Abstract
Despite years of appreciating the potential role of environment to influence the pathogenesis of type 1 diabetes, specific agents or mechanisms serving in such a capacity remain ill defined. This is exceedingly disappointing as the identification of factors capable of modulating the disease, either as triggers or regulators of the autoimmune response underlying type 1 diabetes, would not only provide clues as to why the disorder develops but, in addition, afford opportunities for improved biomarkers of disease activity and the potential to design novel therapeutics capable of disease abatement. Recent improvements in sequencing technologies, combined with increasing appreciation of the role of innate and mucosal immunity in human disease, have stirred strong interest in what is commonly referred to as the ‘gut microbiota’. The gut (or intestinal) microbiota is an exceedingly complex microenvironment that is intimately linked with the immune system, including the regulation of immune responses. After evaluating evidence supporting a role for environment in type 1 diabetes, this review will convey current notions for contributions of the gut microbiota to human health and disease, including information gleaned from studies of humans and animal models for this autoimmune disorder.
Keywords: Bacteria, Environment, Gut microbiome, Immunology, Innate immunity, Microbiota, Mucosal immunity, NOD mice, Review, Type 1 diabetes
Introduction
Type 1 diabetes is a disorder resulting from the autoimmune destruction of pancreatic beta cells in genetically predisposed individuals [1, 2]. According to current thought, environmental factors contribute to the formation of disease through mechanisms that either trigger the initial autoimmune response in genetically susceptible individuals, or modify the destructive processes at various points throughout the natural history of the disease [3, 4]. A variety of environmental factors have been posited as potential candidates serving in this role, including (but not limited to) viral infections, childhood vaccines, early consumption of cereal or cows’ milk proteins and lack of breast feeding [5–7]. Yet, at present, no agent or lifestyle practice has conclusively been implicated as causative for this disease. Despite this void, the potential influence of environment on the development of this disorder finds support through a variety of observations. For example, epidemiological studies have noted a pronounced increase in the frequency of type 1 diabetes over the last half-century [8, 9], as well as the ‘North–South gradient’, a notion used to explain differences in the geographical incidence of this disease [10]. Furthermore, over the last 50 years, there has been a shift in the degree of genetic susceptibility required to develop the disorder, in that type 1 diabetes is increasingly occurring in populations whose genotype would not previously have been considered high risk [11, 12]. These observations also highlight another important point: that type 1 diabetes is likely a collection of different diseases united by a common outcome (destruction of beta cells and increased blood glucose levels) but differing in their pathogenesis. Hence, distinct forms of type 1 diabetes may be influenced by environment to different degrees.
Even in the absence of a specific agent noted to influence the development of this disorder, the type 1 diabetes research field has been extremely active in terms of developing hypothetical models to explain how its pathogenesis is linked to environmental agents [13–18]. These efforts include the ‘hygiene hypothesis’, which attributes the rising incidence of autoimmune disorders to the reduced or altered (because of the increased overall hygiene in a given society) stimulation of the immune system by undefined environmental factors [13, 14]. Somewhat related, the ‘fertile field hypothesis’ proposes that microbial infection induces a physiological state in which other antigens (including self-antigens) are more easily recognised by the immune system, the result allowing for the generation of self-reactive T cells [15]. Another hypothesis that not only recognises a role for microbes but, in addition, implicates the importance of the gut is the ‘old friends’ hypothesis. Here, gastrointestinal microbes introduced through dietary exposure are noted for their ability to serve as direct inducers/regulators of the immune system through alterations of the gut microbiota [16]. The ‘perfect storm’ hypothesis considers a role for three components in type 1 diabetes development: the gut microbiota (i.e. a constituent influenced by environment), genetic abnormalities in the regulation of mucosal immunity and increased gut leakiness [17]. Finally, and most recently, the ‘threshold hypothesis’ has been proposed as a model for type 1 diabetes capable of simultaneously considering the contributions of genetic and environmental determinants as quantifiable variables that predict risk for this disease [18].
In sum, in recent years, an increasing number of models for type 1 diabetes have considered the potential role for environment, including microbes, with particular emphasis of those that reside in the gut, as an important factor in the pathogenesis of this disease. In this review, our goal is to not only update the reader as to current opinions on the role for the gut microbiota in normal physiology but, in addition, to consider efforts in humans and rodent models of type 1 diabetes for the purpose of defining a microbial role in the disease.
Assessing the gut microbiota
The term ‘gut microbiota’ represents a complex microbial community within the body, one capable of affecting health by contributing to nutrition, prevention of colonisation of the host by pathogens, and through influencing the development and maintenance of the immune system (Fig. 1) [19]. To this end, interest in how the intestinal microbes may contribute to autoimmunity has garnered increasing attention. Experimental evidence suggests that alterations in the gut microbiota are associated with the development of a number of disorders attributed to an overly activated immune system/autoimmunity (e.g. ulcerative colitis, Crohn’s disease), including an influence on type 1 diabetes [20]. Such efforts are important as the identification of factors that influence the immune responses underlying the pathogenesis of type 1 diabetes could provide an opportunity for therapeutic measures to prevent and/or treat the disease, as well as allow for the development of improved biomarkers for predicating future cases of the disorder.
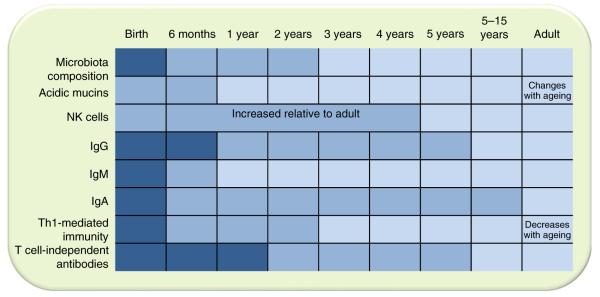
Fig. 1.
