Abstract
Summary: We have established an RNA mapping database (RMDB) to enable structural, thermodynamic and kinetic comparisons across single-nucleotide-resolution RNA structure mapping experiments. The volume of structure mapping data has greatly increased since the development of high-throughput sequencing techniques, accelerated software pipelines and large-scale mutagenesis. For scientists wishing to infer relationships between RNA sequence/structure and these mapping data, there is a need for a database that is curated, tagged with error estimates and interfaced with tools for sharing, visualization, search and meta-analysis. Through its on-line front-end, the RMDB allows users to explore single-nucleotide-resolution mapping data in heat-map, bar-graph and colored secondary structure graphics; to leverage these data to generate secondary structure hypotheses; and to download the data in standardized and computer-friendly files, including the RDAT and community-consensus SNRNASM formats. At the time of writing, the database houses 53 entries, describing more than 2848 experiments of 1098 RNA constructs in several solution conditions and is growing rapidly.
Availability: Freely available on the web at http://rmdb.stanford.edu
Contact: rhiju@stanford.edu
Supplementary information: Supplementary data are available at Bioinformatics Online.
1 INTRODUCTION
Understanding the secondary and tertiary structures of RNAs is critical for dissecting their diverse biological functions, ranging from catalysis in ribosomal RNAs to gene regulation in metabolite-sensing riboswitches and protein-binding elements in RNA messages (Eickbush and Eickbush, 2007; Kudla et al., 2009; Nudler and Mironov, 2004; Spahn et al., 2001; Yanofsky, 2004). RNA structure has therefore been intensely studied with a variety of biophysical and biochemical technologies (Adilakshmi et al., 2006; Getz et al., 2007; Waldsich, 2008; Varani and Tinoco, 1991; Wilkinson et al., 2006). Among these tools, a facile, information-rich and widely used technique is structure mapping (also called structure probing or footprinting), in which the chemical modification, enzymatic cleavage or degradation rate of an RNA nucleotide correlates with the exposure, flexibility or other structural features of the site. Modern methods often reverse transcribe probed RNA molecules into DNA fragments whose lengths can be subsequently analyzed to infer the locations of probe events. These methods permit the single-nucleotide resolution readout of structural data for RNAs as large as ribosomes (Deigan et al., 2009; Culver et al., 1999), and in recent years, investigators have developed high-throughput technologies, such as 96-well capillary electrophoresis (Mitra and Shcherbakova, 2008; Mortimer and Weeks, 2007) and deep sequencing (Lucks et al., 2011) to perform this step. Furthermore, several bioinformatic pipelines have been implemented to rapidly quantify, map and analyze the resulting data (Aviran et al., 2011; Deigan et al., 2009; Low and Weeks 2010; Vasa et al., 2008; Yoon et al., 2011). RNA mapping experiments are now routinely used to improve automated secondary structure modeling (Deigan et al., 2009), probe entire viral genomes (Watts and Dang, 2009), simultaneously map arbitrary RNA mixtures through deep sequencing (Kertesz et al., 2010; Lucks et al., 2011; Underwood et al., 2010; Zheng et al., 2010) and infer an RNA’s ‘contact map’ by coupling to exhaustive single-nucleotide mutagenesis (Kladwang and Das, 2010; Kladwang et al., 2011).
These developments could enable novel methods in RNA structural biology, especially if predictive relationships between RNA sequence/structure and these data can be established. However, unlike existing structural biology fields like nuclear magnetic resonance and crystallography, there is no equivalent of the Biological Magnetic Resonance Bank (Ulrich et al., 2007) or the Protein Data Bank (Bernstein et al., 1977) that stores curated datasets. Structure mapping data are available in the supporting material of papers or self-reported in SNRNASM format (Rocca-Serra et al., 2011), but these formats do not typically include error estimates; are not always normalized or background-subtracted with standardized protocols; are not linked to RNA structures and are not straightforward to visualize, which would enable consistency checks during further analysis. We have therefore created an RNA mapping database (RMDB) and are populating it with curated structure mapping measurements in human and machine-readable formats amenable to inferring relationships between sequence/structure and structure mapping data. Data contained in the RMDB are freely available and can be easily integrated with future repositories such as RNAcentral (Bateman et al., 2011).
