Abstract
Magnetic stain. Bacteria are often classified into Gram-positive and Gram-negative strains by their visual staining properties using crystal violet (CV), a triarylmethane dye. Here we show, that bioorthogonal modification of crystal violet with transcyclooctene (TCO) can be used to render Gram-positive bacteria magnetic with magneto-nanoparticles-Tetrazine (MNP-Tz). This allows for class specific automated magnetic detection, magnetic separation or other magnetic manipulations.
Keywords: Cycloaddition reaction, Bacteria, Nanoparticles, Dyes, Gram Stain
Bacterial cell walls are made up of peptidoglycans (polysaccharides crosslinked by unusual peptides) in addition to other components.[1] Bacteria are often classified into Gram-positive and Gram-negative strains by their visual staining properties using crystal violet (CV), a triarylmethane dye.[2] Here we show that bioorthogonal modification of crystal violet with transcyclooctene can be used to render Gram-positive bacteria magnetic. This allows for class specific automated magnetic detection, magnetic separation or other magnetic manipulations.
The Gram stain is one of the most commonly used tools for detecting and differentiating bacteria. The method is routinely used for clinical diagnostic purposes, identification of a bacterial organism, as well as detecting them in environmental samples. The procedure involves staining bacterial samples with crystal violet, which binds to the peptidoglycan layer of Gram-positive and negative bacteria (Figure 1). Subsequent treatment with iodine solution results in crystal violet to form an insoluble complex. Gram-positive bacteria have a thick peptidoglycan layer, whereas Gram-negative bacteria only have a thin peptidoglycan layer covered by lipopolysaccharides and lipoproteins. Upon decolorization with alcohol or acetone, only Gram-positive bacteria remain purple, while Gram-negatives loose the purple color.[3–5] Despite the simplicity and robustness of the staining procedure, the final detection still relies on optical microscopy which is often susceptible to user-dependent sampling error. Strategies for quantitative and automated detection are highly desirable, especially for the diagnosis of infectious pathogens.
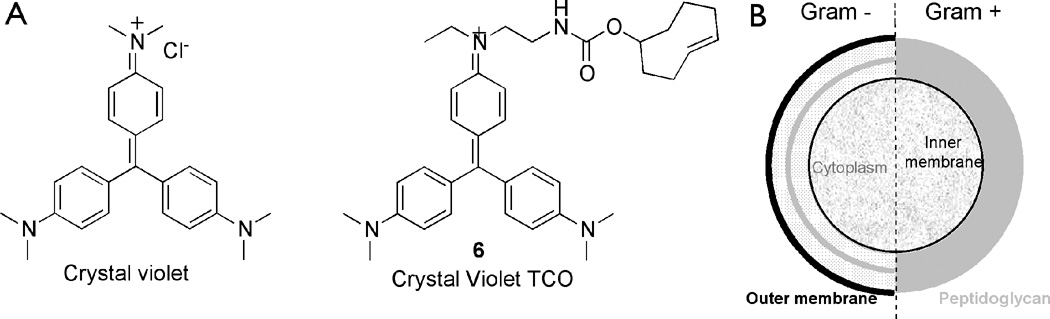
Figure 1.
A) Chemical structure of Crystal violet (left) and the new bioorthogonal crystal violet-TCO (right). B) General composition of Gram-positive and Gram-negative cell wall.
Magnetic, rather than optical, labeling and detection is advantageous because of its high sensitivity and ability to diagnose crude specimens without major purification.[6] For example, one could envision rapid and sensitive detection of bacterial samples in point-of-care settings by using a miniaturized micro-nuclear magnetic resonance (µNMR) device.[7,8] Direct bacterial detection by µNMR is a sensitive diagnostic method[9] and potentially allows the exclusion of culturing steps thus minimize the time required for diagnosis. Alternative magnetic detection devices include giant magnetoresistance,[10] or Hall sensors.[11] Furthermore, rendering bacteria magnetic also has implications for magnetic separation,[11,12] cell sorting,[13] magnetic force microscopy[14] or micromanipulation and force measurements using magnetic tweezers.[15]
We hypothesized that orthogonal triarylmethane dye derivatives could be used as affinity ligands to bioorthogonally couple magnetic nanomaterials onto Gram-positive bacteria. We thus developed a crystal violet modified with transcyclooctene (CV-TCO). We show that this reagent can be used for staining Gram-positive bacteria similar to the native crystal violet. Importantly, the CV-TCO can also serve as an anchor to attach tetrazine (Tz)-modified magnetic nanoparticles (or other Tz derivatized reporters). The developed magnetic Gram stain method was then used to enable highly sensitive detection of Gram-positive pathogens by µNMR.
