Abstract
Sandflies are widely distributed around the Mediterranean Basin. Therefore, human populations in this area are potentially exposed to sandfly-transmitted diseases, including those caused by phleboviruses. Whilst there are substantial data in countries located in the northern part of the Mediterranean basin, few data are available for North Africa. In this study, a total of 1489 sandflies were collected in 2008 in Tunisia from two sites, bioclimatically distinct, located 235 km apart, and identified morphologically. Sandfly species comprised Phlebotomus perniciosus (52.2 %), Phlebotomus longicuspis (30.1 %), Phlebotomus papatasi (12 .0%), Phlebotomus perfiliewi (4.6 %), Phlebotomus langeroni (0.4 %) and Sergentomyia minuta (0.5 %). PCR screening, using generic primers for the genus Phlebovirus, resulted in the detection of ten positive pools. Sequence analysis revealed that two pools contained viral RNA corresponding to a novel virus closely related to sandfly fever Naples virus. Virus isolation in Vero cells was achieved from one pool. Genetic and phylogenetic characterization based on sequences in the three genomic segments showed that it was a novel virus distinct from other recognized members of the species. This novel virus was provisionally named Punique virus. Viral sequences in the polymerase gene corresponding to another phlebovirus closely related to but distinct from sandfly fever Sicilian virus were obtained from the eight remaining positive pools.
INTRODUCTION
According to the eighth report of the International Committee for Taxonomy of Viruses (ICTV), the genus Phlebovirus includes 37 viruses grouped into nine species with 16 additional viruses listed as tentative species, with their taxonomic status still pending (Nichol et al., 2005). Phleboviruses are arthropod-borne RNA viruses with a genomic organization consisting of three segments of single-stranded RNA, designated S (small), M (medium) and L (large), encoding the nucleoprotein and non-structural (NS) protein, envelope glycoproteins and the viral polymerase, respectively. Based on antigenic relationships, the genus Phlebovirus is further subdivided into the sandfly fever and Uukuniemi groups. Most of the viruses in the sandfly fever group are associated with and presumably transmitted by phlebotomine sandflies of the family Psychodidae (Tesh, 1988). Sandflies are widely distributed in all Mediterranean countries and their activity peaks during summer (Chelbi et al., 2007). Sandflies are abundant in peri-urban and rural environments, often close to domestic animals and human populations. Therefore, human populations in this area are potentially exposed to sandfly-transmitted diseases, including those caused by phleboviruses. For example, Toscana virus is transmitted by Phlebotomus perniciosus and Phlebotomus perfiliewi (Verani et al., 1988). This virus was discovered in 1971 in Italy and it is a prominent cause of aseptic acute meningitis in the Mediterranean region (Charrel et al., 2005, 2007; Epelboin et al., 2008; Martinez-Garcia et al., 2007; Sanbonmatsu-Gámez et al., 2005; Santos et al., 2007; Venturi et al., 2007). Seroprevalence studies suggest that asymptomatic or mild febrile illnesses due to Toscana virus are significantly more prevalent than previously suspected (De Lamballerie et al., 2007; de Ory-Manchón et al., 2007; Hukic & Salimovic-Besic, 2009; Santos et al., 2007). To date, most of the data available on sandfly-transmitted phleboviruses have been obtained from countries located in south-western Europe (Italy, Spain, Portugal, France and Greece). However, few studies have been conducted regarding phleboviruses from North Africa. Recently, some of the authors (R. N. Charrel, X. d. Lamballerie and G. Moureau) reported molecular evidence for the presence of sandfly fever Sicilian virus (SFSV) in Algeria and serological evidence of human infection (Izri et al., 2008; Moureau et al., 2010). The aims of our study were to detect, isolate and characterize existing and/or new phleboviruses in sandfly populations of Tunisia.
RESULTS
Sandfly trapping and phlebovirus RNA detection
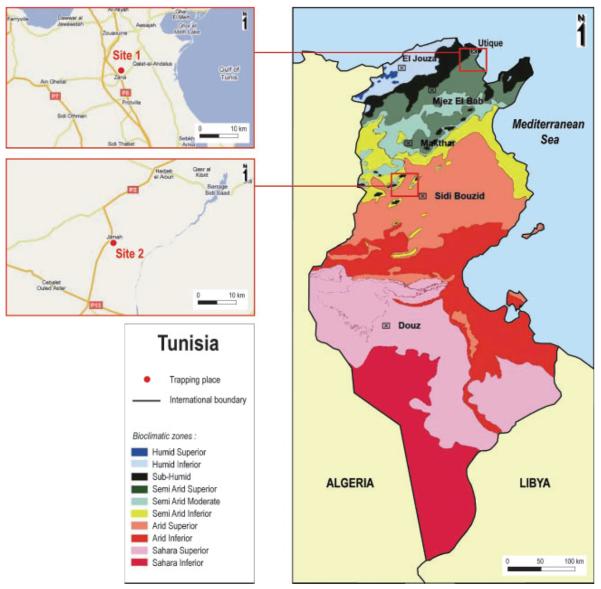
Sandflies were collected from two sites, Utique and Felta, located 235 km apart and corresponding to two different bioclimatic zones (Fig. 1). All sandfly samples trapped during this study were identified individually to the species level (Table 1). A total of 1489 sandflies were collected (778 females, 711 males). P. perniciosus was the most abundant species (52.2 %), followed by Phlebotomus longicuspis (30.1 %), Phlebotomus papatasi (12.0 %) and P. perfiliewi (4.6 %). The remaining species were Phlebotomus langeroni (0.4 %) and Sergentomyia minuta (0.5 %). PCR screening for phlebovirus RNA, using an assay targeting the polymerase gene, showed positive results for ten pools. Of a total of 75 pools, ten contained phlebovirus RNA (Table 1); thus, the overall prevalence of phlebovirus infection among the collected sandflies was 0.67 % (10/1489). Searches using Blastn and Blastx indicated that these ten PCR products corresponded to sequences related to two genetically distinct groups of phleboviruses.