2 DATABASE CONTENT AND STRUCTURE
For a specific RNA in defined solution conditions, each structure mapping experiment can be conceptualized as M × N matrices, where M is the number of measurements made on the RNA, e.g. normalized peak areas calculated by HiTRACE (Yoon et al., 2011), CAFA (Mitra et al., 2008) and ShapeFinder (Vasa et al., 2008); or maximum likelihood parameters (Aviran et al., 2011) and N is the number of nucleotides in the RNA. Entries in the RMDB house these data matrices and are enriched with annotations and free text to describe associated content.
The database currently includes experiments using base methylation by dimethyl sulfate, base adduct formation by 1-cyclohexyl-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate, selective 2′ hydroxyl acylation with primer extension (SHAPE) with either N-methylisatoic anhydride or 1-methyl-7-nitroisatoic anhydride and hydroxyl radical footprinting. The RMDB is further capable of storing data from enzymatic, in-line and other structure mapping experiments. The RNAs probed include riboswitches, tRNAs, ribozyme and ribosomal domains featured in several published studies as well as human-designed sequences accruing in the internet-scale RNA engineering project EteRNA (http://eterna.stanford.edu) (see Supplementary Table S2). For many RMDB entries, the complementary DNA fragment separation and analysis steps were carried out by 96-well format capillary electrophoresis and the HiTRACE pipeline (Yoon et al., 2011), respectively; entries describing data from (Lucks et al., 2011) were read out and processed using the SHAPE-Seq protocol (Aviran et al., 2011).
New experimental data can be uploaded to the RMDB as a spreadsheet in the SNRNASM Isa-Tab format (Rocca-Serra et al., 2011) or in RDAT file format (a simple text file format; detailed in the Supplement Information and at http://rmdb.stanford.edu/repository/specs/). However, like the well-curated Protein DataBank, public release then requires passing review by the RMDB team; the data must include error estimates or replicates, information on estimated or known structure (at least at the level of secondary structure), associated publications or preprints and descriptions of how the data were processed.
3 FEATURES AND EXAMPLE USE CASES
3.1 For experimentalists
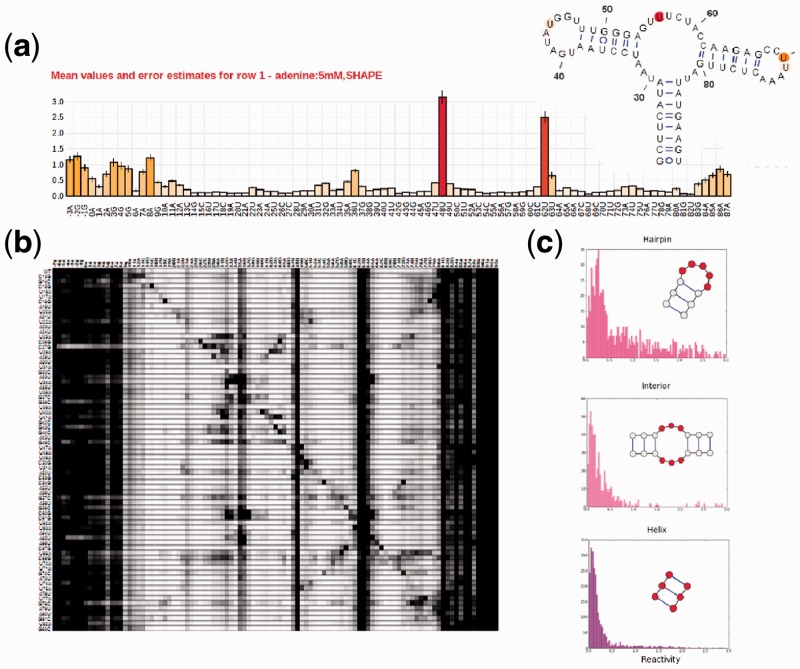
The RMDB is a resource for RNA biochemists and molecular biologists interested in using existing data to guide biological hypotheses, interpret new data or to share their own experimental results. First, users interested in a particular RNA system can quickly find relevant data in the RMDB by using the full-text search field in the upper-right corner of the site. The user can then inspect each entry using the data visualization tools (Fig. 1a and b; Supplementary Material). Second, the integration with the VARNA applet allows for quickly comparing mapping data against structural models. The data can be downloaded in either RDAT or SNRNASM format or exported directly from the VARNA visualization applet for further inspection with other tools. Third, the RMDB also includes a secondary structure prediction server (located at http://rmdb.stanford.edu/structureserver); the server can use structure mapping data to generate sensible secondary structure hypotheses (see Supplementary Material). Finally, experimentalists who wish to submit their data to the RMDB can do so after registering to the site.