Crystal violet (CV; 4,4’,4”-dimethylaminotriphenylmethane), is a deep purple dye. We sought to develop a chromophore derivative where one of the anilino moieties is modified with a transcyclooctene (TCO) orthogonal group. We started the synthesis by the condensation of two equivalents of dimethylaniline with paranitrobenzaldehyde under microwave irradiation at 90°C for 4 min in the presence of a catalytic amount of aniline.[16] The aromatic nitro group was then reduced quantitatively by hydrogenolysis in presence of activated palladium affording the free amine 2 (Figure 2). However, the formed adduct instantaneously oxidizes in presence of air rendering purification and further conjugation difficult. The oxidation process is readily apparent since the oxidized compound has an intense purple color. To avoid oxidation to the cationic dye, the aniline was therefore derivatized twice to be stable under oxygen. We thus explored the synthesis of the disubstituted aniline 4 using multi-step one pot synthetic sequence. The nitro compound 2 was reduced by hydrogenation and reaction progress was followed by LC-MS (Figure S1). After completion of the reaction, the flask was purged with argon and the free aniline was engaged in a reductive amination with Boc-2-aminoacetaldehyde, sodium cyanoborohydride and acetic acid and stirred until completion. The secondary amine 3 was then engaged in a classic reductive amination condition with acetaldehyde, sodium cyanoborohydride and acetic acid for five hours yielding compound 4 in 71% yield over three steps. Structure and purity of 4 was confirmed by 1H NMR showing the characteristic chemical shift of the methylene proton at 5.30 ppm (see supplemental information). Compound 4 was then oxidized with tetrachloroquinone in refluxed ethyl acetate causing the formation of an intense blue indicating the formation of the cationic dye. After Boc deprotection under acidic conditions, compound 5 was isolated and purified on neutral alumina. Finally, the free amine 5 was treated with TCO-NHS, furnishing 6 (Crystal violet-TCO, CV-TCO) with an overall yield of 17% over seven steps (Figure 2A).
Figure 2.
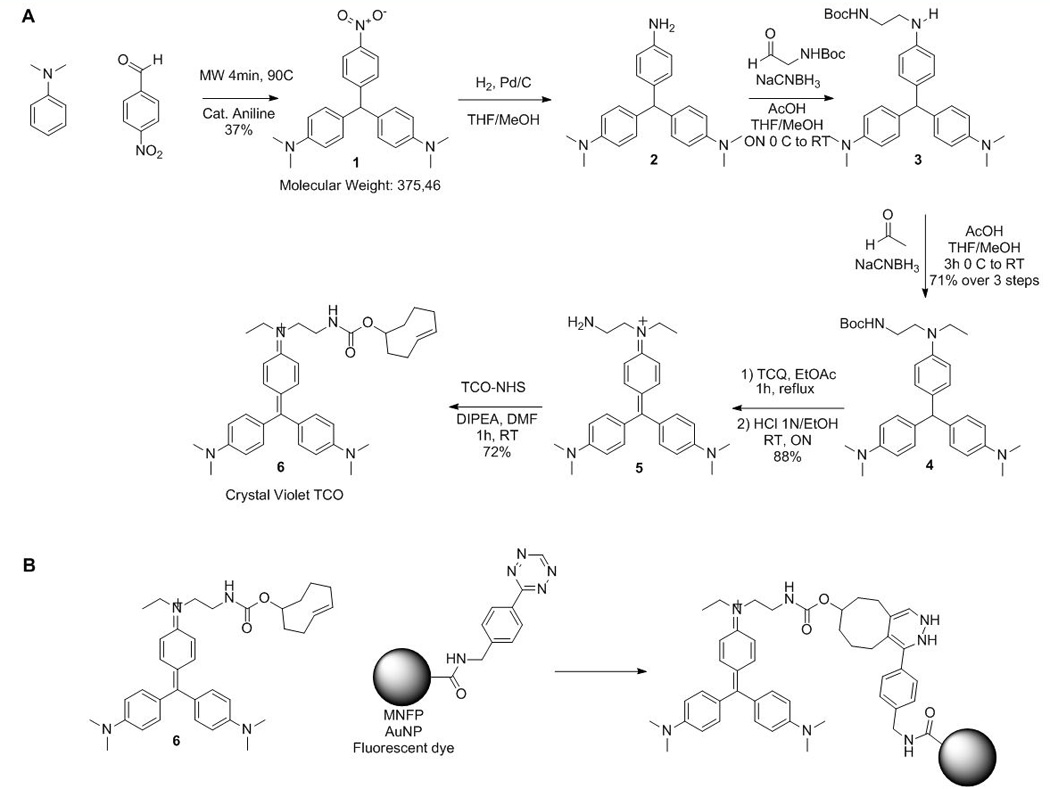
A) Synthesis of the bioorthogonal crystal violet-TCO (6). B) Bioorthogonal reaction between Crystal Violet-TCO (6) and tetrazine-conjugated probes (not all isomers shown).