Fig. 1.
Maps showing the capture sites in Tunisia.
Table 1. Species, sex and collection sites of sandflies processed for the presence of phlebovirus.
Pools shown in bold are those from which virus isolation onto Vero cells was attempted; the underlined pool was the one from which virus was isolated. Sandflies were collected in four independent batches. B1–B3 were preserved in guanidinium thiocyanate, B4 was stored at −80 °C. P, Pool number; B, batch number; M, male; F, female.
| Trapping region |
Species | Sex | No. of sandflies | No. of pools | PCR-positive pool |
GenBank acc. no. |
|
|---|---|---|---|---|---|---|---|
| SFN-like virus | SFS-like virus | ||||||
| Utique | P. perniciosus | M | 306 | 12 | 2 (P13/B2, P4/B4) |
GU233653, GU233648 |
|
| F | 464 | 17 | 1 (P1/B4) | 3 (P6/B1, P15B1, P13/B4) |
FJ848989, GU233649, GU233647, GU233646 |
||
| P. longicuspis | M | 260 | 11 | 1 (P6/B2) | GQ165519 | ||
| F | 186 | 8 | 0 | 1 (P14/B1) | GU233650 | ||
| P. perfiliewi | M | 18 | 2 | ||||
| F | 51 | 5 | |||||
| P. papatasi | M | 5 | 2 | ||||
| F | 3 | 3 | |||||
| P. langeroni | M | 0 | 0 | ||||
| F | 6 | 1 | |||||
| S. minuta | M | 4 | 2 | ||||
| F | 4 | 2 | 0 | 1 (P21/B1) | GU233651 | ||
| Total Utique | 1307 | 65 | 2 | 7 | |||
| Felta | P. perniciosus | M | 7 | 1 | 1 (P23/B3) | GU233652 | |
| F | 1 | 1 | |||||
| P. longicuspis | M | 3 | 1 | ||||
| F | 0 | 0 | |||||
| P. papatasi | M | 108 | 4 | ||||
| F | 63 | 3 | |||||
| Total Felta | 182 | 10 | 1 | ||||
| Total | 1489 | 75 | 2 | 8 | |||
Two pools contained viral RNA most closely related to sandfly fever Naples virus (SFNV): pool #6 of the second batch (P6/B2) containing 30 males of P. longicuspis and pool #1 of the fourth batch (P1/B4) containing 30 females of P. perniciosus (Table 1). Both batches were collected in the vicinity of Utique village. Sequence comparison with sequences available from GenBank indicated that both sequences were most closely related to each other (4.6 % divergence in the polymerase gene), and closely related to but distinct from other viruses within the species Sandfly fever Naples virus such as Toscana, Tehran, Naples and Massilia viruses.
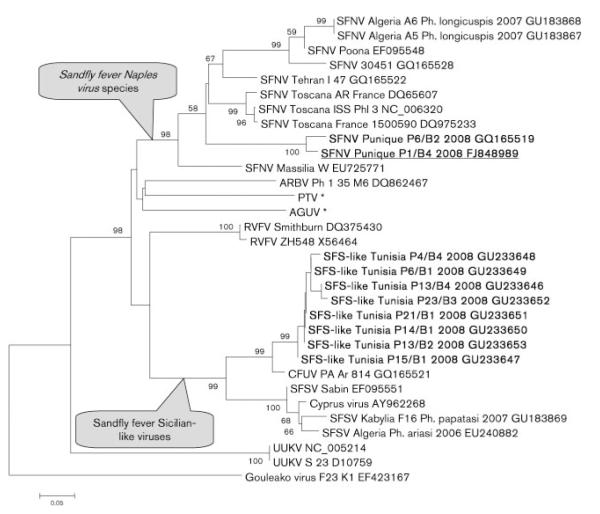
The eight remaining pools (P6/B1, P14/B1, P21/B1, P15/B1, P13/B2, P23/B3, P4/B4 and P13/B4) contained viral RNA (Table 1), with sequences related to SFSV, Corfu virus, SFSV Algeria and SFSV Kabylia (Fig. 2). The eight sequences grouped together with a strong bootstrap support (99 %) and were most closely related to Corfu virus. Out of the eight PCR-positive pools, seven contained sandfly species belonging to the subgenus Larroussius (P. perniciosus, n=6; P. longicuspis, n=1) and one contained a sandfly species of the subgenus Sergentomyia (S. minuta).