Fig. 1.
Different visualization tools for entries in the RNA mapping database. (a) Classic bar plot of 2′-OH acylation (SHAPE) rates across the nucleotides of the adenine-sensing domain of the add riboswitch from Vibrio vulnificus. The data are from a ‘standard state’ study averaging 19 replicates across multiple experiments and estimating the resulting errors (shown as error bars); the RNA’s crystallographic secondary structure, colored by the SHAPE data, is shown in the inset. (b) Through mutate-and-map data, the RMDB also allows exploring the contact map of the same riboswitch. SHAPE data are shown for constructs with single mutations at each RNA position. (c) Histograms for reactivities found in interior loops, hairpin loops and helices. Nucleotides in the motif for which reactivity data were collected are marked in red. Hairpin loops have higher average reactivities than interior loops, bulges and non-helical elements
3.2 For structural bioinformaticists
The RMDB can extract general properties of mapping data including histograms of reactivities for different secondary structure elements (Fig. 1c). Analyses of structure mapping data are facilitated by the Python/MATLAB RDATkit package (http://rdatkit.simtk.org, see Supplementary Material) for RDAT/SNRNASM-IsaTAB parsing. To demonstrate the utility of the RMDB in extracting new information from multiple datasets, we tested whether the SHAPE method can discriminate between interior and hairpin loops. Using the advanced search feature of the database, we downloaded SHAPE data for each secondary structure element (internal loops, hairpins, helices and bulges) collected in standard state experiments for all non-coding RNAs (ncRNAs) with known structure in the database. Interior and hairpin loops of ncRNA have distinct reactivity distributions, suggesting that SHAPE-directed modeling can be made more accurate by taking this effect into account (Fig. 1c).
3.3 For web-app developers
Data stored in the RMDB are exposed through a RESTful API (described in https://sites.google.com/site/rmdbwiki/web-api) in JSON format, simplifying the creation of web applications that use the data contained in the repository. The RMDB also provides RSS feeds that are automatically updated with new entries (see https://sites.google.com/site/rmdbwiki/rss for details). These tools have allowed integration of the entries in the RMDB into the SNRNASM repository (http://snrnasm.bio.unc.edu/browse.html).
4 DISCUSSION
The throughput of structure mapping experiments has taken significant leaps with multiplexed capillary electrophoresis and next-generation sequencing that allow probing of thousands of RNAs at once. These data should, in principle, permit the development of confident structural biology tools that couple structure mapping measurements to secondary and tertiary structure modeling. However, until recently, researchers have had few resources that enable curation and sharing of high-throughput quantified RNA mapping data. It is our hope that the RMDB will make such projects possible.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the authors of RNAstructure, VARNA and the Stanford Visualization Team for making their source code freely available, the Laederach laboratory for aid with SNRNASM integration, the Clote laboratory for data presentation comments and suggestions, R. Astorga for design comments and the members of the Das laboratory and C.C. VanLang for help with data curation and manuscript preparation. The server that houses the RMDB was graciously donated by K. Beauchamp.
Funding: The Burroughs-Wellcome Foundation (CASI to R.D.), a Hewlett-Packard Stanford Graduate Fellowship (to P.C.) and the National Institutes of Health (R01 GM102519 to R.D.).
Conflict of Interest: none declared.
