ATPγS ratio elicited maximal Hsp104 activity (Figure 6B). These activity profiles illustrate the adaptability of the Hsp104 hexamer, which can effectively disaggregate luciferase when diverse ATP:ATPγS mixtures populate its NBDs. By contrast, Hsp104-catalyzed remodeling of NM4 was sharply inhibited by low fractions of ATPγS and NM37 was even more sensitive (Figure 6C, D). Thus, WT Hsp104 uses a distinct mechanism to disaggregate disordered aggregates versus amyloid.
Key middle domain and NBD2 residues enable Hsp104 to switch mechanism
We hypothesized that Hsp104 variants that are functional in thermotolerance but defective in prion propagation in vivo might be unable to switch mechanism. We focused on Hsp104D704N and Hsp104L462R, which confer WT thermotolerance but cannot propagate [PSI+], [RNQ+] or [URE3] (Kurahashi and Nakamura, 2007). D704 is between the NBD2 Walker B and sensor-1 motifs, whereas L462 is in helix 2 of the middle domain. D704 is predicted to contact the middle domain, whereas L462 is predicted to be in proximity to nucleotide in NBD1 (Wendler et al., 2007). Thus, D704 and L462 could mediate the interdomain or intersubunit communication necessary to switch mechanism.
In vitro, Hsp104D704N had reduced ATPase activity, whereas Hsp104L462R had WT levels of ATPase activity (Figure 6E). Both mutants had reduced ability to reactivate luciferase aggregates and could not remodel NM25 (Figure 6E), which explains their ability to confer thermotolerance but not prion propagation in vivo (Kurahashi and Nakamura, 2007). Very little functional Hsp104 is required for thermotolerance (Lindquist and Kim, 1996). Thus, reduced Hsp104D704N or Hsp104L462R activity against disordered aggregates is likely sufficient for thermotolerance, especially when cells are given a conditioning pretreatment.
The limited ability of Hsp104D704N and Hsp104L462R to remodel amyloid is reminiscent of ClpB (Figure 2C, D). Thus, Hsp104D704N and Hsp104L462R subunits might also collaborate differently than WT Hsp104 subunits to dissolve disordered aggregates. To probe how Hsp104D704N and Hsp104L462R subunits collaborate in luciferase reactivation, we doped in mutant Hsp104D704N and Hsp104L462R subunits defective in ATP hydrolysis (DWA) or ATP hydrolysis and substrate binding (DPLDWB). For Hsp104D704N, activity declined sharply upon doping Hsp104D704NDWA consistent with 2–3 mutant subunits inactivating the Hsp104D704N hexamer (Figure 6F). Thus, Hsp104D704N exploits a subglobally cooperative mechanism of ATP hydrolysis to reactivate luciferase, unlike WT Hsp104, which uses a probabilistic mechanism (Figure 1G). Indeed, Hsp104D704N responds to ATPase-defective subunits more like ClpB (Figure 1G), which has limited amyloid-remodeling activity (Figure 2C, D). Hsp104D704NDPLDWB subunits elicited an approximately linear decline in Hsp104D704N luciferase reactivation activity (Figure 6F), rather than the stimulation observed with WT Hsp104 or sharp inhibition observed with ClpB (Figure 1I). Unlike WT Hsp104, Hsp104D704N subunits with defective ATPase and substrate-binding activity do not stimulate adjacent Hsp104D704N subunits. Thus, D704N impairs intersubunit communication and precludes amyloid disaggregation.
Hsp104L462R subunits collaborated differently than Hsp104D704N and WT Hsp104 subunits to disaggregate luciferase. Doping Hsp104L462RDWA caused a roughly linear decline in Hsp104L462R activity, indicating a probabilistic mechanism akin to WT Hsp104 (Figure 1G, 6G). Doping Hsp104L462RDPLDWB elicited a roughly linear decline in Hsp104L462R luciferase reactivation activity, rather than the stimulation observed with WT Hsp104 (Figure 1I, 6G). Thus, Hsp104L462R subunits with defective ATPase and substrate-binding activity do not stimulate adjacent Hsp104L462R subunits. We conclude that L462R disrupts intersubunit communication and ablates amyloid remodeling.