Fig. 2.
Phylogenetic tree of the polymerase gene of selected phleboviruses. Partial sequences determined in this study are shown in bold. The sequence corresponding to the viral strain isolated in cell culture and for which genetic characterization was performed (see Fig. 4) is underlined. The acronyms used in the phylogram are those recommended by the ICTV (Nichol et al., 2005): ARBV, Arbia virus; PTV, Punta Toro virus; AGUV, Aguacate virus; RVFV, Rift Valley fever virus; CFUV, Corfu virus; UUKV, Uukuniemi virus. Sequences indicated by * were kindly provided by Dr Maria-Paz Sánchez-Seco (Instituto de Salud Carlos III, Spain). Bar, 0.05 nucleotide substitutions per site.
Punique virus isolation, and electron microscopy (EM) and antigenic characterization
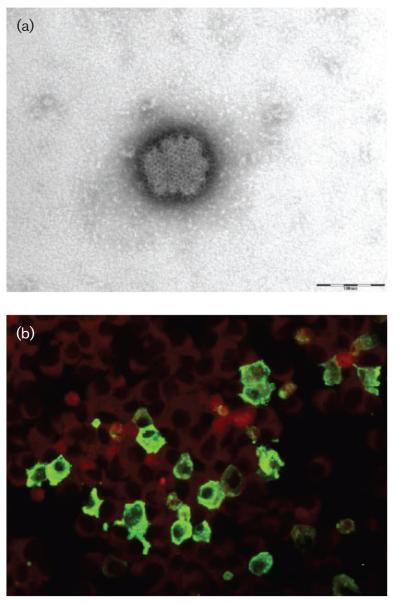
From PCR-positive pools, virus isolation was attempted only from pool P1/B4. Preservation in guanidinium thiocyanate precluded viral isolation from the second PCR-positive pool, P6/B2. Vero cells inoculated with the sandfly homogenate of P1/B4 (female P. perniciosus) showed a cytopathic effect after 4–5 days that was reproduced during at least three serial passages with tenfold serial dilution of the inoculum. The presence of the virus was confirmed by RT-PCR at each passage. Supernatant medium and cells (passage 3) were prepared for EM analysis (Fig. 3a). EM photographs showed spherical structures, 80–120 nm in diameter. Surface projections (5–10 nm long) that evenly covered the surface were clearly visible. Together, these characteristics are compatible with viruses belonging to the genus Phlebovirus within the family Bunyaviridae. Analysis of a serum sample from a convalescent patient who had suffered past infection with Toscana virus, using an indirect immunofluorescence assay with Punique virus-infected Vero cells as antigen, showed cross-reactivity between Toscana and Punique viruses (Fig. 3b).
Fig. 3.
Morphological identification of Punique virus. (a) Negative-staining EM of Vero culture supernatant medium at day 6 post-infection (passage 6). Bar, 100 nm. (b) Vero cells infected with Punique virus react with anti-TOSV human serum.
With regard to the two remaining PCR-positive pools (P4/B4 and P13/B4), with sequences closely related to Corfu virus and genetically related to SFSV, attempts to isolate virus on Vero cells were performed using the protocol described above. No cytopathic effect was observed. EM analysis did not retrieve images indicative of phleboviruses. The RT-PCR performed on Vero cells was negative. Hence, virus isolation was not achieved.
Genetic characterization of Punique virus and comparative analysis
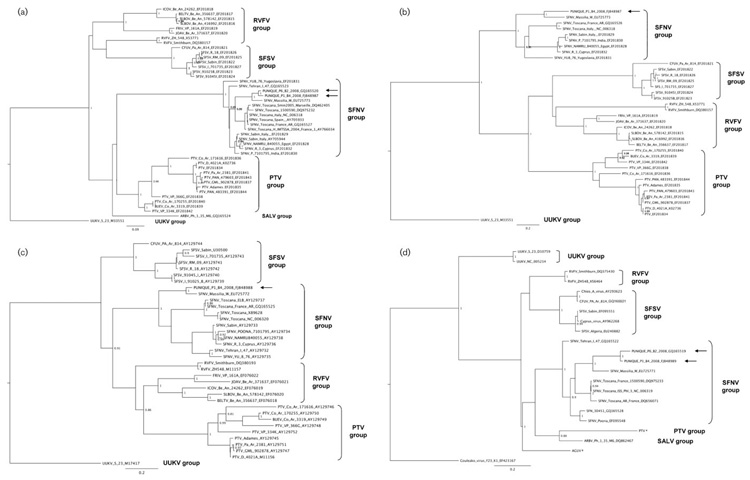
Supernatant from the culture of Vero cells infected with Punique virus was used to sequence partial regions of the S and M RNA segments. Major nodes (representing viruses belonging to the same species or species complex) were clearly distinct and reproduced the topologies reported previously (Charrel et al., 2009). Regardless of the virus protein used for analysis, Punique virus sequences were most closely related to viruses in the SFNV complex. The stable phylogenetic position of Punique virus demonstrated that its genome did not originate from a mechanism involving intersegmental recombination (Fig. 4). Genetic distances between Punique virus and viruses in the SFNV complex were equal to or lower than 17.3/26.8 % (amino acid/nucleotide identity), 46.6/45.6 %, 49.5/43.9 % and 32.8/39.8 % for the nucleoprotein and NS protein (S segment), precursor glycoprotein (M segment) and L proteins, respectively. Comparative analysis of genetic distances with other members of the SFNV complex (Naples, Toscana and Tehran viruses and the newly reported Massilia virus) indicated that Punique virus was most closely related to Massilia virus. The analysis of genetic distances provided convincing evidence that Punique virus may be considered a member of the SFNV complex, but the final decision will be made by the ICTV.
