Abstract
Two transcription factors, C1 (a Myb-domain protein) and B (a basic-helix-loop-helix protein), mediate transcriptional activation of the anthocyanin-biosynthetic genes of maize (Zea mays). To begin to assess the mechanism of activation, the sequences required for C1- and B-mediated induction have been determined for the a2 promoter, which encodes an anthocyanin-biosynthetic enzyme. Analysis of a series of 7- to 13-base-pair substitutions revealed two regions crucial for activation. One region, centered at −99, contained a C1-binding site that abolished C1 binding. The other crucial region was adjacent, centered at −91. C1 binding was not detected at this site, and mutation of this site did not prevent C1 binding at −99. An oligonucleotide dimer containing these two crucial elements was sufficient for C1 and B activation of a heterologous promoter. These data suggest that activation of the anthocyanin genes involves C1 and another factor binding at closely adjacent sites. Mutating a previously postulated anthocyanin consensus sequence within a2 did not significantly reduce activation by C1 and B. However, sequence comparisons of the crucial a2 regions with sequences important for C1- and B-mediated activation in two other anthocyanin promoters led to a revised consensus element shared by these promoters.
Anthocyanins are purple pigments that are ubiquitous in plants, and their production is regulated by a variety of developmental, environmental, and genetic cues (van der Meer et al., 1993). The enzymatic pathway that produces anthocyanins has been studied in a diverse array of plants, with the majority of genetic experiments performed in maize (Zea mays), petunia, and snapdragon (Dooner et al., 1991; Quattrocchio et al., 1993; Holton and Cornish, 1995). The long history of study, along with available transposon systems in these species, have led to the identification and cloning of most of the biosynthetic genes that constitute the anthocyanin pathway, as well as the identification and cloning of many regulatory genes (Dooner et al., 1991; van der Meer et al., 1993).
Regulation of the anthocyanin pathway in maize requires two classes of transcription factors. One class of regulators contains a bHLH motif (B and R), and the other contains a Myb domain (C1 and Pl). To activate the genes of the anthocyanin pathway, a protein from each class must be expressed; neither alone is sufficient for induction (Goff et al., 1990). The C1 and B proteins directly interact with one another via the two-hybrid assay (Goff et al., 1992), suggesting that these proteins physically act together to activate the genes of this pathway. The precise role of the B protein in activating the anthocyanin-biosynthetic genes is uncertain. Experiments have not revealed either specific DNA-binding activity (L.A. Tolar, M.L. Lesnick, and V.L. Chandler, unpublished data) or an activation domain (Goff et al., 1992). In contrast, the C1 protein binds via its Myb domain to the promoter of the a1 anthocyanin-biosynthetic gene (Sainz et al., 1997) and contains an acidic activation domain (Goff et al., 1991). Together with the physical interaction between C1 and B, this suggests that these proteins directly activate transcription of the biosynthetic genes of the pathway.
A key question remaining is what DNA sequences do C1 and B act through to activate the promoters of the anthocyanin pathway? Several promoters of anthocyanin-biosynthetic genes have been studied, including a1, bz1, and bz2 (Goff et al., 1991; Roth et al., 1991; Grotewold et al., 1994; Tuerck and Fromm, 1994; Bodeau and Walbot, 1996; Sainz et al., 1997). All of these promoters appear to be small (less than 200 bp) and most contain redundant regions, each of which is sufficient for C1 and B induction of a heterologous promoter. Putative sites for C1 and B binding have been proposed based on comparisons of the promoter regions with animal Myb and bHLH consensus-binding sites. Mutational analyses suggest that some but not all of these regions are important for activation. However, only in the case of the a1 promoter has C1 binding been tested. The two functionally important sites on the a1 promoter to which C1 binds do not clearly resemble the consensus Myb-binding site in animals (Sainz et al., 1997). Furthermore, analysis of the binding-site preference of the C1 Myb protein via PCR site-selection experiments have revealed that C1 can bind to a variety of sequences that resemble the consensus site A(C/A)C(T/A)A(C/A)C (Sainz et al., 1997), which is distinct from the animal Myb consensus site TAACNG. Thus, it is difficult to identify putative C1-binding sites simply by sequence comparisons.
Certain regions important for C1- and B-mediated induction among these promoters do show sequence similarity. The analysis of the a1 promoter identified a region crucial for activation by C1 and B (Tuerck and Fromm, 1994), which is located between the two C1-binding sites (Sainz et al., 1997). Comparison of this region with the region of the bz1 promoter previously shown to be important for C1- and B-mediated induction (Goff et al., 1990; Roth et al., 1991) revealed sequence similarity (Tuerck and Fromm, 1994). When three other sequenced promoters from the maize anthocyanin pathway were examined for the presence of this putative consensus sequence within 300 bp of the start of transcription, it was found that all three contained such a region, although the sequence identity was lower (Tuerck and Fromm, 1994). This raises the possibility that there is an anthocyanin consensus sequence, which would represent a binding site for either the B protein, the C1 protein, or for some other factor required for activation of these promoters.
There are several compelling reasons to study additional anthocyanin promoters. First, we have only a limited understanding of what sites C1 prefers to bind. As mentioned above, PCR selection of sites bound by the C1 Myb domain revealed a loose consensus site for the C1 Myb protein, and only two functionally important sites have been determined on actual anthocyanin promoters, both of these on a1. This makes it very difficult to predict where a functional C1-binding site may lie within a promoter. Second, function of the postulated anthocyanin consensus sequence has been tested only within two anthocyanin promoters. To determine if this site has relevance for activation of other promoters, these sequences need to be tested.