Doping Hsp104D704N or Hsp104L462R subunits had little effect on luciferase reactivation by WT Hsp104, even though Hsp104D704N and Hsp104L462R are ~5–9-fold less active than WT Hsp104 against luciferase (Figure 6E, H). These data reillustrate the resilience of the Hsp104 hexamer and its capacity to accommodate defective subunits and still effectively resolve disordered aggregates. Even an average of 1 WT subunit per Hsp104D704N or Hsp104L462R hexamer is capable of catalyzing the same amount of disaggregation as a WT hexamer (Figure 6H). By contrast, Hsp104D704N or Hsp104L462R subunits caused a sharp decline in Hsp104-catalyzed NM25 remodeling consistent with 1 mutant subunit disrupting hexamer activity (Figure 6I). Thus, D704 and L462 likely transmit or receive signals that recruit additional Hsp104 subunits for amyloid disaggregation. Impairing intersubunit communication with specific mutations, such as D704N or L462R, yields Hsp104 variants that dissolve disordered aggregates but not amyloid.
Discussion
We have established that Hsp104 employs distinct modes of intersubunit collaboration to resolve disordered aggregates versus amyloid. For disordered aggregates, Hsp104 subunits use probabilistic ATP hydrolysis similar to the mechanism defined for ClpX, a protein unfoldase (Martin et al., 2005). However, unlike ClpX, Hsp104 withstands subunits that cannot bind substrate. ClpX hexamers are severely impaired by 2 subunits that cannot engage substrate (Martin et al., 2008), whereas Hsp104 retains ~70% activity. This sensitivity might explain why ClpX is a poor protein disaggregase (Doyle et al., 2007a).
The permissive nature of Hsp104 hexamers to subunits that cannot hydrolyze ATP or engage substrate enables a highly flexible disaggregase. Thus, 1 WT subunit per hexamer is sufficient to catalyze disaggregation (Figure 7A). Indeed, any opportunely positioned subunit within the hexamer that can hydrolyze ATP and engage the irregular and heterogeneous aggregated structure can promote disaggregation. Individual subunits do not have to co-ordinate ATPase or substrate-binding events with neighboring subunits or wait until all subunits are engaged, which may be sterically improbable. Thus, Hsp104 can resolve the unrelated proteins of the aggregated proteome after stress.
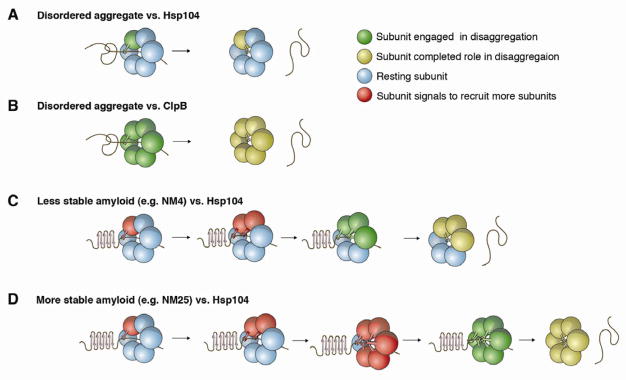
Figure 7. Mechanisms of intersubunit collaboration for Hsp104 and ClpB.
(A–D) Hsp104 (A, C, D) or ClpB
(B) subunits are depicted as spheres and a single aggregated conformer is displayed. Green subunits are engaged in productive disaggregation via substrate binding (depicted by a lever) and/or ATP hydrolysis. Yellow subunits have completed their role in disaggregation. Blue subunits are resting and do not need to hydrolyze ATP or engage substrate for successful disaggregation. Red subunits recruit resting subunits until a sufficient number are recruited to promote disaggregation.