The molar extinction coefficient of CV-TCO 6 was ε592= 133013 L.mol−1.cm−1 as compared to unmodified CV which had ε592= 89146 L.mol−1.cm−1. These results suggest that the TCO linker modification only minimally affects the molar absorptivity of the triarylmethane dye and that the bioorthogonal compound can likewise be used for Gram staining (Figure S2). We then investigated the cycloaddition of 6 with a fluorescently labeled tetrazine, fluorescein-tetrazine (Fluo-Tz). After mixing the two compounds (0.25 mM), stirring for two minutes, the sample was analyzed by high performance liquid chromatography–mass spectrometry (HPLC-MS). HPLC-MS spectra confirmed rapid and quantitative conversion of Fluo-Tz to the cycloaddition-product without any side products (Figure 2B and Figure S3).
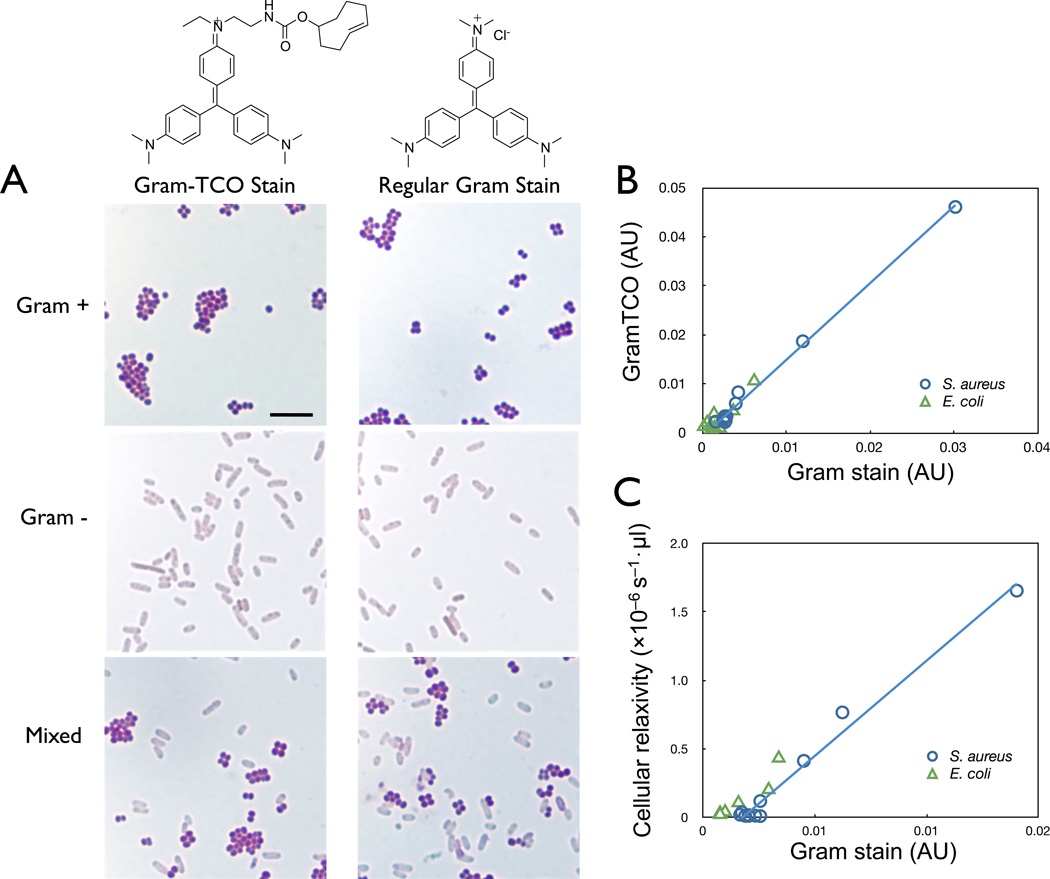
We next evaluated the efficacy of CV-TCO as a staining agent for Gram-positive bacteria. Three representative samples were prepared: Staphylococcus aureus (S. aureus; Gram-positive), Escherichia coli (E. coli; Gram-negative), and the mixture of both bacterial species. Bacterial smears on glass slides were stained with a solution of CV-TCO (1 mM) or CV for three minutes, followed by treatment with Gram’s iodine solution for one minute, decolorization with 95% ethanol, and counterstaining with red Safranin solution. Microscopy revealed that only Gram-positive S. aureus remained purple, while Gram-negative E. coli was decolorized due to dissolution of the outer membrane (Figure 3A). The specificity of CV-TCO was further confirmed by UV-visible spectrometry; only gram-positive bacteria showed an intense absorption at 595 nm (Figure S4). Importantly, there was excellent correlation between CV and CV-TCO staining (r2 > 0.99; Figure 3B).
Figure 3.
A) Gram staining of S. aureus (Gram-positive cocci), E. coli (Gram-negative bacilli), and mixture of S. aureus and E. coli stained with Crystal violet-TCO (left panels) or with Crystal violet (right panels) (scale bar = 10 µm). B) Correlation of absorbance at 595 nm between bacteria stained with Crystal violet and Crystal violet-TCO. C) Correlation between absorbance (595 nm) and magnetic relaxivity values of bacterial cells stained with Crystal violet-TCO and labeled with magnetic MFNP-Tz.