The promoter of the a2 gene was chosen to explore these issues for two reasons. Its location relatively late in the anthocyanin pathway suggests that its regulation might be less complex, since it need not respond to as many regulatory signals as genes earlier in the pathway, the products of which function in other biosynthetic pathways. In addition, because a2 lies between bz1 and a1 in the pathway, the two best-studied anthocyanin promoters at the time this study was undertaken, it was hoped that common themes between the regulation of each of these promoters and the promoter of a2 might be found. In this study we determined the sequences that are necessary and sufficient for C1 and B induction of the a2 promoter, determined the location of the C1-binding sites within this promoter, and addressed the relationship between functionally important sequences, C1-binding sites, and potential anthocyanin consensus sequences.
MATERIALS AND METHODS
Cloning of a 2.2-kb Genomic Clone of the a2 Gene
The maize (Zea mays) a2 promoter was first cloned as a 2.2-kb BamHI fragment by Menssen et al. (1990), who mapped the start of transcription and found that this fragment contained 1.9 kb of upstream sequence. We obtained a plasmid with the BamHI genomic fragment of this gene from Alfons Gierl (Technical University Munchen, Garching, Germany). However, we determined that this clone was deleted for approximately 200 bp near the start of transcription (data not shown). Using an approximately 200-bp fragment from the deleted clone as a probe, we cloned an intact, 2.2-kb BamHI fragment from a maize K55 inbred line. Maize genomic DNA was cut with BamHI and subjected to electrophoresis, and DNA fragments of approximately 2 kb were extracted from the gel. This DNA was ligated into λ Zap (Stratagene) according to the manufacturer's instructions. The packaged phage were plated and screened for hybridization with the aforementioned a2 promoter probe using standard methods. Two independent clones were obtained, which appeared identical by restriction analysis. Further restriction analysis was performed on one clone, as well as sequencing of the first approximately 400 bp closest to the start of transcription. Results from these experiments indicated that the cloned fragment was identical to that previously published (Menssen et al., 1990).
Construction of Plasmids
All promoter constructs were cloned into pABR4 (Sainz et al., 1997). This plasmid contains a polylinker site upstream of the adh1 intron, the coding region of the luciferase gene, followed by the nopaline synthase 3′ polyadenylation site and the 3′ end. The 1.9-kb promoter clone was constructed by digesting the original a2 clone with BamHI and NsiI and cloning into pABR4 cut with PstI and BamHI. Most 5′ deletions were constructed using convenient restriction enzyme sites in the native promoter to clone them into pABR4. The 5′ sites were as follows: −635 (KpnI), −287 (PstI), −161 (PmlI), −112 (NruI), and −17 (XhoI), with the 3′ site NsiI (+5) in all cases. The deletion at −73 was constructed via PCR using primers that amplified the −73 to +5 region. The resulting PCR product was then cloned into pABR4 using the NsiI site at the 3′ end, and the BamHI site was introduced with the primer at the 5′ end. DNA sequencing was carried out to confirm that no additional nucleotide changes were introduced during PCR.
The a2 linker-scanner mutations were constructed via PCR using standard methods (Higuchi et al., 1988). The exact sequence changes introduced in each case are shown in Figure 2. Each mutation includes the introduction of an NheI site. The −73 to −41 internal deletion was made by digesting an a2 promoter plasmid containing mutation 5.5 with NheI and XhoI, generating flush ends with Klenow enzyme, and religating the molecule together. Similarly, the −112 to −73 internal deletion was made by digesting a promoter containing mutation 5 with NheI and NruI, generating flush ends, and ligating. All mutagenized plasmids were sequenced throughout the promoter region using the standard dideoxy method (Sanger et al., 1977).
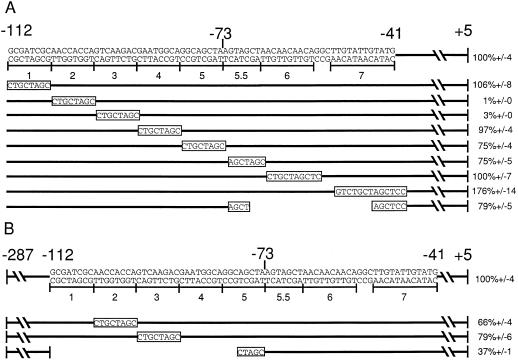
Figure 2.
Activity of a2 promoters with substitution mutations. A, Mutations 1 through 7 were created using PCR to substitute the specific 7- to 13-bp sequence indicated. These mutations maintain the spacing found in the wild-type a2 promoter. The sequence of each of these mutations is shown in the bar below its corresponding number. The last line represents a deletion, with the gap corresponding to the deleted bp. Each of these mutations was assayed for its ability to be activated by C1 and B in the context of the −112-bp promoter. The percentage activation of each of these promoters in transient expression assays relative to the level of activation of the wild-type −112-bp a2 promoter (set at 100%) is shown at the right. Error bars represent se; n = 12. B, Mutations 2 and 3, as well as the deletion shown, were tested in the context of the −287-bp a2 promoter. The percentage activation relative to the −287-bp a2 promoter (set at 100%) is shown at the right. se is indicated for each construct; n = 12.