(A) Hsp104 couples probabilistic ATPase activity and substrate binding to resolve disordered aggregates. Thus, a single subunit within a hexamer that can bind substrate and hydrolyze ATP is sufficient to drive protein disaggregation.
(B) ClpB exploits co-operative ATPase activity and probabilistic substrate binding to resolve disordered aggregates. 5 or 6 ClpB subunits per hexamer must hydrolyze ATP to disaggregate disordered aggregates. Co-operative ATPase activity is not coupled to cooperative substrate handling, as 1 ClpB subunit capable of binding substrate can drive disaggregation provided 5 or 6 subunits can hydrolyze ATP.
(C) Hsp104 switches to a subglobal co-operative mechanism of ATP hydrolysis and substrate binding to resolve NM4 prions. 1 subunit initially engages amyloid but the localized structural stability of the cross-β form antagonizes unfolding, which elicits a signal (red subunit) that recruits additional subunits until a sufficient number are recruited that can together unfold the cross-β structure. For NM4, 3 subunits per hexamer must engage substrate and hydrolyze ATP.
(D) Hsp104 switches to a global co-operative mechanism of ATP hydrolysis and substrate binding to resolve more refractory amyloids, such as NM25 prions. Hsp104 subunits collaborate as in (C) except that the local stability of the amyloid fold is even more antagonistic such that 6 subunits must be recruited to engage substrate and hydrolyze ATP for disaggregation.
Surprisingly, ClpB, the E. coli homolog of Hsp104, is tuned differently to Hsp104. Like Hsp104, ClpB exploits probabilistic substrate binding to dissolve disordered aggregates and tolerates subunits that cannot bind substrate (Figure 7B). This shared feature of ClpB and Hsp104 distinguishes them from the protein unfoldase, ClpX.
Unlike Hsp104, ClpB couples probabilistic substrate binding to highly co-operative ATP hydrolysis (Figure 7B). Unexpectedly, this operating mode enables ClpB to dissolve disordered aggregates more effectively than Hsp104. However, this enhancement comes at the expense of robust disaggregase activity able to accommodate ATPase-defective subunits. Unlike Hsp104, ClpB hexamers cannot tolerate a single ATPase-defective subunit. Our data also suggest that unlike Hsp104, ClpB has limited ability to couple cooperative ATPase activity to co-operative substrate handling, which is necessary to remodel amyloid.
The robustness and plasticity of Hsp104 hexamers is likely an adaptation that enables amyloid remodeling and empowers yeast to exploit prions for beneficial purposes. Indeed, ClpB and E. coli cytosol were unable to remodel amyloid. Amyloid can accumulate in E. coli upon protein overexpression (Wang et al., 2008). Yet, ClpB’s limited amyloid-remodeling activity suggests that E. coli compartmentalizes amyloid rather than disseminating it throughout the cytoplasm. Yeast also partition amyloid, but simultaneously disperse cytosolic prions for beneficial purposes. The profound selective advantages afforded by yeast prions are only made possible by Hsp104’s potent amyloid-remodeling activity (Alberti et al., 2009; Halfmann et al., 2012; Shorter and Lindquist, 2005).
We suggest that Hsp104’s default intersubunit collaboration mechanism is probabilistic (Figure 7A). However, this default-operating mode can be rapidly retuned to a suitable subglobal or global co-operative mechanism upon sensing stable substrates. Thus, amyloid likely antagonizes unfolding and elicits a signal for Hsp104 subunits to work together to engage substrate, hydrolyze ATP and promote disaggregation (Figure 7C, D). For less chemically stable NM4 prions, a subglobal co-operative mechanism that is inactivated by 3 mutant subunits per hexamer is employed (Figure 7C). By contrast, NM25 prions, which are more stable and possess a longer cross-β core, are resolved by a global co-operative mechanism that is inactivated by 1 mutant subunit (Figure 7D). Ure2 prions and α-synA30P amyloid are also resolved in this way (Figure 7D). Cryo-EM reconstructions indicate that Hsp104 might use a co-operative, sequential mechanism of substrate handling (Wendler et al., 2009). However, we suggest that hexamer plasticity enables Hsp104 to adapt a variety of mechanochemical coupling mechanisms that are responsive to the specific physical demands of the aggregated substrate. Thus, Hsp104 is wired do the minimum work necessary to disaggregate any given substrate, i.e. if 2 subunits are sufficient to rapidly disaggregate a substrate, then only 2 will be used. Various multimeric, NTP-fueled ring-translocases with diverse substrate portfolios could use similar adaptable repertoires of intersubunit collaboration.