We further investigated if the bacteria stained with CV-TCO could be magnetically labeled via the TCO group. Bacteria stained with CV-TCO were incubated with magnetofluorescent nanoparticles modified with tetrazine (MFNP-Tz). Control samples were prepared by incubating unstained bacteria with MFNP-Tz. The T2 relaxation values of samples were measured using a miniaturized µNMR system. For comparative analyses, the absorption (at 595 nm) of the same samples was also measured. Cellular relaxivity (r2) was obtained by normalizing the measured 1/T2 values with bacterial concentration, and the r2 differences (Δr2) between targeted and control samples were calculated. We observed an excellent correlation (r2 > 0.9) between the extent of Gram-staining and the cellular relaxivity in Gram-positive species, which confirmed that CV-TCO on the bacterial surface was accessible for reaction with MFNP-Tz.
The labeling strategy was further applied to a panel of different bacterial species (Figure 4). Results showed that all Gram-positive species tested showed significantly higher cellular relaxivity values when compared to Gram-negative bacteria. Such magnetic labeling enabled the performance of highly sensitive and rapid detection of gram-positive bacteria. Titration measurements with serially diluted bacterial samples established that the detection limit with the current experimental setup was ~4,000 bacteria (Figure S5). This is significantly better than standard UV absorption detection, which has a detection limit of approximately 105 bacteria (Figure S6). It is likely that the sensitivity of the magnetic detector could be improved to the level of single-cells by 1) further miniaturizing the µNMR detection coils, 2) implementing fluidic systems for bacterial enrichment (e.g., membrane filters, magnetic separation steps), and 3) employing different types of magnetic readers (e.g., Hall-effect sensors, giant magnetoresistive sensors).[7–9]
Figure 4.