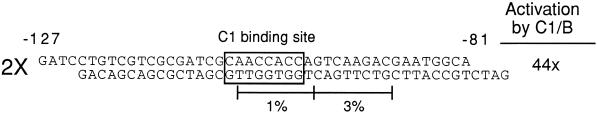
The plasmid used for the sufficiency experiment (see Fig. 6) was made by annealing two complementary oligonucleotides containing DNA from −121 to −81 of the a2 promoter (GATCCTGTCGTCGCGATCGCAACCACCAGTCAAGACGAATGGCA) and ligating into pPHI1960 (Grotewold et al., 1994) cut with BamHI. This plasmid contains a unique BamHI site upstream of a truncated CaMV 35S promoter (from −59 to +2), driving expression of the firefly luciferase gene with the maize adh1 intron. Dideoxy sequencing of resulting constructs was performed to identify a promoter region containing two intact copies of the oligonucleotide in the same orientation as found in the a2 promoter.
Figure 6.
a2-promoter sequences sufficient for C1 and B activation of a heterologous promoter. A synthetic promoter composed of a dimer of an oligonucleotide containing a2 sequences from −121 to −81 in front of a truncated, −59-bp CaMV 35S promoter was tested in the maize transient transformation system. The induction in the presence of C1 and B is shown. The boxed region is the C1-binding site that overlaps the crucial region identified by mutation 2. The lines represent the two regions that when mutated in the context of the −112 region had dramatic effects on C1- and B-mediated activation.
Transient Transformation Assay
Promoter activity of the a2-luciferase constructs was tested in tissue-cultured cells of the maize cv Black Mexican Sweet as described previously (Sainz et al., 1997). All DNA was purified either by CsCl2-gradient centrifugation or by using a Midi-Prep Kit (Qiagen, Chatsworth, CA). Ten micrograms each of the a2 luciferase promoter construct to be tested and a transformation control plasmid (pJB4, which expresses GUS) were mixed with either 1 μg each of p35SBP and p35SC1 (B and C1 expression plasmids, respectively) or 2 μg of pMF6 (an empty vector control plasmid), and precipitated onto 1-μm gold particles. These were then introduced into 0.4 mL of packed maize cells using a biolistic He gun. After approximately 36 h, cells were ground in 0.4 mL of luciferase-grinding buffer (100 mm K2HPO4, pH 7.8, and 1 mm DTT), and luciferase and GUS activities were assayed as described previously (Sainz et al., 1997). Activation was quantified as luciferase reporter activity divided by GUS activity in the presence of B and C1. Background, in which no B or C1 proteins were expressed, was very low (Fig. 1B). The activation observed with the mutant promoters was normalized to a percentage of the activation seen by a wild-type promoter of the same length (wild type was set at 100%).
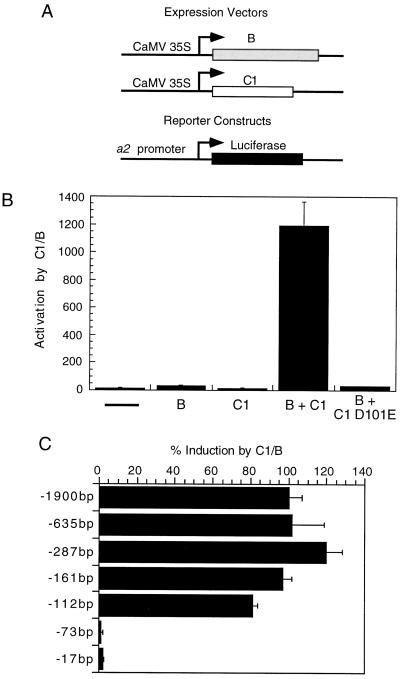
Figure 1.
C1 and B activation of the 1.9-kb a2 promoter or 5′ deletion derivatives in tissue-cultured maize cells. A, Schematic representations of the plasmids used in the transient transformation assay. The CaMV 35S promoter controls the expression of the C1 and B proteins. The reporter plasmid consists of the a2 promoter or mutant derivatives fused to the luciferase-coding region. Details of these constructs are given in Methods. B, The 1.9-kb a2 promoter was cotransformed with either an empty vector control plasmid containing the CaMV 35S promoter without any coding region (–), or with plasmids designed to express B, C1, or a mutant derivative of C1, D101E (defective in DNA binding). Bars represent activation of the a2-luciferase reporter gene, which was obtained by dividing the luciferase activity obtained in each bombardment by the activity from the transformation control included in each bombardment (for details, see Methods). Error bars represent se; n = 12. The −287 a2 promoter, which is equivalent to the 1.9-kb a2 promoter (C), was used for the C1-D101E experiment. Activity is normalized to wild-type C1 and B activation of the same promoter. C, 5′ Deletions of the a2 promoter were generated and tested for activation by C1 and B in transient transformation assays as described in B. Histograms represent the percentage activation in the presence of C1 and B, normalized to the activation observed with the 1.9-kb a2 promoter, set at 100%. Error bars represent se; n = 12.