We establish that D704N or L462R mutations impair intersubunit communication, reduce plasticity and selectively ablate amyloid disaggregation. Indeed, D704 and L462 likely transmit or receive signals to recruit additional Hsp104 subunits during prion disaggregation (Figure 7C, D). Although further studies are needed to gain a structural understanding of how Hsp104 switches mechanism, our findings explain why Hsp104D704N and Hsp104L462R are functional in thermotolerance but defective in prion propagation (Kurahashi and Nakamura, 2007).
Hsp104 might be designed to be more potent and selective against specific proteins, which could empower facile purification of irksome recombinant proteins for basic or therapeutic purposes. Hsp104 could also be developed to target select misfolded proteins in neurodegenerative disease (Vashist et al., 2010). The intrinsic ability of Hsp104 to remodel diverse disease-associated amyloids as well as toxic oligomers suggests that this avenue warrants exploration. Here, it will be key to increase the specificity of the Hsp104 hexamer for a target polypeptide while simultaneously tuning plasticity such that toxic conformers are selectively eradicated. For example, hypomorphic scaffolds based on Hsp104D704N or Hsp104L462R could be useful in settings where amyloids are protective and disordered aggregates are toxic.
Experimental Procedures
Modeling Heterohexamer Ensemble Activity
The bionomial distribution was used to simulate the activity of various heterohexamer ensembles (Werbeck et al., 2008). For more details, see Extended Experimental Procedures.
Proteins
Proteins were purified as described (Shorter and Lindquist, 2004, 2006; Lo Bianco et al., 2008). For more details see Extended Experimental Procedures.
Cytosol preparation
Yeast and bacterial cytosol were prepared as described (Glover and Lindquist, 1998) with modifications as detailed in Extended Experimental Procedures.
Subunit mixing
Statistical subunit mixing using biotinylated, his-tagged or fluorescently labeled Hsp104 was assessed as described (Werbeck et al., 2008). For more details see Extended Experimental Procedures.
ATPase assay
ATPase activity was measured as described (Wendler et al., 2007). For more details see Extended Experimental Procedures.
Disaggregation assays
Disaggregation of luciferase, GFP, CS, various amyloids and α-synA30P oligomers was performed as described (Doyle et al, 2007b; Glover and Lindquist, 1998; Lo Bianco et al., 2008; Shorter and Lindquist, 2004, 2006). For more details see Extended Experimental Procedures.
Thermotolerance, in vivo luciferase reactivation and [PSI+] curing assays
In vivo thermotolerance, luciferase reactivation and [PSI+] curing assays were performed as described (Chernoff et al., 1995; Parsell et al., 1994: Wendler et al., 2007). For more details see Extended Experimental Procedures.
Supplementary Material
Article Highlights.