Development of the human immune system in relation to microbiota composition. Newborns have a limited capacity to initiate immune responses; both innate and adaptive immune responses are compromised. The kinetics of the maturation of the immune system varies for the different components. Dark blue, immature; medium blue, developing; light blue, adult levels. NK, natural killer. Modified from [89] with permission
Studies of mammalian (including human) commensal microbiota are complicated by the enormous variability of bacterial and other microbial lineages colonising the healthy hosts. Inability to culture the majority of these microorganisms [21] indicates that they underwent a long evolutionary process that made them dependent on the other members of the community and on the host for essential metabolites. On the other hand, the problem led to development of new means of assessing the microbial communities—high-throughput sequences and associated methods of statistical analysis. Much like the space programme of the 1960s has been credited with influencing a wide range of products that feature in our daily lives (e.g. microwave ovens, rehydrated beverages), so too has the Human Genome Project influenced studies of the gut microbiome—a combined sequence of microbial DNA. Indeed, improvements in our ability to accurately and rapidly sequence large spans of genomes (regardless of whether the source is human, viral or bacterial) has had a dramatic impact on the field of microbiome research; allowing the identification of not only the presence, but the relative representation of specific microbial genera and species within given samples. One common approach is the analysis of 16S rRNA gene sequences (16S) from bacteria (Fig. 2) [22]. By this method, many phylotypes have been classified within the human gut microbiome [23]. While hundreds of specific genera and species of bacteria can be identified in the human gut by analysis of stool samples, in humans, the majority of populations comprise bacteria from four phyla, namely, Actinobacteria, Bacteroidetes, Firmicutes and Proteobacteria [24]. Whereas 16S sequencing has proven itself extremely useful in terms of typing and determination of the number of phyla; newer methods, especially that of metagenomic sequencing, have provided an increasingly powerful method to not only identify the diversity of the microbiome but, in addition, provide clues as to its function [25]. Metagenomic analysis is based on sequencing the entire bacterial genome and subsequent assembly of short sequences into contigs representing genes. These genes can be ascribed a particularfunction based on homology with known microbial genes. Whole genome sequencing can also provide more information on microbial diversity compared with 16S analysis, because protein-encoding genes are less conserved at the nucleotide level compared with genes encoding ribosomal RNA. The results of metagenomic analysis can also provide some information on the representation of particular microbes (more identical sequences will be found for an expanded lineage). Metagenomic testing has identified microbial sequences involved in a vast array of physiological processes (e.g. metabolism of amino acid, vitamins) [23]. The results of such efforts have been quite dramatic in terms of their potential impact, as metagenomic analysis suggests that there are >100-fold more genes within the gut microbiome compared with the human genome [26]. The limitation of this approach, however, is that we can only assume that the highly represented sequences, and metabolic pathways that they reveal, are meaningful. An alternative approach is high-throughput sequencing of RNA (metatranscriptomic analysis). This method promises to become a high-resolution method for understanding of real physiological processes taking place in the gut under varying conditions. This approach is not being widely used yet since it requires the removal of ribosomal RNA species that represent the majority of microbial RNA. Nevertheless, it promises to become an important alternative to DNA sequencing.
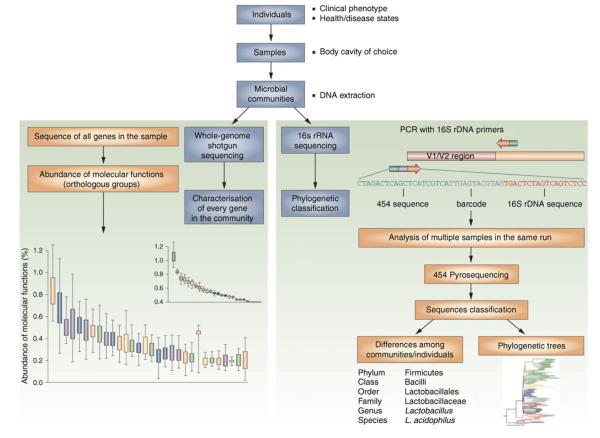
Fig. 2.
Culture-independent genomic analysis of the human microbiome. Culture-independent techniques have advanced our capacity to survey complex microbial communities in human samples. Two sequencing approaches are utilised. As shown on the right of the figure, conserved and variable 16S rRNA genomic regions are amplified and subjected to pyrosequencing. The resulting sequences are then aligned, filtered and compared with publicly available databases of 16S rRNA sequences, enabling taxonomic classification of the bacteria present or absent in a given sample. Whole genome shotgun sequencing (on the left-hand side) provides information that enables identification of genes present and allows for subsequent comparison of enzymatic pathways and functions represented among different samples. Enzymatic databases are also available to assist in the identification of protein function, describing the richness and diversity of functional capacities provided by the analysed microbiome. Adapted by permission from Macmillan Publishers Ltd: Nat Rev Rheumatol [90], copyright 2011
At present, high-throughput DNA sequencing is constantly producing new provocative results that require further investigation of their biological significance. For example, large co-operative efforts and analysis of individuals from diverse geographical regions, led to the development of the term ‘enterotype’. The three enterotypes, which are defined by the predominance of particular phyla in their microbial communities, may, presumably, respond differently to environmental stimuli [27]. However, the dynamic stability of enterotypes has not yet been established. It is worth noting that, although improvements in 16S and metagenomic sequencing (see Fig. 2) have rapidly occurred, a ‘bottleneck’ has also developed. This is related to the means by which these vast quantities of sequence data are subject to analysis [28]; a variety of new statistical methods have been developed for this purpose [29].
Finally, owing to a variety of obvious limitations (e.g. anatomical, means of collection), many gut microbiome-based studies involve sequence analysis of stool samples, subject to a potentially major limitation that the end-product (i.e. stool) does not reflect the microbial–host interactions present within regions of the gut. Indeed, studies of patients with inflammatory bowel disease suggest that sampling from various regions within the intestine can provide dramatically variant results [30]. Nevertheless, at least one effort has suggested that stool samples, even if subjected to various times and temperatures prior to storage/analysis, represent a relatively stable population of microbial signatures [31]. Beyond this, the limitation that many gut microorganisms cannot be cultured using current techniques represents an important hurdle which, left unaddressed, may limit our ability to understand the role of gut microflora in this or other disorders. Hence, this important technological gap should be addressed in future efforts, through the identification of novel isolation and culture techniques.