Gel-Mobility Shift Assays
C1 and C1 Myb proteins were prepared as previously described (Sainz et al., 1997). Proteins were expressed as His-tagged fusion proteins from inducible promoters in Escherichia coli and purified via Ni2+-column chromatography. Protein concentrations were determined using the Lowry assay (Lowry et al., 1951). Gel-mobility shift assays were performed as previously described (Sainz et al., 1997). Radiolabeled oligonucleotides were end labeled with [γ-32P]ATP (ICN) and T4 polynucleotide kinase (New England Biolabs). DNA fragments containing single-stranded overhangs generated by restriction digest were radiolabeled using Klenow enzyme (New England Biolabs) and the appropriate [α-32P]dNTP (DuPont or Amersham). Binding reactions contained from 1 to 10 μg of protein in 50 mm Tris-HCl, pH 7.5, 50 mm NaCl, 1 mm DTT, 1 mm EDTA, 100 μg/mL BSA, and 200 μg/mL polydeoxyinosinic-deoxycytidylic acid (Pharmacia). Reactions were preincubated on ice for 30 min without radiolabeled probe for the purposes of competition or immunodepletion experiments, and then in all cases were incubated on ice for an additional 30 min after the addition of radiolabeled DNA. The reactions were then subjected to electrophoresis on 5% polyacrylamide gels using 0.25× TBE buffer (1× TBE is 0.09 m Tris base, 0.09 m boric acid, and 0.002 m EDTA) for 1 to 2 h at 40 V/cm at 4°C. Gels were then either dried for autoradiography or frozen and exposed overnight to autoradiographic film, and the desired bands were excised and eluted using an electroelution device (Elutrap, Schleicher & Schuell).
Methylation Interference Assays
End-labeled oligonucleotides were subjected to methylation (Maxam and Gilbert, 1980) using dimethylsulfate. The oligonucleotides were then purified by ethanol precipitation to remove the dimethylsulfate. Gel-mobility assays were performed as described above, and free and bound oligonucleotide probes were excised and eluted from the gel. Purified oligonucleotides were subjected to piperidine cleavage, according to the method of Maxam and Gilbert (1980), and run on an 18% denaturing polyacrylamide gel for 8 h at 1700 V, and the gel was exposed to film at −70°C.
RESULTS
C1 and B Activate the a2 Promoter in Vivo
To begin our analysis of the regulation of the a2 promoter, it was necessary to obtain a maize genomic fragment containing a region of the putative promoter large enough to contain the regulatory elements necessary for appropriate expression. To this end, a 2.2-kb BamHI fragment containing approximately 1.9 kb of DNA upstream of the start of transcription was cloned from a size-selected genomic maize library using a probe to the a2 promoter region (see Methods). Extensive restriction analysis of this putative promoter region, as well as sequencing of the first approximately 400 bp upstream of the start of transcription (data not shown), demonstrated that the cloned fragment of the a2 gene was identical to that previously published (Menssen et al., 1990).
To determine whether this 1.9-kb a2 promoter fragment was capable of being activated by the C1 and B proteins in our transient transformation system, it was cloned into a plant expression vector upstream of a firefly luciferase reporter gene and tested in transient He biolistic-transformation assays in tissue-cultured maize cells. Cotransformation of this a2 promoter plasmid with plasmids that express C1 and B (Fig. 1A) resulted in a >1000 activation of luciferase (Fig. 1B) compared with the promoter alone. Significant activation over background was not seen when transformations were performed with either B- or C1-expressing plasmids alone (Fig. 1B). This result demonstrates that the 1.9-kb region includes regulatory signals necessary for C1- and B-mediated induction in our transient assay system. Furthermore, this result indicates that transcriptional regulation of a2 is similar to that of a1, bz1, and bz2 in that each is activated only in the presence of a bHLH (B or R) and a Myb protein (C1 or Pl) (Goff et al., 1990; Tuerck and Fromm, 1994; Bodeau and Walbot, 1996), but not with either class of protein alone.
To address whether the binding of C1 to DNA was required for activation of the a2 promoter, a C1 mutant protein specifically defective in DNA binding was used in activation assays with the a2 promoter. The previously characterized C1 D101E mutation has greatly reduced ability to bind DNA but its ability to interact with B is unaffected (Sainz et al., 1997). This mutant C1 protein, when tested with wild-type B protein in the transient expression assay with the a2 promoter, resulted in background levels of promoter activation (Fig. 1B). This demonstrates that the ability of C1 to bind DNA is crucial for a2 promoter activation.
The Minimal Sequences Required for C1- and B-Mediated Activation Lie between −112 and +5
To begin to determine the minimal sequences necessary for C1- and B-mediated activation of the a2 promoter, a series of 5′ deletions was constructed. As seen in Figure 1C, deletions to −161 bp relative to the start of transcription give the same activation as the 1.9-kb a2 clone, suggesting that there are no regions between −1.9 and −161 crucial for C1- and B-mediated activation. Deletion to −112 bp relative to the start of transcription resulted in a slight reduction in activation compared with the −1.9-kb clone (approximately 80% of full-length activation), suggesting that there may be sequences contributing to C1- and B-mediated activation between −161 and −112. Further deletion from −112 to −73 decreased activation to background levels, suggesting that sequences crucial for activation by C1 and B lie between −112 and −73. These experiments define the minimal C1- and B-inducible promoter region to be within −112 to +5. This size is comparable with the 123-, 134-, and 84-bp sequences shown to be sufficient for C1- and B-mediated activation of the a1, bz1, and bz2 promoters, respectively (Roth et al., 1991; Tuerck and Fromm, 1994; Bodeau and Walbot, 1996).