Fig. 4.
Phylogenetic analysis of Punique virus together with partial sequences from selected phleboviruses. Maximum-likelihood phylogeny for the S (nucleocapsid protein) (a), S (NS protein) (b), M (c) and L (d) genome segments. Sequence information corresponds to virus/country of origin/strain/GenBank accession number. Sequences representing Punique virus are indicated by arrows. Antigenic complexes are indicated on the right: PTV, Punta Toro virus; RVFV, Rift Valley fever virus; SALV, Salehabad virus; UUKV, Uukuniemi virus. Sequences indicated by * were kindly provided by Dr Maria-Paz Sánchez-Seco. Only posterior probabilities greater than or equal to 0.8 are shown. Bars indicate nucleotide substitutions per site.
Phylogenetic study using partial sequences in the polymerase gene
The phylogenetic tree produced using partial amino acid sequence in the polymerase gene (Fig. 2) showed that: (i) six taxonomically recognized species or species complexes were unambiguously distinguished with high bootstrap support, (ii) Punique virus was clearly distinct from other viruses (Naples, Toscana, Tehran, Massilia and Algeria viruses) within the SFNV complex, and (iii) the eight sequences corresponding to Tunisian SFS-like virus that grouped together (99 % bootstrap value) were clearly distinct from other viruses genetically related to SFSV or Corfu virus, and more specifically from the two SFS-like viruses reported from Algeria (Moureau et al., 2010). Interestingly, viruses closely related to SFSV seemed to be split into two sublineages: one included SFS-like Tunisia and Corfu virus, and the other consisted of the prototype SFSV together with the Cyprus, SFS-like Algeria and SFS-like Kabylia viruses. These two sublineages within the SFS-like phleboviruses were supported by high bootstrap values (>99 %). Viruses in the sublineage of SFS-like Tunisia and Corfu viruses were mostly detected in sandflies from the subgenus Larroussius (P. perniciosus, P. longicuspis and Phlebotomus major). In contrast, viruses in the second sublineage were mostly detected and isolated in P. papatasi (Schmidt et al., 1960, 1971).
DISCUSSION
To date, very few studies concerning phleboviruses have been conducted in North Africa, and more specifically in Tunisia. At the outset of our work, the only phlebovirus isolated in Tunisia was Tunis virus (included in the Uukuniemi serogroup), associated with Argas reflexus hermanni ticks (Chastel et al., 1994). Neutralization-based seroprevalence studies in Tunisia reported that 1.3 % of 155 human serum samples contained antibodies that neutralized Sicilian virus; none of these 155 sera tested positive for Naples, Arumowot, Gabek Forest, Karimabad or Salehabad virus (Tesh et al., 1976). It is important to note that the methodology used for trapping, identification, preparation and storage of sandflies has been used previously and shown to be capable of virus detection and isolation (Charrel et al., 2007; Izri et al., 2008).
The infection rate of sandflies with phleboviruses was 0.67 % for phleboviruses, 0.13 % for SFN-like viruses and 0.53 % for SFS-like viruses. Similar results have been reported from other countries of the Mediterranean Basin. Infection rates of sandflies with Toscana virus of 0.22, 0.29 and 0.05 % have been reported from Italy, France and Spain, respectively (Charrel et al., 2007; Sanbonmatsu-Gámez et al., 2005; Verani et al., 1988). In France, the infection rate of sandflies with Massilia virus was 0.37 % (Charrel et al., 2009). The infection rate of sandflies from Algeria with SFS-like virus was 0.4 % (2/460) (Izri et al., 2008; Moureau et al., 2010).
The novel phlebovirus isolated in our study was named Punique virus because it was isolated from sandflies collected from a Punique ruin site. Genetic distances between Punique virus and other relatives clearly indicated that it is most closely related to members of the SFNV complex. In the four phylogenetic trees, Punique virus grouped with viruses within the SFNV complex and was clearly distinct from other phleboviruses. Within this group, it appeared to be most closely related to Massilia virus, regardless of the genome segment used for analysis. However, amino acid distances within the L segment were of the same order of magnitude between Punique virus and all other members of the SFNV complex (25–29 %). The identification of new sequences displaying a high level of heterogeneity with previously documented phleboviruses indicates that Punique virus is a novel phlebovirus. According to the genetic analysis presented here and in a recent article (Charrel et al., 2009), we propose that Punique virus should be included in the SFNV complex with the status of a new genotype genetically distinct from other members of the complex. Punique virus will be proposed to the ICTV for a final decision concerning its classification, as previously conducted for Massilia virus (Charrel et al., 2009).