Mechanisms whereby the gut microbiota influences health
The mutualistic relationship between animals and their commensal microbiota are now well appreciated. Although this was evident to Louis Pasteur, who suggested that microbe-free animals be made, to prove that they cannot survive without their commensal bacteria [32], the true impact of the microbiota on human health only became amenable to thorough investigation relatively recently. As noted previously, the advent of high-throughput sequencing of microbialgenomes, combined with the development of new methods of cultivation of the commensal microbes, when taken together with insights from germ-free (GF) animals, have dramatically changed our ability to address questions related to the gut microbiota.
Pasteur’s prediction was correct for the vertebrates: with-out proper nutritional supplementation, GF animals die. Primitive animals (e.g. Hydra) survive, but show signs of malformation (not obvious in GF mammals) [33]. These findings provide two crucial points for consideration. First, the symbiosis between hosts and gut microbiota is evolutionary conserved. This evolutionary adaptation is mutual— most of the members of mammalian gut microbiota are not currently culturable and likely need other microbial or host factors to thrive. Second, there are likely points of physiological impact that we are still not aware of. Although GF animals under proper nutrition are viable and develop normally, they have multiple distinctions from specific-pathogen-free animals in terms of the development of their immune system [34]. Most of these anomalies disappear upon re-colonisation with commensal bacteria, but some irreversible imprinting of the host immunity may occur. In addition to influencing immune functions, microbes affect other activities including the perception of pain and animal behaviour [35]. One of the major effects that gut microbiota have on the host’s health is that of colonisation resistance—the ability to block a pathogen from creating a foothold within the local gut environment. This property is likely the first line of defence before the immune system reacts and elicits an anti-pathogen response associated with damage to self. Microbiota not only prevent damage caused by pathogens and anti-pathogen immunity, but also directly influence local tissue repair mechanisms (i.e. a homeostatic function) [36].
The influence of the gut microbiota on autoimmunity
Since microbiota influence the immune system through their ability to affect immune responses to pathogens and commensals, it is likely that autoimmunity would (directly or indirectly) be affected as well. Indeed, the very basic questions of whether and how the gut microbiota affects auto-immunity were addressed by experiments using GF (axenic, completely sterile) and gnotobiotic (populated with known microbes) animals [37, 38]. These experiments have been instrumental in terms of revealing four key facts regarding the microbiota: (1) some autoimmune diseases, including type 1 diabetes, can develop in the absence of commensal bacteria, whereas others (e.g. arthritis in IL-1-receptor antagonist-deficient mice) are dependent on the presence of commensal bacteria [39]; (2) the severity of some auto-immune lesions depends on the presence of commensal bacteria [40]; (3) sensitivity to the influence of the microbiota can be linked to specific bacterial lineages [41]; and (4) the intestinal microbiota can influence autoimmune responses in tissues distant from the gut [40]. Thus, autoimmune diseases could even be classified on the basis of their dependence on microbiota, suggesting new insights into the pathogenesis of these maladies. Beyond this, it also became clear that, even in the diseases that do not require microbiota for their development, the microbiota can play the role of a regulator/facilitator [37].
With the importance of the gut microbiota for autoimmunity established, the mechanisms by which microbes specifically influence the pathogenesis of such disorders has became the focus of much in the way of immunological research. Given the complexity of the microbiota (both in numbers and variation) and the current lack of in vitro cultivation capability for the majority of the microbes, much attention is given to specific microbial lineages that seem to have unique properties. For example, Bacteroidetes has been shown to reduce intestinal inflammation [42]. On the other hand, segmented filamentous bacteria (SFB) were reported to have a unique ability to induce a special form of immune activity known as a T helper 17 (Th17) response [43, 44]. Moreover, Th17 cells were reported to affect autoimmune responses in organs both proximal [45], as well as distant [46], from the gut.
Why such findings are important resides in our knowledge of Th17 immunity. Upon encountering an antigen, T cells bearing CD4 molecules proliferate and become functionally polarised. During this process, specific cytokines are produced that signal T cells to become more specialised. What results is the production of T cell subsets, such as Th1, Th2, Th17, amongst others. Th17 lymphocytes are noted for their ability to produce IL-17. The production of this cytokine stimulates stromal cells to produce a number of inflammatory cytokines (e.g. IL-6, IL-8). Through promotion of inflammation and attraction of neutrophils, the primary function of Th17 cells appears to be the clearance of extra-cellular pathogens during infections. It is possible, however, that through triggering an excessive inflammatory response, Th17 cells could contribute to autoimmunity. From what is known about Th17 immunity and the observations on the role of bacteria in their formation, it is likely that many other lineages can substitute for these essential functions. SFB are not normally found in humans, but humans do sustain Th17 responses in the gut. Moreover, a relatively random consortium of mouse intestinal bacteria, termed ‘altered Schaedler flora’ (ASF), are also capable of eliciting Th17 responses in the gut of GF mice populated with such bacteria [46]. One of the more interesting asides of this report was that Th17 induction was dependent on the genetic background of the host.
Thus, understanding the importance of specific lineages having particular effects vs more general influences of microbiota that can be attributed to multiple lineages, is paramount to understanding the complex interactions between hosts and their commensal microbes in the regulation of immunity and autoimmunity. As noted previously, the host’s genetics is also a major player in these interactions. Whereas these considerations are applicable to the studies of the role of gut microbiota in autoimmunity in general, the following section addresses the importance of host–microbial interactions in type 1 diabetes.
