SUMMARY
Lipid composition can differ widely among organelles and even between leaflets of a membrane. Lipid homeostasis is critical because disequilibrium can have disease outcomes. Despite their importance, mechanisms maintaining lipid homeostasis remain poorly understood. Here, we establish a model system to study the global effects of lipid imbalance. Quantitative lipid profiling was integral to monitor changes to lipid composition and for system validation. Applying global transcriptional and proteomic analyses, a dramatically altered biochemical landscape was revealed from adaptive cells. The resulting composite regulation we term the “membrane stress response” (MSR) confers compensation, not through restoration of lipid composition, but by remodeling the protein homeostasis network. To validate its physiological significance, we analyzed the unfolded protein response (UPR), one facet of the MSR and a key regulator of protein homeostasis. We demonstrate that the UPR maintains protein biogenesis, quality control, and membrane integrity—functions otherwise lethally compromised in lipid dysregulated cells.
INTRODUCTION
Although seemingly uniform in appearance, biological membranes are composed of hundreds of distinct lipids (van Meer et al., 2008). The major groups include phospholipids, sphingolipids, and sterols. The most abundant, phospholipids, are subdivided further according to head group structures (Wenk, 2005). Additionally, phospholipids of the same class can have acyl chains of 14 to 26 carbon atoms and containing 0 to 6 double bonds (Henry et al., 2012; Hermansson et al., 2011). Lipid composition can vary dramatically between organelles and cell types (van Meer et al., 2008). In most cases, the biological significance of these differences remains unclear. Despite gaps in our present understanding, numerous links between disease states and lipid disequilibrium underscores the importance of membrane homeostasis (Sanyal, 2005; Wenk, 2005).
The complex organization of cellular membranes suggests the need for sophisticated homeostatic regulatory mechanisms. Although little is known, links were made with an endoplasmic reticulum (ER) stress pathway called the unfolded protein response (UPR) (Fu et al., 2011; Lee et al., 2008; Oyadomari et al., 2008; Rutkowski et al., 2008). For example, hepatic lipid metabolism in mammals is UPR regulated through Ire1p. High dietary carbohydrates activate the UPR, which controls the expression of genes involved in fatty acid and cholesterol biosynthesis (Lee and Glimcher, 2009). In obese mice, chronic UPR activation leads to suppression of the insulin-signaling pathway, which could contribute to type II diabetes (Ozcan et al., 2004). In budding yeast, a large-scale screen uncovered several lipid metabolic mutants that activate the UPR, suggesting a compensatory role when these pathways are disrupted (Jonikas et al., 2009). Together, these studies indicate an intimate relationship between the UPR and lipid homeostasis.
How the UPR responds to internal cues of lipid disequilibrium is unclear. This is partly due to the paucity of information regarding its role here and inherent limitations of classical lipid analytical methods. Recently, it was shown that the obese mouse liver has a phosphatidylcholine (PC)/phosphatidylethanolamine (PE) ratio that is slightly higher than normal liver. This difference was correlated with a disturbance of calcium homeostasis through the SERCA transporter that leads to ER stress (Fu et al., 2011). This study shows that the UPR can respond to the consequences of lipid disequilibrium but how it alleviates this form of stress remains unanswered.
The importance of the PC/PE ratio to health and disease was already established by earlier studies. In mammals, PC can be synthesized using the CDP-choline pathway or through three PE methylation steps catalyzed by PE N-methyltransferase (PEMT) (Li and Vance, 2008). PEMT-deficient mice (Pemt−/−) develop steatohepatitis and liver failure within three days after switching to choline deficient diets (Walkey et al., 1998). This is caused by a reduction of the PC to PE ratio that leads to loss of membrane integrity in hepatocytes (Li et al., 2006). The mouse model is relevant to human nonalcoholic steatohepatitis (NASH) because patients have similarly reduced PC/PE ratios in the liver. Despite these important advances, the biological effects of PC/PE disequilibrium are not well understood. In particular, the cellular mechanisms used to detect and buffer such homeostatic shocks remain unexplored.