The public health importance of Punique virus remains unknown at this time. However, the fact that this new phlebovirus is a member of the SFNV complex (most members of which can infect humans) and that it is associated with P. perniciosus, a proven vector of visceral leishmaniasis and Toscana virus infection in the Mediterranean region (Dujardin et al., 2008), indicates the need for studies aimed at determination of the public health impact of Punique virus.
Punique virus was isolated from P. perniciosus and detected in P. longicuspis. Notably, Punique virus was isolated from the Utique region, a well-known focus of human visceral leishmaniasis, which is transmitted in Tunisia principally by P. perniciosus (Zhioua et al., 2007). Toscana virus has been isolated only from P. perniciosus and P. perfiliewi (Charrel et al., 2007; Verani et al., 1988). Massilia virus has been isolated from P. perniciosus (Charrel et al., 2009). This is the first report of a phlebovirus within the SFNV complex being detected in P. longicuspis. However, further studies are needed to confirm the transmission of this phlebovirus by P. longicuspis. Despite the fact that Toscana virus has also been detected in S. minuta (Charrel et al., 2006), viruses in the SFNV group such as Toscana, Punique and Massilia viruses appear to be transmitted mostly by sandfly species of the subgenus Larroussius such as P. perniciosus, P. perfiliewi and P. longicuspis. This finding is corroborated by the fact that, in Felta, where P. papatasi is the most abundant sandfly species and belongs to the subgenus Phlebotomus, Punique virus was not detected. Punique virus was detected in both male and female sandflies, suggesting venereal or transovarial transmission, as for Toscana virus (Ciufolini et al., 1991), although other routes of transmission should be considered.
The majority of pools containing phlebovirus sequences genetically related to SFSV or SFS-like viruses corresponded to sandflies belonging to the subgenus Larroussius. SFS-like virus was detected in P. papatasi, Phlebotomus ariasi and P. major (Izri et al., 2008; Liu et al., 2003; Tesh, 1988). In our study, we report for the first time the detection of SFS-like virus in P. perniciosus, P. longicuspis and S. minuta. It is also of major importance to point out that two phleboviruses within distinct genetic clusters were detected in a single sandfly species (P. perniciosus) collected from the same geographical area. In south-eastern France, Toscana and Massilia viruses, which are genetically related and belong to the same antigenic and genetic group, were isolated from the same sandfly species: P. perniciosus (Charrel et al., 2009).
In conclusion, a novel phlebovirus, provisionally named Punique virus, was isolated from sandflies from northern Tunisia. Its antigenic and genetic characterization through complete or partial sequencing of the four virus genes revealed that Punique virus is closely related to other phleboviruses members of the species Sandfly fever Naples virus. In addition, another novel phlebovirus, most closely related to Corfu virus and to SFSV, was detected (but not isolated) in eight pools of sandflies collected from the same region in Tunisia. Further investigations (seroprevalence studies and medical investigations of patients with fever of unknown origin and of patients with meningitis and encephalitis) are necessary to determine the public health impact of these phleboviruses in Tunisia.
METHODS
Study sites and collection of sandflies
Sandflies were collected from two sites located 235 km apart corresponding to two different bioclimatic zones (Utique 37° 2′ N 10° 2′ E, subhumid; Felta, 35° 16′ N 9° 25′ E, arid) (Fig. 1). These two sites, Utique and Felta, are well known foci of visceral and cutaneous leishmaniasis, respectively (Chelbi et al., 2007, 2009; Zhioua et al., 2007). Sandflies were collected inside houses and in animal shelters located in peri-domestic areas using CDC light traps. Trapping was conducted from dusk to dawn on 1 night per site per month from June to September 2008. Sandflies were dissected under a stereo-microscope on ice to remove the genitalia for species identification and were morphologically identified to the species level (Croset et al., 1978). Sandflies were pooled with a maximum of 30 individuals per pool, based on trapping location, species and sex, and were placed in 1.5 ml tubes and stored either in guanidinium isothiocyanate (RNA Now; Biogentex) (batches 1–3) or at −80 °C (batch 4).
Detection of phleboviruses in sandfly pools by RT-PCR
Pools of sandflies were homogenized using a Mixer Mill MM300 (Qiagen) with one 3 mm tungsten bead at a frequency of 30 cycles s−1 for 3–5 min depending on the viscosity of the material. A treatment protocol was then conducted depending on the method of preservation: (i) total RNA from RNA Now-preserved pools was purified following the manufacturer’s recommendations and eluted in 50 μl distilled water; (ii) frozen pools were crushed in the presence of 600 μl L15 medium supplemented with 3 % decomplemented calf serum, 5 % tryptose phosphate broth and 100 IU penicillin G ml−1, 100 mg kanamycin ml−1, 100 mg streptomycin ml−1 and 2.5 μg amphotericin B ml−1. The resulting mixture was clarified by centrifugation at 5800 g for 10 min and the supernatant fluid was aliquotted (six aliquots of 100 μl for each pool) and stored at −80 °C.