Lessons from animal models of type 1 diabetes
While our understanding of the role of the gut microbiota in health and autoimmune disease has improved, its role in type 1 diabetes remains relatively unexplored. What evidence does exist has been indirect and largely derived from two rodent models of the disease: the NOD mouse and the BioBreeding diabetes-prone (BB-DP) rat [47–66]. In both models, GF animals acquired the disease at high rates, indicating that the microbes are entirely dispensable for type 1 diabetes development. Yet, microbiota can profoundly modify the incidence of the disease in susceptible animals. At the same time, some studies found that feeding antibiotics to either NOD mice or BB-DP rats can prevent disease development [48, 49]. The simplest explanation for these results is that antibiotic treatments have selected microbial lineages capable of preventing autoimmune diabetes. In another study, accidental contamination of GF NOD mice with a spore-forming bacterium reduced the rate of type 1 diabetes [50]. Thus, indirect evidence supports the idea that some bacterial lineages can be protective against type 1 diabetes. Bacterial preparations such as complete Freund’s adjuvant, which contains killed Mycobacterium, or streptococcal preparations, protect NOD mice from diabetes [51–53]. Feeding so-called ‘probiotic’ bacterial strains (e.g. lactic acid bacteria) to NOD mice or BB-DP rats can prevent or delay disease development [54, 55]. Moreover, conventionally raised NOD mice lacking an adaptor protein for multiple Toll-like receptors known to bind to bacterial ligands fail to develop diabetes, indicating that interactions between the intestinal microbiota and the innate immune system are critical for disease development [56].
Microbial influences can be indirect and linked to dietary changes. Dietary modifications (e.g. use of casein hydrolysate, avoidance of wheat) prevent or reduce the incidence of diabetes in these animal models [57–60]. However, evidence that this is linked to the composition of the intestinal microflora is lacking in most instances. Beyond this, the propensity for a leaky gut, described earlier as part of the ‘perfect storm’ hypothesis, has been observed in pre-diabetic NOD mice infected with the enteric bacterial pathogen Citrobacter rodentium, correlating with specific acceleration of insulitis [61]. Evidence for increased intestinal permeability also finds support in BB rats [62, 63].
Studies (culture independent) of the gut microbiome in BB-DP and BB diabetes-resistant (BB-DR) rats indicated that, at disease onset, bacterial communities were quite different [64]. Specifically, stool samples from BB-DR rats contained much higher proportions of the so-called probiotic-like bacteria, such as Lactobacillus and Bifidobacterium, whereas BB-DP rats had higher numbers of Bacteroides, Eubacterium and Ruminococcus. Quite strikingly, one strain, Lactobacillus johnsonii, noted for its increased presence in stools from BB-DR rats (relative to BB-DP rats) prevents the development of diabetes when administered (post-weaning) to BB-DP rats [65]. Recent efforts using this same strain of L. johnsonii, but in NOD mice, revealed its capacity to modulate Th17 immunity [66]. These findings, taken together with the aforementioned studies suggesting that dietary alterations including the introduction of probiotics, as well as antibiotic feeding, support the notion that pro-pathogenic bacteria are either diminished, or beneficial bacteria enhanced, through these actions.
These approaches are not free of caveats, one being the inability of probiotics to stably colonise the gut, requiring repetitive administration through oral gavage, a procedure that is hardly innocuous to the host and may result in systemic delivery of bacteria. The results of colonisation of GF type 1 diabetes-prone animals with different bacteria should shed light on their anti-diabetic properties.
Lessons from humans with type 1 diabetes
For a variety of reasons (e.g. the need to study large numbers of individuals, complex data analysis, potential disease heterogeneity), the studies of microbiota in humans are extremely difficult to perform. On average, humans carry 1014 bacteria that colonise not only the large intestine (the largest depot by number) but in addition, other mucosal surfaces (e.g. oral cavity, upper airways, small intestine) [21]. While individuals are born with epithelial surfaces free of microbes, the process of colonisation begins within hours of birth [67]. This event represents quite a dynamic process within the first years of life but, in the end, eventually results in the establishment of the adult microbiota [68, 69]. The neonatal period is subject to a variety of environmental influences (e.g. diet, fever, antibiotic use), and even at its earliest stages is influenced by the mode of child delivery (i.e. vaginal vs Caesarean), as well as the source of early nutrition (i.e. breast vs formula feeding) [70, 71]. Interestingly, neonates subject to vaginal delivery develop a microbiome that is reflective of the vaginal flora while those subject to Caesarean delivery see microbial domination by genera normally reflective of skin. This variance leads to delayed microbial colonisation by Bacteroides, Bifidobacterium and Lactobacillus. In terms of importance for type 1 diabetes, the disorder has been noted to occur more frequently in offspring born by Caesarean delivery [72, 73] (Fig. 3). While data describing the natural history of microbiota development in neonates, children, as well as in adults, remain somewhat limited, there is evidence to suggest that the microbiota in adults (unlike neonates) appears to represent some-what of a relatively stable community, with far more limited variance throughout adult life [22]. Clearly, additional studies are required to coalesce these still unrelated concepts/observations into a unified understanding of their role (if any) in the pathogenesis of type 1 diabetes. Because of the more intimate (and obvious) potential link between the gut microbiota and diseases of the intestine, namely, inflammatory bowel disease, the majority of investigations within the gut microbiome field have occurred in patients afflicted by those disorders [74]. The results have been quite interesting if not striking. Studies of the gut microbiota in patients with type 1 diabetes are extremely limited compared with those of human inflammatory bowel disease or in animal models of the disease. However, certain physiological features of the intestine, such as ‘leakiness’ show similarities in human type 1 diabetes and animal models of the disease [75–78]. Interestingly, this property (both in animal models and in humans at increased risk for the disease) occurs prior to the onset of overt disease (i.e. hyperglycaemia), consistent with, but not proving, the notion that such a defect could be a component within the natural history of this disease. Electron microscopy studies have noted that structural changes within the intestine of patients with type 1 diabetes are present, as reflected by alterations in tight junctions as well as in microvilli [79]. Levels of zonulin, a molecule resident in tight junctions are upregulated in patients with type 1 diabetes, and this upregulation is associated with increased intestinal permeability [80]. In addition to increased intestinal permeability, patients with type 1 diabetes display a heightened number of intestinal inflammatory cells [81, 82] and a reduced number of, FoxP3+CD4+CD25+ T cells, a presumed master regulator of the immune system [83]. In addition, it is possible that altered mucosal immunity (noted for its association with type 1 diabetes), as well as mucosal pathogens, might directly influence the microbiota and, as a result, influence the pathogenesis of this disease [17]. It needs to be emphasised that these observations are still no more than correlations, and that it is not clear whether they are pre-existent or induced by the disease. However, they provide good ideas for future experiments.
