Abstract
Summary: NetworkView is an application for the display and analysis of protein·RNA interaction networks derived from structure and/or dynamics. These networks typically model individual protein residues and nucleic acid monomers as nodes and their pairwise contacts as edges with associated weights. NetworkView projects the network onto the underlying 3D molecular structure so that visualization and analysis of the network can be coupled to physical and biological properties. NetworkView is implemented as a plugin to the molecular visualization software VMD.
Availability and implementation: NetworkView is included with VMD, which is available at http://www.ks.uiuc.edu/Research/vmd/. Documentation, tutorials and supporting programs are available at http://www.scs.illinois.edu/schulten/software/.
Contact: networkview@scs.illinois.edu
1 INTRODUCTION
The usefulness of 2D contact network visualization in the study of protein function has already been demonstrated (Rahat et al., 2009; Sethi et al., 2009) leading to the development of software to display these networks (Doncheva et al., 2011). The NetworkView extension to the visual molecular dynamics (VMD) program (Humphrey et al., 1996) was developed to help structural biologists study allostery and molecular signaling through network models of protein·RNA complexes in translation (Black Pyrkosz et al., 2010; Trabuco et al., 2010). Residues essential for molecular recognition and reaction mechanisms at different states of the system can be determined by using molecular dynamics (MD) simulations to calculate variations in contacts and displaying the resulting interaction networks and their properties directly onto the 3D structure (Alexander et al., 2010). As networks are increasingly used to study the function and dynamics of biological macromolecules, the informative display of network data directly onto 3D representations of biomolecules is needed (Bhattacharyya and Vishveshwara, 2011; Csermely et al., 2010; Daily et al., 2008; Süel et al., 2002).
2 NETWORK ANALYSIS CONCEPTS
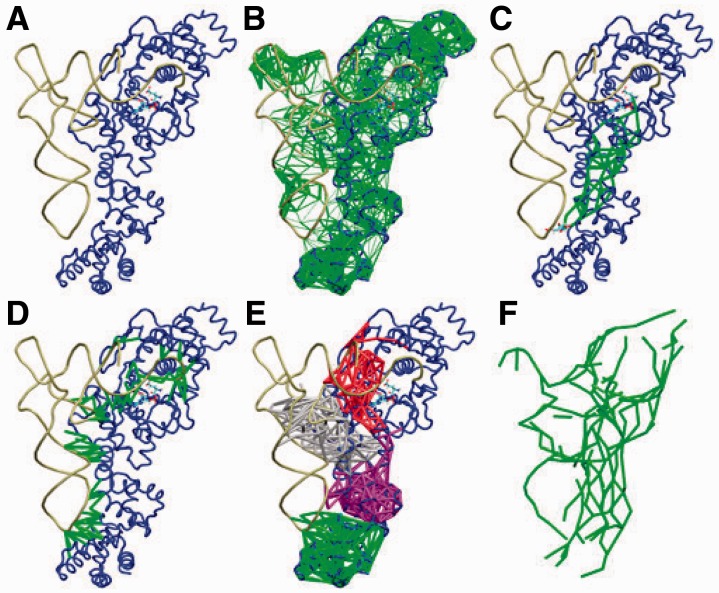
Networks are sets of nodes and edges between pairs of these nodes. Nodes here are located at specific atoms and represent amino acids (C ), nucleobases (N1/N9) or the sugar phosphates (P) of RNA. Edges are drawn if the average distance observed in an MD simulation (or contact distance in the X-ray structure) is less than a pre-defined cutoff. Individual edges may have associated weights or lengths based on properties such as correlated motions (see Fig. 1B), energies or physical distance. A path between two nodes is simply a set of nodes and edges connecting one node to the other, and the path length is the sum of weights for edges in the path. If two nodes lie within a connected network, there exists at least one optimal, shortest path between them, and slightly longer paths are referred to as suboptimal (see Fig. 1C).
), nucleobases (N1/N9) or the sugar phosphates (P) of RNA. Edges are drawn if the average distance observed in an MD simulation (or contact distance in the X-ray structure) is less than a pre-defined cutoff. Individual edges may have associated weights or lengths based on properties such as correlated motions (see Fig. 1B), energies or physical distance. A path between two nodes is simply a set of nodes and edges connecting one node to the other, and the path length is the sum of weights for edges in the path. If two nodes lie within a connected network, there exists at least one optimal, shortest path between them, and slightly longer paths are referred to as suboptimal (see Fig. 1C).
Fig. 1.
Various network visualizations for GluRS tRNAGlu
tRNAGlu Glu-AMP. (A) GluRS (blue) and tRNA (tan). (B) Network (green) weighted by correlated motion from MD simulations. Edge thickness is greater between more highly correlated nodes. (C) Optimal and suboptimal paths (green) between Glu-AMP in the active site and U35 in the tRNA anticodon. (D) Edges (green) bridging the interface between GluRS and the tRNA. (E) The four communities (red, gray, purple and green) that contain both protein and RNA nodes. (F) Subnetwork (green) consisting of the top 10% of edges with the highest betweenness values
Glu-AMP. (A) GluRS (blue) and tRNA (tan). (B) Network (green) weighted by correlated motion from MD simulations. Edge thickness is greater between more highly correlated nodes. (C) Optimal and suboptimal paths (green) between Glu-AMP in the active site and U35 in the tRNA anticodon. (D) Edges (green) bridging the interface between GluRS and the tRNA. (E) The four communities (red, gray, purple and green) that contain both protein and RNA nodes. (F) Subnetwork (green) consisting of the top 10% of edges with the highest betweenness values
Major communication pathways are identified by edge betweenness, the count of all pairwise optimal paths that cross a given edge (see Fig. 1F). Using the Girvan–Newman (GN) algorithm (Girvan and Newman, 2002), networks are divided into disjoint subnetworks called communities in which nodes have stronger and more connections to other nodes within the same community than they have to those outside the community. These communities, which can contain both amino acids and nucleotides, are similar to structural domains but are defined by the dynamics of the biomolecules (see Fig. 1E). A more detailed explanation of the network concepts and their associated algorithms as applied to dynamic networks is available in Sethi et al. (2009).