REFERENCES
- Adilakshmi T, et al. Hydroxyl radical footprinting in vivo: mapping macromolecular structures with synchrotron radiation. Nucleic Acids Res. 2006;34:e6. doi: 10.1093/nar/gkl291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviran S, et al. Modeling and automation of sequencing-based characterization of RNA structure. Proc. Natl Acad. Sci. 2011;108:11069–11074. doi: 10.1073/pnas.1106541108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, et al. RNAcentral: a vision for an international database of RNA sequences. RNA. 2011;17:1941–1946. doi: 10.1261/rna.2750811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein FC, et al. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Culver G, et al. Identification of an RNA–protein bridge spanning the ribosomal subunit interface. Science. 1999;285:2133–2135. doi: 10.1126/science.285.5436.2133. [DOI] [PubMed] [Google Scholar]
- Darty K, et al. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, et al. Atomic accuracy in predicting and designing noncanonical RNA structure. Nat. Methods. 2010;7:291–294. doi: 10.1038/nmeth.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deigan KE, et al. Accurate SHAPE directed RNA structure determination. Proc. Natl Acad. Sci. 2009;106:97–100. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush TH, Eickbush D. Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics. 2007;175:477–485. doi: 10.1534/genetics.107.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getz M, et al. Review NMR studies of RNA dynamics and structural plasticity using NMR residual dipolar couplings. Biopolymers. 2007;86:384–402. doi: 10.1002/bip.20765. [DOI] [PubMed] [Google Scholar]
- Kertesz M, et al. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;267:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladwang W, et al. A mutate-and-map strategy accurately infers the base pairs of a 35-nucleotide model RNA. RNA. 2011;17:522–534. doi: 10.1261/rna.2516311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladwang W, Das R. A mutate-and-map strategy for inferring base pairs in structured nucleic acids: proof of concept on a DNA/RNA helix. Biochemistry. 2010;49:7414–7416. doi: 10.1021/bi101123g. [DOI] [PubMed] [Google Scholar]
- Kladwang W, et al. Two-dimensional chemical mapping of non-coding RNAs. Nat. Chem. 2011;3:954–962. doi: 10.1038/nchem.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladwang W, et al. Understanding the errors of SHAPE-directed RNA structure modeling. Biochemistry. 2011;50:8049–8056. doi: 10.1021/bi200524n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G, et al. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low JT, Weeks KM. SHAPE-directed RNA secondary structure prediction. Methods. 2010;52:150–158. doi: 10.1016/j.ymeth.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucks JB, et al. Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc. Natl. Acad. Sci. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews D, Turner D. Prediction of RNA secondary structure by free energy minimization. Curr. Opin. Struct. Biol. 2006;16:270–278. doi: 10.1016/j.sbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Mitra S, et al. High-throughput single-nucleotide structural mapping by capillary automated footprinting analysis. Nucleic Acids Res. 2008;36:e63. doi: 10.1093/nar/gkn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE Chemistry. J. Am. Chem. Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Rocca-Serra P, et al. Sharing and archiving nucleic acid structure mapping data. RNA. 2011;17:1204–1212. doi: 10.1261/rna.2753211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CMT, et al. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- Underwood JG, et al. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat. Methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich EL, et al. BioMagResBank. Nucleic Acids Res. 2007;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G, Tinoco I. RNA structure and NMR spectroscopy. Quart. Rev. Biophys. 1991;24:479–532. doi: 10.1017/s0033583500003875. [DOI] [PubMed] [Google Scholar]
- Vasa SM, et al. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldsich C. Dissecting RNA folding by nucleotide analog interference mapping (NAIM) Nat. Protoc. 2008;3:811–823. doi: 10.1038/nprot.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JM, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, et al. Selective 29-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. The different roles of tryptophan transfer RNA in regulating trp operon expression in E. coli versus B. subtilis. Trends Genet. 2004;20:367–374. doi: 10.1016/j.tig.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Yoon S, et al. HiTRACE: high-throughput robust analysis for capillary electrophoresis. Bioinformatics. 2011;27:1798–805. doi: 10.1093/bioinformatics/btr277. [DOI] [PubMed] [Google Scholar]
- Zheng Q, et al. Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genet. 2010;6:e1001141. doi: 10.1371/journal.pgen.1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.