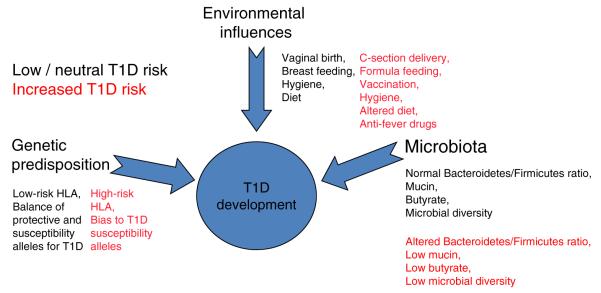
Fig. 3.
A hypothetical model for how microbiota, based on genetic and environmental influences, may contribute to type 1 diabetes. In this model, the end result is the autoimmune destruction of pancreatic beta cells. However, for this to occur, there must be significant variations from the normal setting of development (i.e. healthy microbiota). Here, molecules produced by a given microbiota network, in response to a combination of environmental exposures and genetic susceptibility, influence proneness to type 1 diabetes. In a healthy microbiome, there is an optimal proportion of organisms, which provide signals to the developing immune system (controlled by genetic susceptibility and environmental events) that lead to a balance in immune regulatory activities that avoid, or provide susceptibility to, type 1 diabetes. At present, the list of potential means by which environment can influence microbiota development is large; with a variety of putative (i.e. unproven but previously noted) candidates listed. T1D, type 1 diabetes
In terms of attempts to use 16S or metagenomic analysis to identify a potential role for the microbiome in human type 1 diabetes, one initial reported effort identified specific phyla that not only differed between Finnish individuals with type 1 diabetes in comparison with case-matched (non-diabetic) healthy controls but, in addition and of most interest, longitudinal analysis of those who developed type 1 diabetes demonstrated an increased percentage of Bacteroidetes alongside a lower proportion of Firmicutes [84] (Fig. 3). What made these observations of special interest is that the ratio of the two phyla differed (i.e. the relative levels were inverse to each other) with increasing age. Beyond this, there was variation in the diversity of microbiota in those destined to develop type 1 diabetes relative to controls (i.e. colonisation with far fewer phyla were observed in those who developed disease). More recent studies from this same group switched techniques from 16S to metagenomic analysis, efforts that represent movement from a species-based approach towards one of function [85].
It is presently unknown whether the observed correlations have any causative role or result from disease development itself [86]. Longitudinal studies of the dynamics of the microbiota in large cohorts of potentially sensitive volunteers, as well as simultaneous examination of immune responses (including mucosal immunity), should shed light on this intriguing question and provide important guidance to investigations seeking to utilise microbes or microbial products to attenuate type 1 diabetes [87].
Conclusions
An increasing body of evidence supports the notion that the gut microbiota may influence the pathogenesis of type 1 diabetes. This said, much in the way of additional studies is required to progress this from an interesting hypothesis to one with undeniable support. To meet this goal, future investigations—both in humans and in animal models of the disease—must address a series of key voids in our knowledge. For example, are there microbiome ‘signatures’ for disease progression, by what immune mediated mechanisms does the microbiota influence type 1 diabetes, how does genetic susceptibility for the disorder influence microbiota formation, and is their validity for the aforementioned hypotheses tying the microbiome to the development of this disease? In humans, additional studies across a broad range of geographically different populations that vary in their risk for type 1 diabetes are certainly required. In addition, for individuals studied as part of such investigations, the quality of data/data interpretation would certainly be improved by increased sampling (i.e. more and more often), as well as analyses in which results are interpreted in combination with a series of additional measures (e.g. antibiotic use, means of delivery at birth, time of first fever). Consortium-based efforts such as the US National Institutes of Health programme the Environmental Determinants of Diabetes in the Young (TEDDY), as well as large independent trials (e.g. BABYDIAB in Germany, Diabetes Prediction and Prevention [DIPP] in Finland, Trial to Reduce IDDM in the Genetically at Risk [TRIGR]) should be beneficial to this cause [reviewed in 88]. Data from these studies could, in theory, be beneficial to the design of a prevention-based probiotic approach to therapy in which microbiota beneficial to disease attenuation could be introduced/expanded, or those capable of promoting disease development could be diminished.
Acknowledgements
The authors wish to thank their colleagues for their collective insights and microbiota based research efforts whose concepts were considered in this review, including Ezio Bonifacio, Michael Burrows, Joseph Larkin III, Graciela Lorca, Josef Neu, Joseph Pickard, Desmond Schatz, Olli Simell, Eric Triplett, Clive Wasserfall, Leonid Yurkovetsky and Anette Ziegler.
Funding This work was supported, in part, by funding obtained from The National Institutes of Health, The Juvenile Diabetes Research Foundation, The Brehm Coalition for Type 1 Diabetes Research, and the Jeffrey Keene Family Professorship.
Abbreviations
- ASF
Altered Schaedler flora
- BB
BioBreeding
- GF
Germ free
- SBF
Segmented filamentous bacteria
- Th17
T helper 17
Footnotes
Duality of interest The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement The authors were responsible for the conception, design and drafting of the manuscript, and approved the final version for publication.