Once the physical network of nodes and weighted edges is generated by the program networkSetup, both optimal and suboptimal paths are calculated by subopt, and the edge betweenness and community structure are determined by gncommunities. NetworkView simplifies the viewing of the above network properties superimposed onto large biomolecular complexes.
3 IMPLEMENTATION
VMD is available for Windows, MacOS X and Linux/UNIX. NetworkView is implemented as a Tcl plugin to VMD, which allows direct access to VMD’s powerful atomselection language and wrapper procedures for the creation and display of OpenGL objects. A graphical user interface with access to the most commonly used features in NetworkView is available through the VMD menu system: Extensions/Analysis/NetworkView.
Three different types of data files can be loaded into NetworkView: network, community and suboptimal path. The data files are ASCII text and were designed to be human readable. These files can be generated by the programs networkSetup, gncommunities and subopt. NetworkView is not technically tied to these programs, however, so it can be used to visualize and analyze network data generated by other means.
NetworkView provides an application programming interface (API) for selecting nodes and edges and then viewing the values associated with them. Documentation for the NetworkView API is available on the web at http://www.scs.illinois.edu/schulten/software/. A detailed introduction to the use of NetworkView has been provided in a tutorial with data files hosted at the same location.
4 USAGE
The necessary data files can be generated from MD trajectories or from PDB structures. To calculate dynamic networks, a trajectory file containing selected atoms is processed into a correlation matrix made with the MD analysis program Carma (Glykos, 2006) through networkSetup. This matrix is then used to create the local-contact, weighted, adjacency matrix. If no trajectory data are available, structure networks with unweighted edges can also be generated from PDB files. To take full advantage of NetworkView’s capabilities, further processing can be done on the adjacency matrix to calculate the network’s community structure, high-betweenness paths and suboptimal paths between residues known to participate in molecular signaling.
Aside from visualization, NetworkView is also intended for quantitative analysis. Edge weights and other, user-assigned values associated with nodes and edges can be accessed through the API. For example, edge weights along a specified optimal path or at the interface between two molecules can be extracted for statistical analysis. By projecting network information onto the atoms within VMD, features related to active sites, molecular binding interfaces and areas with high sequence or structure conservation (Roberts et al., 2006) can be correlated with the calculated network properties to identify functionally important residues and structures.
5 CONCLUSION
NetworkView provides high-level, interactive views of dynamic and/or structure networks that enable the analysis and interpretation of molecular signaling. Projection of these networks onto 3D biomolecular structures is useful for rapidly identifying residues which can be targeted in mutagenesis experiments to test mechanistic hypotheses.
ACKNOWLEDGEMENTS
The authors thank VMD developers John Stone and Kirby Vandivort for helpful discussions and incorporation of NetworkView into VMD. We would also like to thank Alexis Pyrkosz, Anurag Sethi and Li Li for suggestions and help with testing.
Funding: The National Center for Supercomputing Applications (MCA03T027); the National Institutes of Health (P41-RR05969) and the National Science Foundation (MCB-0844670 and PHY-0822613).
Conflict of Interest: none declared.
REFERENCES
- Alexander RW, et al. Experimental and computational determination of tRNA dynamics. FEBS Lett. 2010;584:376–386. doi: 10.1016/j.febslet.2009.11.061. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya M, Vishveshwara S. Probing the allosteric mechanism in pyrrolysyl-tRNA synthetase using energy-weighted network formalism. Biochemistry. 2011;50:6225–6236. doi: 10.1021/bi200306u. [DOI] [PubMed] [Google Scholar]
- Black Pyrkosz A, et al. Exit strategies for charged tRNA from GluRS. J. Mol. Biol. 2010;397:1350–1371. doi: 10.1016/j.jmb.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csermely P, et al. Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends Biochem. Sci. 2010;35:539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daily, et al. Contact rearrangements form coupled networks from local motions in allosteric proteins. Prot. Struct. Func. Bioinf. 2008;71:455–466. doi: 10.1002/prot.21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncheva N, et al. Analyzing and visualizing residue networks of protein structures. Trends Biochem. Sci. 2011;36:179–182. doi: 10.1016/j.tibs.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Girvan M, Newman M. Community structure in social and biological networks. Proc. Natl. Acad. Sci. USA. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glykos NM. Software news and updates. CARMA: a molecular dynamics analysis program. J. Comp. Chem. 2006;27:1765–1768. doi: 10.1002/jcc.20482. [DOI] [PubMed] [Google Scholar]
- Humphrey W, et al. VMD—visual molecular dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Rahat O, et al. Understanding hydrogen-bond patterns in proteins using network motifs. Bioinformatics. 2009;25:2921–2928. doi: 10.1093/bioinformatics/btp541. [DOI] [PubMed] [Google Scholar]
- Roberts E, et al. MultiSeq: unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics. 2006;7:382. doi: 10.1186/1471-2105-7-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi A, et al. Dynamical networks in tRNA: protein complexes. Proc. Natl. Acad. Sci. USA. 2009;106:6620–6625. doi: 10.1073/pnas.0810961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süel G, et al. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Mol. Biol. 2002;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- Trabuco LG, et al. The role of L1 stalk-tRNA interaction in the ribosome elongation cycle. J. Mol. Biol. 2010;402:741–760. doi: 10.1016/j.jmb.2010.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]