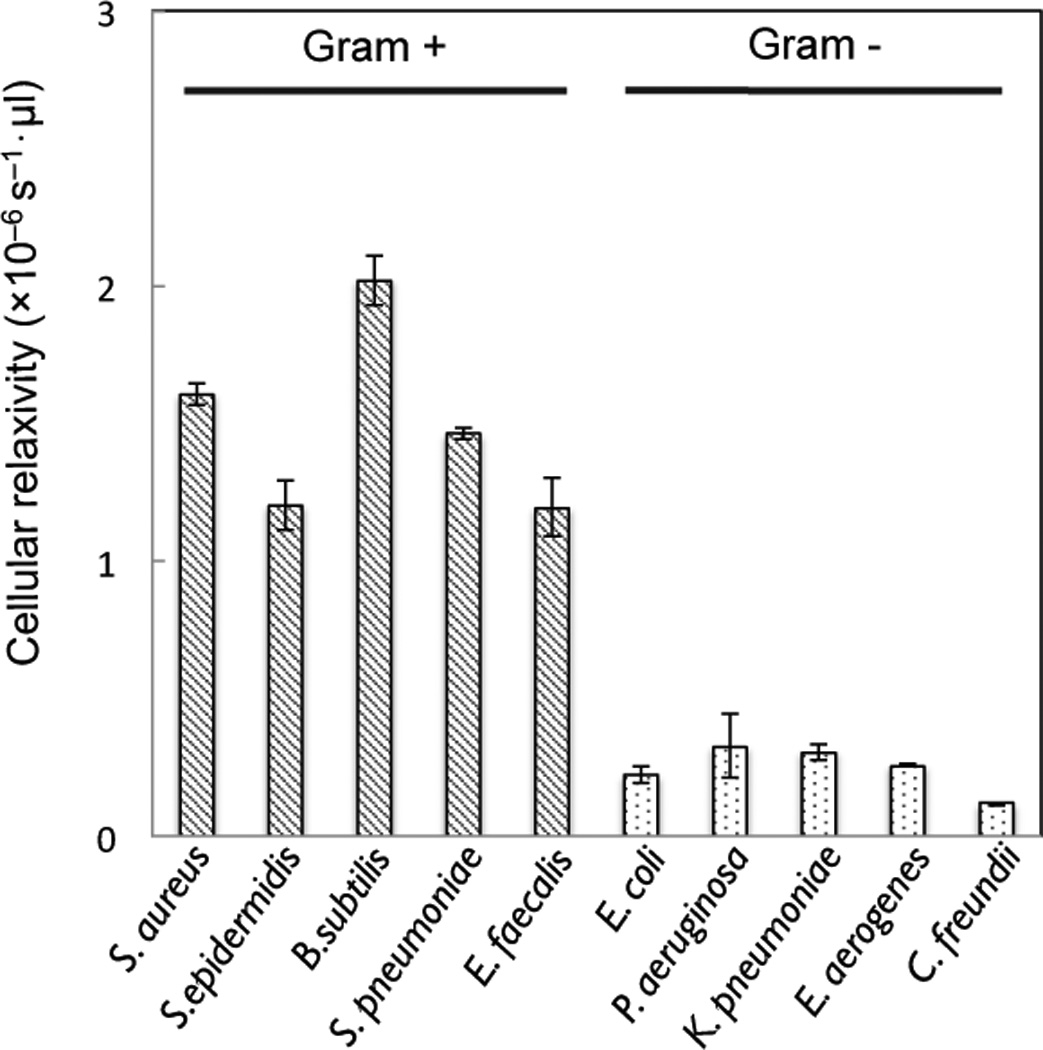
Magnetic detection by µNMR of different species of Gram-positive and Gram-negative bacteria labeled using crystal violet-TCO and MFNP-Tz.