Contributor Information
M. A. Atkinson, Department of Pathology, University of Florida, College of Medicine, 1600 SW Archer Road, Gainesville, FL 32610-0275, USA
A. Chervonsky, A. Chervonsky Department of Pathology, University of Chicago, College of Medicine, Chicago, IL, USA
References
- 1.Atkinson MA, Eisenbarth GS. Type 1 diabetes; new perspectives on disease pathogenesis and treatment. Lancet. 2001;358:221–229. doi: 10.1016/S0140-6736(01)05415-0. [DOI] [PubMed] [Google Scholar]
- 2.Bluestone JA, Herold K, Eisenbarth G. Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature. 2010;464:1293–1300. doi: 10.1038/nature08933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trucco M. Gene–environment interaction in type 1 diabetes mellitus. Endocrinol Nutr. 2009;56:56–59. doi: 10.1016/S1575-0922(09)73521-1. [DOI] [PubMed] [Google Scholar]
- 4.Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32:457–467. doi: 10.1016/j.immuni.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Mathieu C, Gysemans C, Giulietti A, Bouillon R. Vitamin D and diabetes. Diabetologia. 2005;48:1247–1257. doi: 10.1007/s00125-005-1802-7. [DOI] [PubMed] [Google Scholar]
- 6.Knip M, Virtanen SM, Becker D, Dupré J, Krischer JP, Åkerblom HK. Early feeding and risk of type 1 diabetes: experiences from the trial to reduce insulin-dependent diabetes mellitus in the genetically at risk (TRIGR) Am J Clin Nutr. 2011;94(Suppl):1814S–1820S. doi: 10.3945/ajcn.110.000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coppieters KT, Boettler T, von Herrath M. Virus infections in type 1 diabetes. Cold Spring Harb Perspect Med. 2012;2:a007682. doi: 10.1101/cshperspect.a007682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gale EA. Type 1 diabetes in the young: the harvest of sorrow goes on. Diabetologia. 2005;48:1435–1438. doi: 10.1007/s00125-005-1833-0. [DOI] [PubMed] [Google Scholar]
- 9.Soltesz G, Patterson CC, Dahlquist G, EURODIAB Study Group Worldwide childhood type 1 diabetes incidence—what can we learn from epidemiology? Pediatr Diabetes. 2007;8:6–14. doi: 10.1111/j.1399-5448.2007.00280.x. [DOI] [PubMed] [Google Scholar]
- 10.Karvonen M, Viik-Kajander M, Moltchanova E, Libman I, La Porte R, Tuomilehto J. Incidence of childhood type 1 diabetes worldwide: Diabetes Mondiale (DiaMond) Project Group. Diabetes Care. 2000;23:1516–1526. doi: 10.2337/diacare.23.10.1516. [DOI] [PubMed] [Google Scholar]
- 11.Gillespie KM, Bain SC, Barnett AH, et al. The rising incidence of childhood type 1 diabetes and reduced contribution of high-risk HLA haplotypes. Lancet. 2004;364:1699–1700. doi: 10.1016/S0140-6736(04)17357-1. [DOI] [PubMed] [Google Scholar]
- 12.Fourlanos S, Varney MD, Morahan G, et al. The rising incidence of type 1 diabetes is accounted for by cases with lower-risk human leukocyte antigen genotypes. Diabetes Care. 2008;31:1546–1549. doi: 10.2337/dc08-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bach JF. Six questions about the hygiene hypothesis. Cell Immunol. 2005;233:158–161. doi: 10.1016/j.cellimm.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 14.Cooke A. Review series on helminths, immune modulation and the hygiene hypothesis: how might infection modulate the onset of type 1 diabetes? Immunology. 2009;126:12–17. doi: 10.1111/j.1365-2567.2008.03009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Herrath MG, Fujinami RS, Whitton JL. Microorganisms and autoimmunity: making the barren field fertile? Nat Rev Microbiol. 2003;1:151–157. doi: 10.1038/nrmicro754. [DOI] [PubMed] [Google Scholar]
- 16.Rook GA, Adams V, Hunt J, et al. Mycobacteria and other environmental organisms as immunomodulators for immunoregulatory disorders. Springer Semin Immunopathol. 2004;25:237–255. doi: 10.1007/s00281-003-0148-9. [DOI] [PubMed] [Google Scholar]
- 17.Vaarala O, Atkinson MA, Neu J. The “perfect storm” for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes. 2008;57:2555–2562. doi: 10.2337/db08-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wasserfall C, Nead K, Mathews C, Atkinson MA. The threshold hypothesis: solving the equation of nurture vs nature in type 1 diabetes. Diabetologia. 2011;54:2232–2236. doi: 10.1007/s00125-011-2244-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boermer BP, Sarvetnick NE. Type 1 diabetes: role of intestinal microbiome in humans and mice. Ann N Y Acad Sci. 2011;1243:103–118. doi: 10.1111/j.1749-6632.2011.06340.x. [DOI] [PubMed] [Google Scholar]
- 21.Ley RE, Lozupone CA, Hamady M, et al. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu GD, Lewis JD, Hoffmann C, et al. Sampling and pyro-sequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010;10:206–210. doi: 10.1186/1471-2180-10-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker A, Tarraga AC, Bentley S. Faecal matters. Nat Rev Microbiol. 2006;4:572–573. doi: 10.1038/nrmicro1483. [DOI] [PubMed] [Google Scholar]
- 27.Arumugam M, Raes J, Pelletier E, et al. Enterotype of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sul WJ, Cole JR, Jesus EDC, et al. Bacterial community comparisons by taxonomy-supervised analysis independent of sequence alignment and clustering. Proc Natl Acad Sci USA. 2011;108:14637–14642. doi: 10.1073/pnas.1111435108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamady M, Knight R. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Momozawa Y, Deffontaine LE, Medrano JF. Characterization of bacteria in biopsies of colon and stools by high throughput sequencing of the V2 region of bacterial 16S rRNA Gene in Human. PLoS One. 2011;6:e16952. doi: 10.1371/journal.pone.0016952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roesch LFW, Casella G, Simell O, Krischer J, et al. Influence of fecal sample storage on bacterial community diversity. Open Microbiol J. 2009;3:40–46. doi: 10.2174/1874285800903010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pasteur L. Observations relatives à la note précedénte de M. Duclaux. C R Acad Sci (Paris) 1885;100:68. [Google Scholar]