One 100 μl aliquot was used for viral RNA purification using a BioRobot EZ1 (Viral RNA Mini kit: Qiagen) and stored directly at −80 °C. In both cases, 10 μl RNA suspension was used for RT-PCR. A variety of primers targeting different genes were used in independent reactions: (i) phlebovirus consensus primers targeting the polymerase gene in the L RNA segment (Sánchez-Seco et al., 2003), and (ii) primers specific for phleboviruses within the SFNV complex and targeting the nucleoprotein gene in the S RNA segment (Charrel et al., 2007). RT-PCR was performed using an Access RT-PCR kit (Promega), according to the manufacturer’s recommendations. L and S RNA primers were used at 0.8 and 0.4 μM per reaction, respectively. The cycling program of the RT-PCR consisted of 48 °C for 45 min and 94 °C for 2 min, followed by 40 cycles at 94 °C for 30 s, 45 °C for 1 min and 68 °C for 45 s, with a final elongation step at 68 °C for 7 min. Nested PCRs were performed using the same conditions with 1.25 U Taq DNA polymerase (Invitrogen). PCR products were visualized in a 2 % TAE/agarose electrophoresis gel and sequenced in both directions.
Virus isolation, and morphological and antigenic studies
Sandfly homogenates from PCR-positive pools preserved previously at −80 °C (i.e. those not in guanidinium thiocyanate) were used to inoculate Vero cells. Briefly, 100 μl each homogenate was diluted with 900 μl Eagle’s minimal essential medium without fetal bovine serum (FBS), but enriched with antibiotics (100 IU penicillin G ml−1, 100 mg streptomycin ml−1, 100 mg kanamycin ml−1 and 2.5 μg amphotericin B ml−1) and used to seed Vero monolayers under a 12.5 cm2 flask. After incubation at room temperature for 1 h, 4 ml fresh 3 % FBS medium was added. The flask was incubated at 37 °C in an atmosphere containing 5 % CO2. A negative-control flask was prepared under identical conditions. Flasks were examined daily for the presence of a cytopathic effect, and 400 μl supernatant medium was extracted and tested by RT-PCR.
Tissue culture samples showing a cytopathic effect were prepared for EM examination. Negatively stained EM specimens were prepared by drying culture supernatant medium, mixed 1 : 1 with 2.5 % paraformaldehyde, onto Formvar/carbon-coated grids and staining with 2 % methylamine tungstate.
Punique virus-infected cells were used to prepare slides for an indirect immunofluorescence assay after cytospin centrifugation. Slides were air dried and fixed with 100 % cold acetone for 20 min. The assay was performed as described previously (Fulhorst et al., 1997) using the serum of a patient who suffered from Toscana virus infection with a high titre of IgG (Hemmersbach-Miller et al., 2004), in conjunction with a fluorescein isothiocyanate-conjugated goat anti-human IgG (Fluoline G; bioMérieux).
Sequence analysis and molecular characterization of Punique virus
Viral RNA obtained from infected Vero cells was used to extend sequences obtained at the detection stage. Primers and protocols are available upon request from the corresponding author. Consensus sequences for each gene in both directions were collated using Sequencher 4.9 (Gene Codes Corporation) and submitted to the NCBI Blastn and Blastx programs (www.ncbi.nlm.nih.gov/BLAST/) for identification. Datasets were prepared using Se-Al (available at http://tree.bio.ed.ac.uk/software/seal/). Multiple sequence alignment was conducted using Muscle (Edgar, 2004) together with sequences from other phleboviruses retrieved from GenBank. The accession numbers of GenBank sequences used for genetic analyses are indicated in the phylogenetic trees (Figs 2 and 4). Regions of ambiguous alignment were removed via GBlocks (Talavera & Castresana, 2007). Phylogenetic analyses were conducted on amino acid alignments via Bayesian methods in MrBayes v3.1.2 (Huelsenbeck & Ronquist, 2001) with a minimum of 107 generations, a burn-in of 10 % and assessment of stationarity at an effective sample size of >400 using Tracer v1.4.1 (part of the Beast package; Drummond & Rambaut, 2007). Matrices of amino acid p distances were calculated using Mega 4.0 (Tamura et al., 2007).
Phylogenetic study using partial sequences in the polymerase gene
The ten PCR products obtained using primers located in the polymerase gene (L RNA segment), as described by Sánchez-Seco et al. (2003), were cloned and sequenced resulting in sequences of 201 nt. The corresponding amino acid sequences were used for genetic and phylogenetic analyses together with homologous sequences of genetically related phleboviruses. Distances and groupings were determined by the pairwise-distance algorithm and neighbour-joining method within Mega4, and the robustness of the groups was tested using 500 bootstrap pseudoreplicates (Fig. 2).
ACKNOWLEDGEMENTS
This work has been supported in part (i) by SERST (Secretariat de l’Enseignement et de la Recherche Scientifique et Technique de Tunisie), (ii) by the European Commission through RiVigene project and the European Virus Archive (GA no. 228292) in the FP7 Capacities, and (iii) by the French authorities through the PhleboMED project (funded by Agence National de la Recherche and AIRD) and the Institute of Research for Development. The authors thank Bernard Campagna for excellent technical assistance with EM.