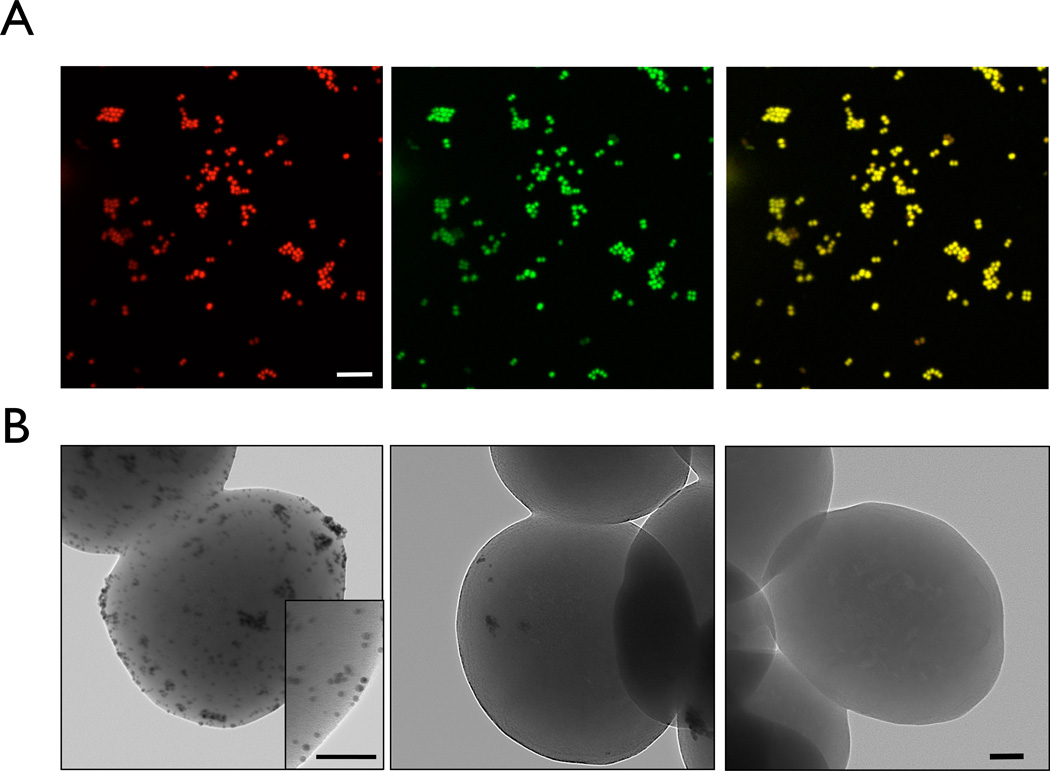
Bioorthogonally labeled bacteria were also analyzed by confocal microscopy using MFNP-Tz (Figure 5A). CV-TCO stained bacteria showed uniform and high fluorescence signals in the bacterial cell wall, while the control experiments without CV-TCO showed no signal (Figure S7). Similarly, transmission electron microscopy was performed in CV-TCO treated bacteria but which were incubated with tetrazine modified gold nanoparticles. Gold nanoparticles were used instead of magnetic nanoparticles to obtain higher contrast. Gold nanoparticles were found distributed throughout the bacterial surface treated with CV-TCO, while bacteria without CV-TCO labeling showed a smooth surface devoid of nanoparticles (Figure 5B).
Figure 5.
A) Fluorescence confocal microscopy of S. aureus stained with Crystal violet-TCO and labeled with MFNP-Tz. Left, middle, and right shows images of red channel, green channel, and merged from red and green, respectively (Red: propidium iodide for nuclear staining; Green: MNFP-Tz staining; Scale bar = 10 µm). B) Transmission electron microscopy of S. aureus stained with Crystal violet-TCO and labeled with GNP-Tz (left), GNP-Tz alone (middle), and without any treatment (right) (Scale bar = 100 nm).
By modifying the above procedure, the detection strategy can be applied to detect both Gram-positive and Gram-negative bacteria. Performing the staining without the decolorization process would result in labeling both Gram-positive and negative species, since the Gram-negative species would also retain the CV-TCO (Figure S8A). This is in analogy to the conventional Grams stain where the first staining step “colors” all bacteria and the second decolorization step allows differentiation between the two Gram classes. µNMR measurements showed that before decolorization, both Gram-positive and negative bacteria could be magnetically labeled and detected, while after decolorization, only Gram-positive species retained their signals (Figure S8B). Through these sequential measurements, it is thus possible to obtain total bacterial counts (i.e. detection before decolorization) as well as their Gram-negative and Gram-positive composition (i.e. detection after decolorization).