Hsp104 switches mechanism to disaggregate disordered aggregates versus amyloid
ClpB operates with reduced plasticity and has limited amyloid-disaggregase activity
Hsp104 remodels diverse toxic oligomers and amyloids linked to neurodegeneration
Hsp104 plasticity enables yeast to harness prions for advantageous purposes
Acknowledgments
We thank Sue Lindquist, Sabine Kedzierska-Mieszkowska and Virginia Lee for reagents. We thank Sandra Maday, Mark Lemmon, Aaron Gitler, Walter Englander and Nancy Bonini for critiques and Lili Guo for artwork. Our work was funded by NIH training grant (T32GM071339) and NRSA predoctoral fellowship (F31NS079009) (M.E.D.); NIH training grant (T32GM008275) (E.A.S. & M.A.S.); AHA predoctoral (E.A.S.) and postdoctoral fellowships (M.E.J); NIH training grant (T32AG000255) and NRSA predoctoral fellowship (F31NS067890) (M.C-N.); NIH Director’s New Innovator Award (DP2OD002177), Ellison Medical Foundation New Scholar in Aging Award, Penn Institute of Aging, Alzheimer Disease Core Center, and Diabetes Research Center Awards (J.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosl B, Grimminger V, Walter S. Substrate Binding to the Molecular Chaperone Hsp104 and Its Regulation by Nucleotides. J Biol Chem. 2005;280:38170–38176. doi: 10.1074/jbc.M506149200. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [PSI+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- Cushman M, Johnson BS, King OD, Gitler AD, Shorter J. Prion-like disorders: blurring the divide between transmissibility and infectivity. J Cell Sci. 2010;123:1191–1201. doi: 10.1242/jcs.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandoy-Dron F, Bogdanova A, Beringue V, Bailly Y, Tovey MG, Laude H, Dron M. Infection by ME7 prion is not modified in transgenic mice expressing the yeast chaperone Hsp104 in neurons. Neurosci Lett. 2006;405:181–185. doi: 10.1016/j.neulet.2006.05.066. [DOI] [PubMed] [Google Scholar]
- Doyle SM, Hoskins JR, Wickner S. Collaboration between the ClpB AAA+ remodeling protein and the DnaK chaperone system. Proc Natl Acad Sci USA. 2007a;104:11138–11144. doi: 10.1073/pnas.0703980104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SM, Shorter J, Zolkiewski M, Hoskins JR, Lindquist S, Wickner S. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat Struct Mol Biol. 2007b;14:114–122. doi: 10.1038/nsmb1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SM, Wickner S. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem Sci. 2009;34:40–48. doi: 10.1016/j.tibs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Duennwald ML, Echeverria A, Shorter J. Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol. 2012;10:e1001346. doi: 10.1371/journal.pbio.1001346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity SJ, Sivanathan V, Dong J, Lindquist S, Hochschild A. Conversion of a yeast prion protein to an infectious form in bacteria. Proc Natl Acad Sci USA. 2010;107:10596–10601. doi: 10.1073/pnas.0913280107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482:363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattendorf DA, Lindquist SL. Cooperative kinetics of both Hsp104 ATPase domains and interdomain communication revealed by AAA sensor-1 mutants. EMBO J. 2002;21:12–21. doi: 10.1093/emboj/21.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins JR, Doyle SM, Wickner S. Coupling ATP utilization to protein remodeling by ClpB, a hexameric AAA+ protein. Proc Natl Acad Sci USA. 2009;106:22233–22238. doi: 10.1073/pnas.0911937106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles TP, Fitzpatrick AW, Meehan S, Mott HR, Vendruscolo M, Dobson CM, Welland ME. Role of intermolecular forces in defining material properties of protein nanofibrils. Science. 2007;318:1900–1903. doi: 10.1126/science.1150057. [DOI] [PubMed] [Google Scholar]
- Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H, Nakamura Y. Channel mutations in Hsp104 hexamer distinctively affect thermotolerance and prion-specific propagation. Mol Microbiol. 2007;63:1669–1683. doi: 10.1111/j.1365-2958.2007.05629.x. [DOI] [PubMed] [Google Scholar]