Mutagenesis of the Minimal Promoter Region
Directed mutations within the minimal promoter were made to identify the sequences important for C1- and B-mediated activation. Mutations were created using PCR mutagenesis (Higuchi et al., 1988), in which small sections of DNA were replaced with a specific sequence. Eight mutations were created, spanning −112 to −40, the border of the putative TATA box (Fig. 2A). Mutations 4 and 5 spanned the putative anthocyanin consensus sequence proposed by Tuerck and Fromm (1994), located at −88 to −74. Mutations were tested in the transient transformation assay as described above.
Because redundant regulatory sequences upstream might diminish the effect of any particular mutation, the mutations were first tested in the context of the minimal −112-bp promoter (Fig. 2A). In this context, mutations 2 and 3 had a dramatic effect on C1- and B-mediated activation of the promoter, respectively. Both mutations decreased activation to near background levels, to 1 and 3% of wild-type activation, respectively. Mutation 7 raised promoter activity slightly, but this effect was not further investigated. The other mutations had either no or only a very modest effect on activation (Fig. 2A). The previously proposed anthocyanin consensus sequence (Tuerck and Fromm, 1994) is not crucial for a2 regulation because mutations 4 and 5 spanning this region did not dramatically affect activation by C1 and B (Fig. 2A).
To determine if the −74 to −40 region contained redundant elements important for activation by C1 and B that might have been missed by studying each mutation individually, a deletion of this region was tested. This deletion had only a modest reduction in activation by C1 and B, suggesting that sequences in this region are not required for activation by C1 and B. Furthermore, this deletion demonstrates that the activity of those regions between −104 and −88 that do mediate activation by C1 and B is not significantly altered by moving them 30 bp closer to the start of transcription. Together, these experiments demonstrate that the only region between −112 and the putative TATA box crucial for C1- and B-mediated activation is the region between −104 and −88, defined by mutations 2 and 3.
The 5′ deletion analysis suggested that there might be sequences between −161 and −112 that were involved in C1- and B-mediated activation. To determine if upstream sequences could compensate for mutations in the crucial regions, we tested the effects of mutations 2 and 3 in a longer promoter context, from −287 to the start of transcription. In this context, each mutation had only a slight effect on activation mediated by C1 and B, 66 and 79%, respectively. This suggests that there are redundant sequences upstream of −112 that can partially compensate for mutations in the −104 to −88 region defined by mutations 2 and 3 (Fig. 2B).
To determine if these redundant regulatory sequences could compensate for the loss of the entire −112 to −73 region, a deletion of this region was made in the context of the −287 promoter construct (Fig. 2B). Deletion of −112 to −73 reduced activation to 37% of the level of the intact −287 promoter, demonstrating that although the −112 to −73 region was absolutely required in the shorter −112 context, regions upstream can partially compensate for their loss.
Comparison of the a2 Promoter with Two Other Characterized Anthocyanin Promoters
A computer alignment program (Devereux et al., 1984) and visual inspection were used to identify the regions of the a2 promoter that had the highest sequence similarity to the important sequences from a1 and bz1. The region with the highest similarity to these sequences was not the site proposed by Tuerck and Fromm (1994), but an adjacent region that encompasses mutations 2 and 3. This is the same region that is most crucial for C1 and B activation of the −112 a2 promoter (Fig. 2A). The new consensus site is shown in Figure 3A, and an alignment of this sequence with the functionally important regions of a1 (Tuerck and From, 1994; Sainz et al., 1997) and bz1 (Goff et al., 1990; Roth et al., 1991) is shown in Figure 3B. Note that the orientation of this site is reversed in the a2 promoter relative to that in a1 and bz1. This new alignment is larger and more conserved than that previously proposed. To distinguish the newly identified element from the previously proposed anthocyanin consensus sequence, it is called the anthocyanin-regulatory element (ARE).
Figure 3.
An ARE overlaps with regions crucial for C1 and B induction of the a2 promoter. A, The sequence of the −112 to −41 a2 promoter is shown, with results from the linker-scanner mutagenesis shown below. Bars above the sequence indicate the anthocyanin consensus sequence suggested by Tuerck and Fromm (1994) (Previous Consensus), or as proposed here (New Consensus). B, Computer alignment (Devereux et al., 1984) of three characterized anthocyanin promoters reveals common sequences among all three. The consensus site derived from this sequence similarity is shown above. WT, Wild type.
C1 Myb Binds to the a2 Promoter in at Least Three Locations
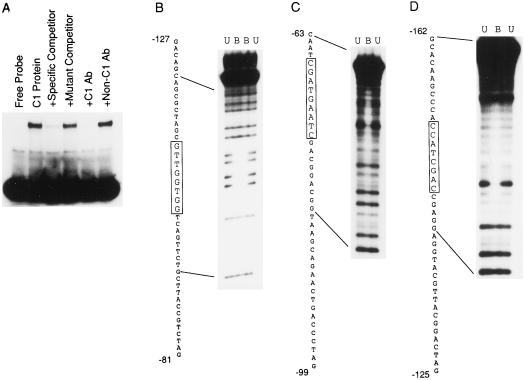
To determine whether the a2 promoter contains sites that are capable of binding the C1 protein, gel-mobility shift assays were performed using purified C1 protein expressed in E. coli and radiolabeled a2 promoter fragments from −161 to +5. This region is capable of mediating full activation by C1 and B. C1 does bind specifically to the a2 promoter in the region between −161 and +5 (Fig. 4A). An unlabeled oligonucleotide containing a high-affinity C1-binding site from the a1 promoter competed for C1 binding with the a2 site (−76 to −47 of the a1 promoter). In contrast, the same oligonucleotide specifically mutated in its C1-binding site (Sainz et al., 1997) failed to compete when used at the same concentration as the wild-type a1 oligonucleotide. Gel-shift assays with an increasing protein concentration of purified C1 or purified C1 Myb revealed multiple bands with lower mobility, suggesting that there might be multiple binding sites on the a2 promoter (data not shown).