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the sequences of the partial regions of the S, M and L RNA segments of Punique virus are FJ848987, FJ848988 and FJ848989, respectively. Other sequences determined in this study are GU165519, GU165520, GU233646–GU233653, GU183869, GQ165521, GQ165522, GQ165528 and DQ65607.
REFERENCES
- Charrel RN, Gallian P, Navarro-Mari JM, Nicoletti L, Papa A, Sánchez-Seco MP, Tenorio A, de Lamballerie X. Emergence of Toscana virus in Europe. Emerg Infect Dis. 2005;11:1657–1663. doi: 10.3201/eid1111.050869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrel RN, Izri A, Temmam S, de Lamballerie X, Parola P. Toscana virus RNA in Sergentomyia minuta flies. Emerg Infect Dis. 2006;12:1299–1300. doi: 10.3201/eid1208.060345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrel RN, Izri A, Temmam S, Delaunay P, Toga I, Dumon H, Marty P, de Lamballerie X, Parola P. Cocirculation of 2 genotypes of Toscana virus, southeastern France. Emerg Infect Dis. 2007;13:465–468. doi: 10.3201/eid1303.061086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrel RN, Moureau G, Temmam S, Izri A, Marty P, Parola P, da Rosa AT, Tesh RB, de Lamballerie X. Massilia virus, a novel Phlebovirus (Bunyaviridae) isolated from sandflies in the Mediterranean. Vector Borne Zoonotic Dis. 2009;9:519–530. doi: 10.1089/vbz.2008.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastel C, Bach-Hamba D, Karabatsos N, Bouattour A, Le Lay G, Le Goff F, Vermeil C. Tunis virus: a new Phlebovirus from Argas reflexus hermanni ticks in Tunisia. Acta Virol. 1994;38:285–289. [PubMed] [Google Scholar]
- Chelbi I, Derbali M, Al-Ahmadi Z, Zaafouri B, El Fahem A, Zhioua E. Phenology of Phlebotomus papatasi (Diptera: Psychodidae) relative to the seasonal prevalence of zoonotic cutaneous leishmaniasis in central Tunisia. J Med Entomol. 2007;44:385–388. doi: 10.1603/0022-2585(2007)44[385:poppdp]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Chelbi I, Kaabi B, Bejaoui M, Derbali M, Zhioua E. Spatial correlation between Phlebotomus papatasi Scopoli (Diptera: Psychodidae) and incidence of zoonotic cutaneous leishmaniasis in Tunisia. J Med Entomol. 2009;46:400–402. doi: 10.1603/033.046.0229. [DOI] [PubMed] [Google Scholar]
- Ciufolini MG, Maroli M, Verani P. Laboratory reared sandflies (Diptera: Psychodidae) and studies on phleboviruses. Parassitologia. 1991;33(Suppl.):137–142. [PubMed] [Google Scholar]
- Croset H, Rioux JA, Maistre M, Bayar N. The phlebotomines of Tunisia (Diptera-Phlebotominae). A revision of the systematics, distribution and behaviour. Ann Parasitol Hum Comp. 1978;53:711–749. (in French) [PubMed] [Google Scholar]
- De Lamballerie X, Tolou H, Durand JP, Charrel RN. Prevalence of Toscana virus antibodies in volunteer blood donors and patients with central nervous system infections in southeastern France. Vector Borne Zoonotic Dis. 2007;7:275–277. doi: 10.1089/vbz.2006.0637. [DOI] [PubMed] [Google Scholar]
- de Ory-Manchón F, Sanz-Moreno JC, Aranguez-Ruiz E, Ramírez-Fernández R. Age-dependent seroprevalence of Toscana virus in the Community of Madrid: 1993–1994 and 1999– 2000. Enferm Infecc Microbiol Clin. 2007;25:187–189. doi: 10.1157/13099371. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. Beast: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin JC, Campino L, Canavate C, Dedet JP, Gradoni L, Soteriadou K, Mazeris A, Ozbel Y, Boelaert M. Spread of vector-borne diseases and neglect of leishmaniasis, Europe. Emerg Infect Dis. 2008;14:1013–1018. doi: 10.3201/eid1407.071589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epelboin L, Hausfater P, Schuffenecker I, Riou B, Zeller H, Bricaire F, Bossi P. Meningoencephalitis due to Toscana virus in a French traveler returning from central Italy. J Travel Med. 2008;15:361–363. doi: 10.1111/j.1708-8305.2008.00221.x. [DOI] [PubMed] [Google Scholar]
- Fulhorst CF, Monroe MC, Salas RA, Duno G, Utrera A, Ksiazek TG, Nichol ST, de Manzione NM, Tovar D, Tesh RB. Isolation, characterization and geographic distribution of Caño Delgadito virus, a newly discovered South American hantavirus (family Bunyaviridae) Virus Res. 1997;51:159–171. doi: 10.1016/s0168-1702(97)00091-9. [DOI] [PubMed] [Google Scholar]