- 33.Rahat M, Dimentman C. Cultivation of bacteria-free Hydra viridis: missing budding factor in nonsymbiotic hydra. Science. 1982;216:67–68. doi: 10.1126/science.7063873. [DOI] [PubMed] [Google Scholar]
- 34.Kelly D, King T, Aminov R. Importance of microbial colonization of the gut in early life to the development of immunity. Mutat Res. 2001;622:58–69. doi: 10.1016/j.mrfmmm.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 35.Fagundes CT, Amaral FA, Teixeira AL, Souza DG, Teixeira MM. Adapting to environmental stresses: the role of the microbiota in controlling innate immunity and behavioral responses. Immunol Rev. 2012;245:250–264. doi: 10.1111/j.1600-065X.2011.01077.x. [DOI] [PubMed] [Google Scholar]
- 36.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Chervonsky AV. Influence of microbial environment on autoimmunity. Nat Immunol. 2010;11:28–35. doi: 10.1038/ni.1801. [DOI] [PubMed] [Google Scholar]
- 38.Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat Rev Immunol. 2010;10:735–744. doi: 10.1038/nri2850. [DOI] [PubMed] [Google Scholar]
- 39.Abdollahi-Roodsaz S, Joosten LA, Koenders MI, et al. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J Clin Invest. 2008;118:205–216. doi: 10.1172/JCI32639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu HJ, Ivanov II, Darce J, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinkorova Z, Capkova J, Niederlova J, Stepankova R, Sinkora J. Commensal intestinal bacterial strains trigger ankylosing enthesopathy of the ankle in inbred B10.BR (H-2k) male mice. Hum Immunol. 2008;69:845–850. doi: 10.1016/j.humimm.2008.08.296. [DOI] [PubMed] [Google Scholar]
- 42.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 43.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 45.Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci USA. 2011;108:11548–11553. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geuking MB, Cahenzli J, Lawson MA. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 47.Hoorfar J, Buschard K, Dagnaes-Hansen F. Prophylactic nutritional modification of the incidence of diabetes in autoimmune non-obese diabetic (NOD) mice. Br J Nutr. 1993;69:597–607. doi: 10.1079/bjn19930059. [DOI] [PubMed] [Google Scholar]
- 48.Brugman S, Klatter FA, Visser JT, et al. Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia. 2006;49:2105–2108. doi: 10.1007/s00125-006-0334-0. [DOI] [PubMed] [Google Scholar]
- 49.Schwartz RF, Neu J, Schatz D, Atkinson MA, Wasserfall C. Comment on: Brugman S et al. (2006) Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat. Is the gut flora involved in the development of type-1 diabetes? Diabetologia. 2007;49:2105–2108. doi: 10.1007/s00125-006-0334-0. [DOI] [PubMed] [Google Scholar]
- 50.King C, Sarvetnick N. The incidence of type-1 diabetes in NOD mice is modulated by restricted flora not germ-free conditions. PLoS One. 2011;6:e17049. doi: 10.1371/journal.pone.0017049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Satoh J, Shintani S, Oya K, Tanaka S, et al. Treatment with streptococcal preparation (OK-432) suppresses anti-islet autoimmunity and prevents diabetes in BB rats. Diabetes. 1988;37:1188–1194. doi: 10.2337/diab.37.9.1188. [DOI] [PubMed] [Google Scholar]
- 52.Qin HY, Singh B. BCG vaccination prevents insulin-dependent diabetes mellitus (IDDM) in NOD mice after disease acceleration with cyclophosphamide. J Autoimmun. 1997;10:271–278. doi: 10.1006/jaut.1997.0136. [DOI] [PubMed] [Google Scholar]
- 53.McInerney MF, Pek SB, Thomas DW. Prevention of insulitis and diabetes onset by treatment with complete Freund’s adjuvant in NOD mice. Diabetes. 1991;40:715–725. doi: 10.2337/diab.40.6.715. [DOI] [PubMed] [Google Scholar]
- 54.Calcinaro F, Dionisi S, Marinaro M, et al. Oral probiotic administration induces interleukin-10 production and prevents spontaneous autoimmune diabetes in the non-obese diabetic mouse. Diabetologia. 2005;48:1565–1575. doi: 10.1007/s00125-005-1831-2. [DOI] [PubMed] [Google Scholar]
- 55.Matsuzaki T, Nagata Y, Kado S, et al. Prevention of onset in an insulin-dependent diabetes mellitus model, NOD mice, by oral feeding of Lactobacillus casei. APMIS. 1997;105:643–649. doi: 10.1111/j.1699-0463.1997.tb05066.x. [DOI] [PubMed] [Google Scholar]
- 56.Wen L, Ley RE, Volchkov PY, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmid S, Koczwara K, Schwinghammer S, et al. Delayed exposure to wheat and barley proteins reduces diabetes incidence in non-obese diabetic mice. Clin Immunol. 2004;111:108–118. doi: 10.1016/j.clim.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 58.Hansen AK, Ling F, Kaas A, et al. Diabetes preventive gluten-free diet decreases the number of caecal bacteria in non-obese diabetic mice. Diabetes Metab Res Rev. 2006;22:220–225. doi: 10.1002/dmrr.609. [DOI] [PubMed] [Google Scholar]
- 59.Funda DP, Kaas A, Tlaskalova-Hogenova H, Buschard K. Gluten-free but also gluten-enriched (gluten+) diet prevent diabetes in NOD mice; the gluten enigma in type 1 diabetes. Diabetes Metab Res Rev. 2008;24:59–63. doi: 10.1002/dmrr.748. [DOI] [PubMed] [Google Scholar]
- 60.Maurano F, Mazzarella G, Luongo D, et al. Small intestinal enteropathy in non-obese diabetic mice fed a diet containing wheat. Diabetologia. 2005;48:931–937. doi: 10.1007/s00125-005-1718-2. [DOI] [PubMed] [Google Scholar]
- 61.Lee AS, Gibson DL, Zhang Y, Sham HP, Vallance BA, Dutz JP. Gut barrier disruption by an enteric bacterial pathogen accelerates insulitis in NOD mice. Diabetologia. 2010;53:741–748. doi: 10.1007/s00125-009-1626-y. [DOI] [PubMed] [Google Scholar]