Figure 4.
C1 binds specifically to the a2 promoter in at least three locations. A, A radiolabeled fragment of the a2 promoter, spanning −161 to +5, was mixed with purified C1 protein in the presence or absence of competitor oligonucleotides or C1 and non-C1 antibodies. The specific competitor is a C1-binding site from the a1 promoter (−76 to −47). The mutant competitor is the same fragment of a1, but with 2-bp substitutions that reduce activation (Grotewold et al., 1994) and C1 DNA binding (Sainz et al., 1997). B through D, Methylation interference experiments using purified C1 Myb protein and the indicated end-labeled oligonucleotides of the a2 promoter (see Methods). Lanes U contain methylation-interference reactions purified from unbound labeled probe. Lanes B contain reactions purified from bound probe. The sequence to the left of each gel corresponds to the sequence of the oligonucleotide, with the protected region boxed.
Methylation interference and DNAse I footprinting experiments were performed to determine the precise location of C1-binding sites within the a2 promoter. Using double-stranded oligonucleotides containing the region to be studied and an affinity-purified protein comprising the Myb domain of C1 (Sainz et al., 1997), these assays identified three binding sites on the a2 promoter within −161 and +5. The data shown in Figure 4B are from representative methylation interference experiments demonstrating C1-Myb binding to each of these sites.
The C1-binding site at −99 coincides with mutation 2, which dramatically lowered activation mediated by C1 and B. This suggests that the basis of the reduction in activation caused by mutation 2 is the abolishment of a critical C1-binding site. This binding site also lies within the ARE. The binding site centered at −70 overlaps with mutation 5.5, which did not have a dramatic effect on C1- and B-mediated activation (Fig. 2A), suggesting that this site is less important.
The C1-binding site centered at −147 was upstream of the region mutagenized during the initial set of experiments. However, the 5′ deletion series and the analysis of mutations 2 and 3 in the −287 context suggested that there were regulatory sequences upstream of −112 contributing to activation. It was possible that the C1-binding site centered at −147 played a role in this regulation. To test this possibility, a promoter construct containing site-directed mutations designed to abolish C1 binding to both the −99 and −147 sites was constructed in the context of the −287 a2 promoter. The −99 mutation used was mutation 2 (Fig. 2A) and the −147 mutation replaced −152 to −145 with the same sequence used in mutation 2. The promoter with both mutations was activated to 22% (± se) of wild-type levels, compared with 66% for the −99 mutation alone (Fig. 2). This demonstrates that the upstream C1-binding site at −147 contributes to C1 and B activation of the a2 promoter.
The −102 to −86 Region Contains Two Distinct Types of Regulatory Elements
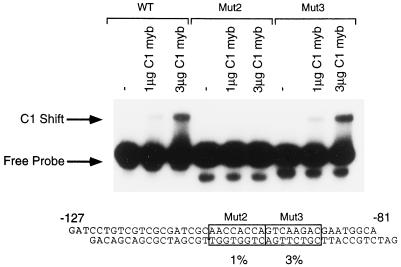
Mutagenesis experiments determined that two adjacent mutations, 2 and 3, spanning −104 to −88, have a dramatic effect on promoter activation. Methylation-interference experiments identified a C1-binding site that coincided with mutation 2, but no binding was detected to the adjacent region that coincided with mutation 3. One possibility is that the change in sequence within mutation 3 altered C1 binding to the adjacent site, defined by mutation 2. To test this possibility, the ability of C1 Myb to bind to a double-stranded oligonucleotide containing either the wild-type region from −127 to −81 or a similar oligonucleotide containing either mutation 2 or mutation 3 was examined. C1 Myb was unable to bind to an oligonucleotide containing mutation 2, but could bind at wild-type levels to an oligonucleotide containing mutation 3 (Fig. 5). Thus, the defect in activation by C1 and B seen with mutation 2 can be explained by the lack of C1 binding to this mutated site. In contrast, the inability of mutation 3 to be activated by C1 and B cannot be explained by an effect on C1 DNA binding, suggesting that this region is important for other reasons.
Figure 5.
The binding of C1 to radiolabeled oligonucleotides containing either the wild-type (WT) sequence or mutations 2 (Mut2) or 3 (Mut3). Each labeled probe was used in three binding reactions, with probe alone and with 1 and 3 μg of C1 Myb protein, as indicated above each lane. The diagram below shows the wild-type oligonucleotide sequence, with the location of the two mutations boxed. Figure 2 contains the sequence changes in the mutant derivatives. The levels of activation of these mutations by C1 and B in the context of the −112-bp a2 promoter is shown below.
The −127 to −81 Region Is Sufficient for C1- and B-Mediated Activation
Because the region between −104 and −88 had been shown to contain elements important for C1- and B-mediated activation of the minimal a2 promoter, we wanted to determine if this region was sufficient to mediate activation of a heterologous promoter by C1 and B. To test this, we created a double-stranded oligonucleotide centered on this crucial region spanning −121 to −81 relative to the start of transcription. This oligonucleotide was dimerized in direct repeats in the same orientation as that of the native promoter and fused to a minimal CaMV 35S promoter (see Methods). The minimal promoter provides a TATA box, as well as any other elements close to the start of transcription necessary for basic promoter function. This construct was then tested in transient assays to determine if it was capable of being induced by C1 and B. This construct was induced 44-fold over background in the presence of C1 and B (Fig. 6). The minimal CaMV promoter alone was not activated by C1 and B, indicating that this −121 to −81 region of the a2 promoter contains elements sufficient for regulation by C1 and B.