- Hemmersbach-Miller M, Parola P, Charrel RN, Paul Durand J, Brouqui P. Sandfly fever due to Toscana virus: an emerging infection in southern France. Eur J Intern Med. 2004;15:316–317. doi: 10.1016/j.ejim.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. Mrbayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hukic M, Salimovic-Besic I. Sandfly – Pappataci fever in Bosnia and Herzegovina: the new–old disease. Bosn J Basic Med Sci. 2009;9:39–43. doi: 10.17305/bjbms.2009.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izri A, Temmam S, Moureau G, Hamrioui B, de Lamballerie X, Charrel RN. Sandfly fever Sicilian virus, Algeria. Emerg Infect Dis. 2008;14:795–797. doi: 10.3201/eid1405.071487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DY, Tesh RB, Travassos Da Rosa AP, Peters CJ, Yang Z, Guzman H, Xiao SY. Phylogenetic relationships among members of the genus Phlebovirus (Bunyaviridae) based on partial M segment sequence analyses. J Gen Virol. 2003;84:465–473. doi: 10.1099/vir.0.18765-0. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia FA, Moreno-Docon A, Lopez-Lopez M, Albert-Lacal L, Martinez-Toldos MC, Segovia-Hernandez M, Fernandez-Barreiro A. A case of meningitis due to Toscana virus in Murcia. Rev Neurol. 2007;45:317–318. (in Spanish) [PubMed] [Google Scholar]
- Moureau G, Bichaud L, Salez N, Ninove L, Hamrioui B, Belazzoug S, de Lamballerie X, Izri A, Charrel RN. Molecular and serological evidence for the presence of novel phleboviruses in sandflies from Northern Algeria. Open Virol J. 2010;4 doi: 10.2174/1874357901004010015. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichol ST, Beaty BJ, Elliott RM, Goldbach R, Plyusnin A, Schmaljohn CS, Tesh RB. Genus Phlebovirus. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses. Academic Press; San Diego: 2005. pp. 709–711. [Google Scholar]
- Sanbonmatsu-Gámez S, Pérez-Ruiz M, Collao X, Sánchez-Seco MP, Morillas-Márquez F, de la Rosa-Fraile M, Navarro-Mari JM, Tenorio A. Toscana virus in Spain. Emerg Infect Dis. 2005;11:1701–1707. doi: 10.3201/eid1111.050851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Seco MP, Echevarría JM, Hernández L, Estévez D, Navarro-Marí JM, Tenorio A. Detection and identificationof Toscana and other phleboviruses by RT-nested-PCR assays with degenerated primers. J Med Virol. 2003;71:140–149. doi: 10.1002/jmv.10465. [DOI] [PubMed] [Google Scholar]
- Santos L, Simões J, Costa R, Martins S, Lecour H. Toscana virus meningitis in Portugal, 2002–2005. Euro Surveill. 2007;12:E3–E4. doi: 10.2807/esm.12.06.00715-en. [DOI] [PubMed] [Google Scholar]
- Schmidt JR, Schmidt ML, McWilliams JG. Isolation of phlebotomus fever virus from Phlebotomus papatasi. Am J Trop Med Hyg. 1960;9:450–454. doi: 10.4269/ajtmh.1960.9.450. [DOI] [PubMed] [Google Scholar]
- Schmidt JR, Schmidt ML, Said MI. Phlebotomus fever in Egypt: isolation of phlebotomus fever viruses from Phlebotomus papatasi. Am J Trop Med Hyg. 1971;20:483–490. doi: 10.4269/ajtmh.1971.20.483. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. Mega4: Molecular Evolutionary Genetics Analysis (Mega) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tesh RB. The genus Phlebovirus and its vectors. Annu Rev Entomol. 1988;33:169–181. doi: 10.1146/annurev.en.33.010188.001125. [DOI] [PubMed] [Google Scholar]
- Tesh RB, Saidi S, Gajdamovic SJ, Rodhain F, Vesenjak-Hirjan J. Serological studies on the epidemiology of sandfly fever in the Old World. Bull World Health Organ. 1976;54:663–674. [PMC free article] [PubMed] [Google Scholar]
- Venturi G, Madeddu G, Rezza G, Ciccozzi M, Pettinato ML, Cilliano M, Fiorentini C, Mura MS, Ciufolini MG. Detection of Toscana virus central nervous system infections in Sardinia Island, Italy. J Clin Virol. 2007;40:90–91. doi: 10.1016/j.jcv.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Verani P, Ciufolini MG, Caciolli S, Renzi A, Nicoletti L, Sabatinelli G, Bartolozzi D, Volpi G, Amaducci L. Ecology of viruses isolated from sand flies in Italy and characterized of a new Phlebovirus (Arabia virus) Am J Trop Med Hyg. 1988;38:433–439. doi: 10.4269/ajtmh.1988.38.433. other authors. [DOI] [PubMed] [Google Scholar]
- Zhioua E, Kaabi B, Chelbi I. Entomological investigations following the spread of visceral leishmaniasis in Tunisia. J Vector Ecol. 2007;32:371–374. doi: 10.3376/1081-1710(2007)32[371:eiftso]2.0.co;2. [DOI] [PubMed] [Google Scholar]