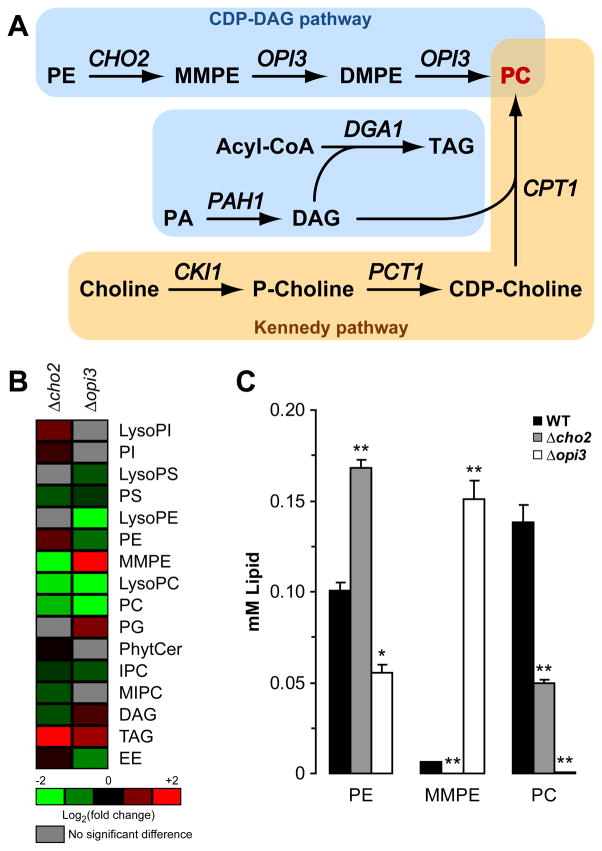
In budding yeast, de novo synthesis of PC is similar to mammals except PE methylation is catalyzed sequentially by two distinct phosphatidyl N-methyltransferases (Figure 1A) (Daum et al., 1999; Moreau and Cassagne, 1994; van Meer et al., 2008; Zinser et al., 1991). First, Cho2p methylates PE to generate N-monomethyl phosphatidylethanolamine (MMPE). Next, Opi3p methylates MMPE to N,N-dimethyl phosphatidylethanolamine (DMPE) in one step and to PC in the second (Kodaki and Yamashita, 1987, 1989). Alternatively, yeast cells can synthesize PC through the Kennedy pathway when choline is available (Figure 1A) (Carman and Henry, 1989). Thus, in principle, a yeast lipid homeostatic model can be established based on the mouse NASH system.
Figure 1. Lack of Cho2p or Opi3p causes severe lipid imbalance.
(A) Metabolic pathways for the synthesis of phosphatidylcholine in S. cerevisiae. PE, phosphatidylethanolamine; MMPE, N-monomethyl phosphatidylethanolamine; DMPE, N,N-dimethyl phosphatidylethanolamine; PC, phosphatidylcholine; PA, phosphatidic acid; DAG, diacylglycerol; TAG, triacylglycerol; P-choline, phosphate-choline. (B) Heat maps reflecting all significant lipid changes to WT. Cells were grown to early log phase at 23°C in selective synthetic complete media. Lipids were extracted with chloroform:methanol mixture from lyophilized cells. Organic extractions were concentrated and analyzed by HPLC-MS. PI, phosphatidylinositol; PS, phosphatidylserine; PG, phosphatidylglycerol; PhytCer, phytoceramide; IPC, inositolphosphoceramide; MIPC, manosylinositolphosphoceramide; EE, ergosterol ester. p < 0.05, Student’s t-test compared to WT. (C) Comparison of total PE, MMPE, and PC as prepared in (B). *p < 0.01, **p < 0.005, Student’s t-test compared to WT.
In this study, classical genetic and biochemical approaches were combined with lipidomic, genomic, and proteomic technologies to understand the maintenance and regulation of membrane homeostasis. Specifically, we focus on how cells respond to and cope with PC/PE disequilibrium. The data reveal a broad response that remodels the proteome to buffer the otherwise lethal effects of lipid disequilibrium. Surprisingly, the major changes reconfigure the protein homeostasis network to regulate protein synthesis, folding, and quality control. Of one facet, we demonstrate that activation of the unfolded protein response is essential to support protein translocation, transport, ER quality control, and membrane integrity. Lipidomic analysis shows that the UPR program compensates for these effects without altering the lipid composition of membranes.
RESULTS
Cho2p or Opi3p deficiency causes severe but non-lethal imbalances in lipid composition
Cho2p and Opi3p are required for synthesis of PC from PE in budding yeast (Figure 1A) (Daum et al., 1999; van Meer et al., 2008; Zinser et al., 1991). Despite these important roles, cells are viable if either of their genes is deleted suggesting alternative means of PC synthesis, or more radically, mechanisms that bypass the need for the phospholipid altogether. Indeed, cho2 and opi3 mutants displayed sharp declines of PC by thin layer chromatography (Daum et al., 1999; McGraw and Henry, 1989).