DISCUSSION
We have carried out an extensive analysis of the a2 promoter, identifying the key sequences involved in mediating activation by C1 and B, and demonstrating that C1 is crucial for promoter activation. The importance of C1 binding is demonstrated by the observation that a C1 mutant specifically defective in DNA binding is unable to activate the a2 promoter. In addition, activation was reduced to background levels when the C1-binding site at −99 was mutated within a minimal fragment sufficient for activation. Comparison of our results with a2 against those obtained with three other anthocyanin promoters (a1, bz1, and bz2) revealed common themes and differences. For all of these promoters, the minimal regions needed for promoter activation by C1 and B are small (less than 200 bp), yet even within this small region there are multiple regions important for induction. Within each of the four promoters there is a conserved region, which we refer to as the ARE. In the two promoters tested, a1 and a2, C1 binds to multiple sites within each promoter. However, within the a1 and a2 promoters there are differences with respect to the positions of the ARE and the C1-binding sites.
The C1-binding sites within the a2 promoter vary dramatically in their contribution to activation. Although C1 binds to three sites on the a2 promoter, only two of these, at −99 and −147, contribute to C1 and B activation of a2. Mutation of the −70 site does not dramatically alter activation and this site does not compensate for mutations at the −99 site. In contrast, the −147 site does partially compensate for mutations at −99. The presence of dispensable C1-binding sites differs from the results with a1, in which the two C1-binding sites both contribute to C1- and B-mediated activation (Sainz et al., 1997). The differences seen in the importance of the binding sites within a2 could be the result of at least two factors. The affinity of C1 for these sites might vary such that sites that are less important are bound more weakly by C1. It is also possible that the context of these sites within the promoters might be a crucial variable. For example, the C1-binding sites that are most important for activation may lie either near an ARE or near binding sites for another factor(s). A potential role for the C1-binding site, which is not crucial for activation, might be to increase the local concentration of the C1 protein on the DNA, thereby increasing the occupancy at the nearby −99 binding site that plays a more crucial role in activation.
The most important sequences of the a2 promoter responsible for activation by C1 and B within the minimal −112-bp promoter are composed of a C1-binding site and an adjacent site. A mutation at the second important site does not affect C1 binding to the neighboring site, suggesting that the activation mediated by this region is not through C1 DNA binding. This site, centered at −93, could be a binding site for another transcription factor. B is an obvious candidate, but one line of evidence suggests that B does not bind to this site. A deletion derivative of B missing the bHLH domain activates the a2 promoter at 50% of the level observed with the wild-type protein (data not shown). Thus, the removal of the putative DNA-binding domain of B causes a less severe reduction in activation (50% of wild type) than removal of its putative binding site (3% of wild type). Caveats to this interpretation are that B may bind the site at −93 through an uncharacterized DNA-binding motif present in another part of the protein or through protein-protein interactions with an adjacent C1 molecule. Another possibility is that the site at −93 might interact with another transcription factor that has not yet been identified.
The two regions most important for C1 and B activation of the a2 promoter, the C1-binding site and the adjacent site, share sequence similarity with key sequences within the a1 and bz1 promoters. These sequences within a1 and bz1 overlap with an anthocyanin consensus sequence previously suggested by Tuerck and Fromm (1994). In contrast, the sequences within a2 predicted to be a consensus sequence are neither important for activation nor the best match. Comparing the minimal promoter sequences, as defined by activation assays, for each of these promoters reveals a more conserved, larger consensus element, which we call ARE. The new alignment predicts an ARE different from that proposed by Tuerck and Fromm (1994) for the bz2 promoter at positions −88 to −72. A mutation spanning part of this putative ARE reduces activation of bz2 by C1 and B to approximately 30% of wild type (Bodeau and Walbot, 1996), consistent with its importance in bz2 regulation. However, further experiments need to be done with bz2, because the bz2 assays were done with electroporation of maize protoplasts and the a1, a2, and bz1 assays were done with microprojectile bombardment of maize callus. It will be important to assay bz2-promoter mutations in the same assay system used with the other promoters.
Although the ARE is conserved in multiple promoters, it may not function identically in each of these promoters. Unlike a2, the a1 ARE does not contain a high-affinity C1-binding site (Sainz et al., 1997), but is instead between the two C1-binding sites. The observation for both a1 and a2 that there are functionally important parts of the ARE to which C1 does not bind with high affinity suggests that another factor is involved in activation through these regions. It is not known whether the ARE in bz1 or the putative ARE in bz2 contains a C1-binding site. C1-binding sites have been suggested for both bz1 and bz2 based on sequence similarity to animal Myb consensus sites (Roth et al., 1991; Bodeau and Walbot, 1996). However, site-selection studies have shown that C1 prefers to bind to sites that do not resemble an animal Myb consensus (Sainz et al., 1997). Thus, further studies on bz1 and bz2 will be necessary to determine whether the AREs within these promoters contain C1-binding sites.