In summary, we show that an orthogonal CV can be used to detect and broadly classify bacteria in biological samples. Staining bacteria with CV-TCO using the standard Gram stain procedure, followed by labeling with MFNP-Tz allows the detection and characterization of bacteria both by µNMR as well as by optical imaging. The “magnetic Gram stain” could be potentially implemented into automated point-of-care diagnostics, bacterial enrichment for subsequent analysis, as well as into therapeutic applications that utilize the antibacterial, antifungal, and antihelminthic properties of CV. The method could also be used to label bacteria in vivo for various imaging applications.[9] Moreover, the staining strategy presented could be further extended to other small molecule affinity ligands (e.g., bioorthogonal carbol fuchsin or trehalose for Mycobacterial species) to enable either universal or specific detection of other bacterial targets. This ability will not only facilitate the clinical diagnosis of a range of bacterial infections but will also promote advances in basic microbiological research.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH) grant number P50CA086355. We thank Yoshi Iwamoto, B.S. and Alex Zaltsman for image processing.
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Ghyslain Budin, Center for Systems Biology, Massachusetts General Hospital, 185 Cambridge Street, Boston, MA 02114 (USA).
Hyun Jung Chung, Center for Systems Biology, Massachusetts General Hospital, 185 Cambridge Street, Boston, MA 02114 (USA).
Hakho Lee, Center for Systems Biology, Massachusetts General Hospital, 185 Cambridge Street, Boston, MA 02114 (USA).
Ralph Weissleder, Email: rweissleder@mgh.harvard.edu, Center for Systems Biology, Massachusetts General Hospital, 185 Cambridge Street, Boston, MA 02114 (USA); Department of Systems Biology, Harvard Medical School, 200 Longwood Avenue, Boston, MA 02115, (USA).
References
- 1.Van Heijenoort J. Glycobiology. 2001;11:25R–36R. doi: 10.1093/glycob/11.3.25r. [DOI] [PubMed] [Google Scholar]
- 2.Coico R. Current Protocols in Immunology. 2001;23:A.3O.1–A.3O.2. doi: 10.1002/0471142735.ima03os23. [DOI] [PubMed] [Google Scholar]
- 3.Beveridge TJ. Biotech. Histochem. 2001;76:111–118. [PubMed] [Google Scholar]
- 4.Bartholomew JW, Mittwer T. Bacteriol Rev. 1952;16:1–29. doi: 10.1128/br.16.1.1-29.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bottone EJ. Lab. Med. 1988;19:288–291. [Google Scholar]
- 6.Issadore D, Shao H, Chung J, Newton A, Pittet M, Weissleder R, Lee H. Lab Chip. 2011;11:147–151. doi: 10.1039/c0lc00149j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung HJ, Reiner T, Budin G, Liong M, Issadore D, Lee H, Weissleder R. ACS Nano. 2011;5:8834–8841. doi: 10.1021/nn2029692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liong M, Fernandez-Suarez M, Issadore D, Min C, Tassa C, Reiner T, Fortune SM, Toner M, Lee H, Weissleder R. Bioconjug. Chem. 2011;22:2390–2394. doi: 10.1021/bc200490r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panizzi P, Nahrendorf M, Figueiredo J-L, Panizzi J, Marinelli B, Iwamoto Y, Keliher E, Maddur AA, Waterman P, Kroh HK, Leuschner F, Aikawa E, Swirski FK, Pittet MJ, Hackeng TM, Fuentes-Prior P, Schneewind O, Bock PE, Weissleder R. Nat Med. 2011;17:1142–1146. doi: 10.1038/nm.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Castro M, Im H, Yao X, Oh S-H, Hu W-S, Wang J-P. J. Med. Devices. 2008;2:27529. [Google Scholar]
- 11.Lönnbro P, Nordenfelt P, Tapper H. BMC Cell Biology. 2008;9:35. doi: 10.1186/1471-2121-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang K, Jenkins DM, Su WW. Journal of Microbiological Methods. 2011;86:69–77. doi: 10.1016/j.mimet.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 13.Guillebault D, Laghdass M, Catala P, Obernosterer I, Lebaron P. Appl. Environ. Microbiol. 2010;76:7352. doi: 10.1128/AEM.00621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Yang M, Ozkan M, Ozkan CS. Biotechnol. Prog. 2009;25:923. doi: 10.1002/btpr.215. [DOI] [PubMed] [Google Scholar]
- 15.Chaves RC, Bensimon D, Freitas PP. J. of Applied Physics. 2011;109:064702. [Google Scholar]
- 16.Guzmán-Lucero D, Guzmán J, Likhatchev D, Martínez-Palou R. Tetrahedron Letters. 2005;46:1119–22. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.