- Lee S, Sielaff B, Lee J, Tsai FT. CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc Natl Acad Sci USA. 2010;107:8135–8140. doi: 10.1073/pnas.1003572107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S, Kim G. Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. Proc Natl Acad Sci USA. 1996;93:5301–5306. doi: 10.1073/pnas.93.11.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Bianco C, Shorter J, Regulier E, Lashuel H, Iwatsubo T, Lindquist S, Aebischer P. Hsp104 antagonizes alpha-synuclein aggregation and reduces dopaminergic degeneration in a rat model of Parkinson disease. J Clin Invest. 2008;118:3087–3097. doi: 10.1172/JCI35781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum R, Niggemann M, Glover JR. Peptide and Protein Binding in the Axial Channel of Hsp104: Insights into the mechanism of protein unfolding. J Biol Chem. 2008;283:30139–30150. doi: 10.1074/jbc.M804849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubimov AY, Strycharska M, Berger JM. The nuts and bolts of ring-translocase structure and mechanism. Curr Opin Struct Biol. 2011;21:240–248. doi: 10.1016/j.sbi.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Rebuilt AAA+ motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Protein unfolding by a AAA+ protease is dependent on ATP-hydrolysis rates and substrate energy landscapes. Nat Struct Mol Biol. 2008;15:139–145. doi: 10.1038/nsmb.1380. [DOI] [PubMed] [Google Scholar]
- Moreau MJ, McGeoch AT, Lowe AR, Itzhaki LS, Bell SD. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol Cell. 2007;28:304–314. doi: 10.1016/j.molcel.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- Reidy M, Miot M, Masison DC. Prokaryotic Chaperones Support Yeast Prions and Thermotolerance and Define Disaggregation Machinery Interactions. Genetics. 2012 doi: 10.1534/genetics.112.142307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer EC, Ware DM, Queitsch C, Kowal AS, Lindquist SL. Subunit interactions influence the biochemical and biological properties of Hsp104. Proc Natl Acad Sci USA. 2001;98:914–919. doi: 10.1073/pnas.031568098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PloS One. 2011;6:e26319. doi: 10.1371/journal.pone.0026319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell. 2006;23:425–438. doi: 10.1016/j.molcel.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarz P, Mogk A, Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol. 2008;68:87–97. doi: 10.1111/j.1365-2958.2008.06135.x. [DOI] [PubMed] [Google Scholar]
- Vacher C, Garcia-Oroz L, Rubinsztein DC. Overexpression of yeast Hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington’s disease. Hum Mol Genet. 2005;14:3425–3433. doi: 10.1093/hmg/ddi372. [DOI] [PubMed] [Google Scholar]
- Vashist S, Cushman M, Shorter J. Applying Hsp104 to protein-misfolding disorders. Biochem Cell Biol. 2010;88:1–13. doi: 10.1139/o09-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Maji SK, Sawaya MR, Eisenberg D, Riek R. Bacterial inclusion bodies contain amyloid-like structure. PLoS Biol. 2008;6:e195. doi: 10.1371/journal.pbio.0060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Schubert D, Sawaya MR, Eisenberg D, Riek R. Multidimensional structure-activity relationship of a protein in its aggregated states. Angew Chem Int Ed Engl. 2010;49:3904–3908. doi: 10.1002/anie.201000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibezahn J, Schlieker C, Bukau B, Mogk A. Characterization of a trap mutant of the AAA+ chaperone ClpB. J Biol Chem. 2003;278:32608–32617. doi: 10.1074/jbc.M303653200. [DOI] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Wendler P, Shorter J, Plisson C, Cashikar AG, Lindquist S, Saibil HR. Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell. 2007;131:1366–1377. doi: 10.1016/j.cell.2007.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler P, Shorter J, Snead D, Plisson C, Clare DK, Lindquist S, Saibil HR. Motor mechanism for protein threading through Hsp104. Mol Cell. 2009;34:81–92. doi: 10.1016/j.molcel.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werbeck ND, Schlee S, Reinstein J. Coupling and Dynamics of Subunits in the Hexameric AAA+ Chaperone ClpB. J Mol Biol. 2008;378:178–190. doi: 10.1016/j.jmb.2008.02.026. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Bateman D, Kelly AC, Gorkovskiy A. The yeast prions [PSI+] and [URE3] are molecular degenerative diseases. Prion. 2011;5:258–262. doi: 10.4161/pri.5.4.17748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.