To date, sequences from three anthocyanin promoters have been shown to be sufficient to mediate robust activation of a heterologous promoter by C1 and B. The a2 −121 to −81 fragment, which mediates a 44-fold activation by C1 and B, contains both an ARE and a C1-binding site (this study). Similarly, the upstream region of a1, which is sufficient for a 200-fold activation by C1 and B (Tuerck and Fromm, 1994), also contains both an ARE and a C1-binding site (Sainz et al., 1997). In contrast, the downstream region of a1, which mediates a 44-fold activation by C1 and B (Grotewold et al., 1994), contains no ARE, but does contain the highest-affinity C1-binding site known (Sainz et al., 1997). This indicates that an ARE is not absolutely necessary for activation. The −78 to −47 bz1 fragment, which mediates a 44-fold activation by C1 and B (Roth et al., 1991), contains an ARE, but it is not known whether it contains a C1-binding site. A modest induction by C1 and B (3-fold) was mediated by an upstream fragment of bz2 (Bodeau and Walbot, 1996); however, it is difficult to compare this result with those of the other promoters because the region within bz2 with the largest effect when mutated was not tested for its ability to activate a heterologous promoter.
All of the fragments that mediate large inductions by C1 and B, including the a1 fragment without an ARE, are absolutely dependent on both C1 and B (Grotewold et al., 1994; Tuerck and Fromm, 1994; Sainz et al., 1997). This may be because B and/or other factors can still interact with this promoter region through protein-protein interactions with C1. It is possible that interaction with additional factors could influence C1-binding affinity or specificity in vivo, such that in vitro affinity may not be a good predictor of contribution to activation. Future work comparing the affinity of C1 for its binding sites with the importance of these binding sites in anthocyanin-biosynthetic gene activation, together with studies to determine the role of the bHLH factor and why C1 is absolutely dependent on it for activation, should further our understanding of anthocyanin gene regulation.
ACKNOWLEDGMENTS
We thank Alice Barkan and Diane Hawley for critical reading of an early draft of the manuscript. We also thank Erich Grotewold, John Bodeau, and Steve Goff for providing plasmids used in the transient transformation assays, and Alfons Gierl for providing the a2 plasmid.
Abbreviations:
- ARE
anthocyanin-regulatory element
- bHLH
basic-helix-loop-helix protein
- CaMV
cauliflower mosaic virus
Footnotes
This research was supported by a National Science Foundation grant (no. MCB 9248180 to V.L.C.) and by a National Institutes of Health predoctoral training grant (no. 5T32HD07348 to M.L.L.).
LITERATURE CITED
- Bodeau JP, Walbot V. Structure and regulation of the maize Bronze2 promoter. Plant Mol Biol. 1996;32:599–609. doi: 10.1007/BF00020201. [DOI] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner HK, Robbins TP, Jorgensen RA. Genetic and developmental control of anthocyanin biosynthesis. Annu Rev Genet. 1991;25:173–199. doi: 10.1146/annurev.ge.25.120191.001133. [DOI] [PubMed] [Google Scholar]
- Goff SA, Cone KC, Chandler VL. Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 1992;6:864–875. doi: 10.1101/gad.6.5.864. [DOI] [PubMed] [Google Scholar]
- Goff SA, Cone KC, Fromm ME. Identification of functional domains in the maize transcriptional activator C1: comparison of wild-type and dominant inhibitor proteins. Genes Dev. 1991;5:298–309. doi: 10.1101/gad.5.2.298. [DOI] [PubMed] [Google Scholar]
- Goff SA, Klein TM, Roth BA, Fromm ME, Cone KC, Radicella JP, Chandler VL. Transactivation of anthocyanin biosynthetic genes following transfer of B regulatory genes into maize tissues. EMBO J. 1990;9:2517–2522. doi: 10.1002/j.1460-2075.1990.tb07431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E, Drummond BJ, Bowen B, Peterson T. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell. 1994;76:543–553. doi: 10.1016/0092-8674(94)90117-1. [DOI] [PubMed] [Google Scholar]
- Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton TA, Cornish EC. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell. 1995;7:1071–1083. doi: 10.1105/tpc.7.7.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Maxam A, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Menssen A, Hohmann S, Martin W, Schnable PS, Peterson PA, Saedler H, Gierl A. The En/Spm transposable element of Zea mays contains splice sites at the termini generating a novel intron from a dSpm element in the a2 gene. EMBO J. 1990;9:3051–3057. doi: 10.1002/j.1460-2075.1990.tb07501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, Leppen HTC, Mol JNM, Koes RE. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell. 1993;5:1497–1512. doi: 10.1105/tpc.5.11.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth BA, Goff SA, Klein TM, Fromm ME. C1- and R-dependent expression of the maize Bz1 gene requires sequences with homology to mammalian myb and myc binding sites. Plant Cell. 1991;3:317–325. doi: 10.1105/tpc.3.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz MB, Grotewold E, Chandler VL. Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell. 1997;9:611–625. doi: 10.1105/tpc.9.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulsen AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerck JA, Fromm ME. Elements of the maize a1 promoter required for transactivation by the anthocyanin B/C1 or phlobaphene P regulatory genes. Plant Cell. 1994;6:1655–1663. doi: 10.1105/tpc.6.11.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer IM, Stuitje AR, Mol JNM. Regulation of general phenylpropanoid and flavonoid gene expression. In: Verma DPS, editor. Control of Plant Gene Expression. Boca Raton, FL: CRC Press; 1993. pp. 125–155. [Google Scholar]