- 62.Neu J, Reverte CM, Mackey AD, et al. Changes in intestinal morphology and permeability in the BioBreeding rat before the onset of type 1 diabetes. J Pediatr Gastroenterol Nutr. 2005;40:589–595. doi: 10.1097/01.mpg.0000159636.19346.c1. [DOI] [PubMed] [Google Scholar]
- 63.Watts T, Berti I, Sapone A, et al. Role of the intestinal tight junction modulator zonulin in the pathogenesis of type I diabetes in BB diabetic-prone rats. Proc Natl Acad Sci USA. 2005;102:2916–2921. doi: 10.1073/pnas.0500178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roesch LF, Lorca GL, Casella G, et al. Culture-independent identification of gut bacteria correlated with the onset of diabetes in a rat model. ISME J. 2009;3:536–548. doi: 10.1038/ismej.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Valladares R, Sankar D, Li N, et al. Lactobacillus johnsonii N6.2 mitigates the development of type 1 diabetes in BB-DP rats. PLoS One. 2010;6:e10507. doi: 10.1371/journal.pone.0010507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lau K, Benitez P, Ardissone A, et al. Inhibition of type 1 diabetes correlated to a Lactobacillus johnsonii N6.2-mediated Th17 bias. J Immunol. 2011;186:3538–3546. doi: 10.4049/jimmunol.1001864. [DOI] [PubMed] [Google Scholar]
- 67.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moore TA, Hanson CK, Anderson-Berry A. Colonization of the gastrointestinal tract in neonates. ICAN: infant, Child Adolesc Nutr. 2011;3:291–295. [Google Scholar]
- 70.Gronlund MM, Lehtonen OP, Eerola E, Kero P. Fecal microflora in healthy infants born by different methods of delivery: permanent changes in intestinal flora after cesarean delivery. J Pediatr Gastroenterol Nutr. 1999;28:19–25. doi: 10.1097/00005176-199901000-00007. [DOI] [PubMed] [Google Scholar]
- 71.Salminen S, Gibson GR, McCartney AL, Isolauri E. Influence of mode of delivery on gut microbiota composition in seven year old children. Gut. 2004;53:1388–1389. doi: 10.1136/gut.2004.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cardwell CR, Stene LC, Joner G, et al. Caesarean section is associated with an increased risk of childhood-onset type 1 diabetes mellitus: a meta-analysis of observational studies. Diabetologia. 2008;51:726–735. doi: 10.1007/s00125-008-0941-z. [DOI] [PubMed] [Google Scholar]
- 73.Vehik K, Dabelea D. Why are C-section deliveries linked to childhood type 1 diabetes? Diabetes. 2012;61:36–37. doi: 10.2337/db11-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 75.Meddings JB, Jarand J, Urbanski SJ, et al. Increased gastrointestinal permeability is an early lesion in the spontaneously diabetic BB rat. Am J Physiol. 1999;276:G951–G957. doi: 10.1152/ajpgi.1999.276.4.G951. [DOI] [PubMed] [Google Scholar]
- 76.Carratu R, Secondulfo M, de Magistris L, et al. Altered intestinal permeability to mannitol in diabetes mellitus type I. J Pediatr Gastroenterol Nutr. 1999;28:264–269. doi: 10.1097/00005176-199903000-00010. [DOI] [PubMed] [Google Scholar]
- 77.Kuitunen M, Saukkonen T, Ilonen J, et al. Intestinal permeability to mannitol and lactulose in children with type 1 diabetes with the HLA-DQB1*02 allele. Autoimmunity. 2002;35:365–368. doi: 10.1080/0891693021000008526. [DOI] [PubMed] [Google Scholar]
- 78.Bosi E, Molteni L, Radaelli MG, et al. Increased intestinal permeability precedes clinical onset of type 1 diabetes. Diabetologia. 2006;49:2824–2827. doi: 10.1007/s00125-006-0465-3. [DOI] [PubMed] [Google Scholar]
- 79.Secondulfo M, Iafusco D, Carratu R, et al. Ultrastructural mucosal alterations and increased intestinal permeability in non-celiac, type I diabetic patients. Dig Liver Dis. 2004;36:35–45. doi: 10.1016/j.dld.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 80.Sapone A, de Magistris L, Pietzak M, et al. Zonulin upregulation is associated with increased gut permeability in subjects with type 1 diabetes and their relatives. Diabetes. 2006;55:1443–1449. doi: 10.2337/db05-1593. [DOI] [PubMed] [Google Scholar]
- 81.Tiittanen M, Westerholm-Ormio M, Verkasalo M, Savilahti E, Vaarala O. Infiltration of forkhead box P3-expressing cells in small intestinal mucosa in coeliac disease but not in type 1 diabetes. Clin Exp Immunol. 2008;152:498–507. doi: 10.1111/j.1365-2249.2008.03662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Badami E, Sorini C, Coccia M, et al. Defective differentiation of regulatory FoxP3+ T cells by small-intestinal dendritic cells in patients with type 1 diabetes. Diabetes. 2011;60:2120–2124. doi: 10.2337/db10-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Giongo A, Gano KA, Crabb DB, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2010;5:82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brown CT, Davis-Richardson AG, Giongo A, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6:e25792–e25801. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vaarala O. The gut as a regulator of early inflammation in type 1 diabetes. Curr Opin Endocrinol Diabetes Obes. 2011;18:241–247. doi: 10.1097/MED.0b013e3283488218. [DOI] [PubMed] [Google Scholar]
- 87.Ljungberg M, Korpela R, Ilonen J, et al. Probiotics for the prevention of beta cell autoimmunity in children at genetic risk of type 1 diabetes—the PRODIA study. Ann NYAcad Sci. 2006;1079:360–364. doi: 10.1196/annals.1375.055. [DOI] [PubMed] [Google Scholar]
- 88.Rewers M, Gottlieb P. Immunotherapy for the prevention and treatment of type 1 diabetes. Diabetes Care. 2009;32:1769–1782. doi: 10.2337/dc09-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martin R, Nauta AJ, Amor KB, Knippels LMJ, Knol J, Garssen J. Early life: gut microbiota and immune development in infancy. Benefic Microbes. 2010;1:367–382. doi: 10.3920/BM2010.0027. [DOI] [PubMed] [Google Scholar]
- 90.Scher JU, Abramson SB. The microbiome and rheumatoid arthritis. Nat Rev Rheumatol. 2011;7:569–578. doi: 10.1038/nrrheum.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]