The robustness of PC deficient cells hints of mechanisms that can buffer sudden and chronic changes to lipid homeostasis. To validate the use of PC biosynthetic mutants to study the effects and response to lipid disequilibrium, quantitative lipid profiles of Δcho2 and Δopi3 strains were determined. Total lipid extracts from logarithmic cells were analyzed using high performance liquid chromatography coupled to mass spectrometry (LC-MS). Consistent with earlier studies, PC is reduced 2.8 fold in Δcho2 cells (Figure 1B and 1C). The reduction but not elimination of PC suggests that Opi3p might partly compensate for Cho2p because its homolog, PEMT, alone catalyzes all three methylation reactions in mammals (Kanipes and Henry, 1997; Preitschopf et al., 1993). Accordingly, PE is proportionally elevated which exacerbates further the PC/PE imbalance. In Δopi3 cells, a similar reduction of PC was not observed. Instead, PC is effectively absent (173-fold reduction compared to wild type) and the inability to process MMPE causes its abnormal accumulation (Figure 1C). Other changes include higher triacylglycerol (TAG) levels, resulting in the appearance of lipid droplets (Figure S1A) (Fei et al.). Significant changes to the levels of several less abundant lipids were also observed (Figure 1B) while other lipid concentrations remained unchanged (Table S1). Although PC/PE ratios are dramatically altered in mutant cells as expected, the data show that lipid homeostasis is more broadly impacted. Despite this, internal membranes of mutant cells are morphologically indistinguishable from wild type (Figure S1A and S1B).
The above phenotypes are consistent with the steatosis effect of Pemt−/− mice fed choline deficient diets (Li et al., 2006). To determine whether choline supplementation can fully ameliorate Cho2p and Opi3p deficiencies as in Pemt−/− mice, lipid profiling was performed on Δcho2 and Δopi3 cells grown in 1 mM choline. PE, PC, and TAG levels were restored to normal levels, in line with normal growth of cells (Figure S1C and Table S1). This experiment shows the preferred compensatory response is through alternative lipid metabolic pathways, if available. Interestingly, MMPE levels stayed depressed in Δcho2 cells and elevated in Δopi3 cells under these conditions (Figure S1C). This shows that the CDP-DAG pathway only partially attenuates when the Kennedy pathway provides all the needed PC (Figure 1A). Nevertheless, the symptoms of stress are largely relieved by restoration of the correct PC/PE ratio (see Figure 4C).
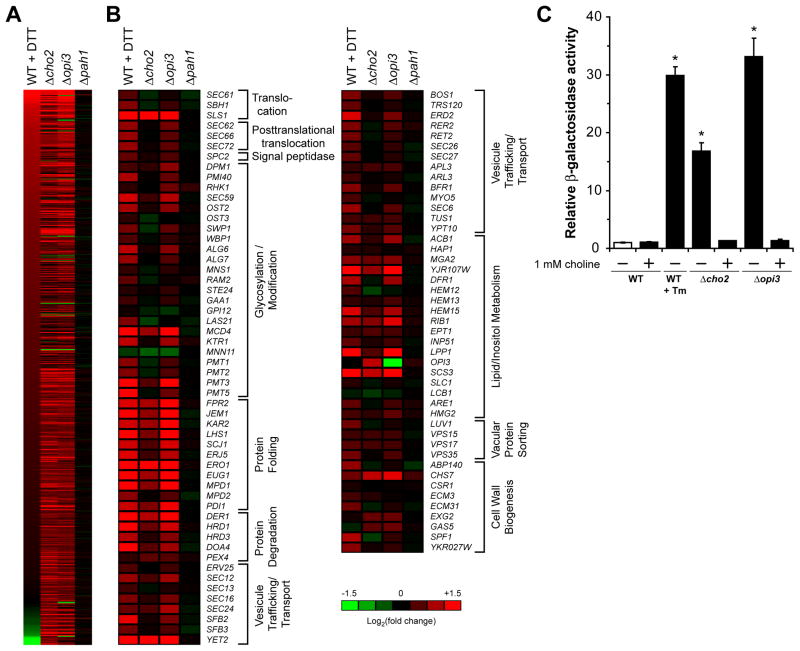
Figure 4. The UPR is strongly activated in Δcho2 and Δopi3 cells.
(A) Heat map representing genes with Log2 (fold change) ≥ 1 for Δcho2, Δopi3, or Δpah1 cells. (B) Heat maps of UPR regulated genes previously listed by Travers et al. (Travers et al., 2000) (A–B) Total RNA was extracted from cells grown to early log phase at 30°C. Probes were prepared using Low Input Quick Amp Labeling System with 100 ng of total RNA as starting material following manufacturer instruction. Based 2 logarithmic fold changes in genes expression were normalized to untreated WT cells. (C) Cells were grown to early log phase at 30°C in selective synthetic complete media in the absence or presence of 1 mM choline. UPR induction was measured using a UPRE-LacZ reporter assay. Tm, tunicamycin. Data shown is the mean ± SD of three independent experiments. *p < 0.005, Student’s t-test compared to WT.
These data show that disrupting PC synthesis leads to dramatic lipid imbalances that are surprisingly well tolerated (Figure 1B). Synthetic genetic array analysis (SGA) performed by the Andrews and Boone laboratories revealed extensive interactions for Δcho2 and Δopi3 mutants, suggesting the existence of broad compensatory mechanisms (Costanzo et al., 2010). Taken together, these data validate the use of cho2 and opi3 mutants as experimental models of lipid disequilibria.
Recalibration of the protein homeostasis network in response to lipid disequilibrium
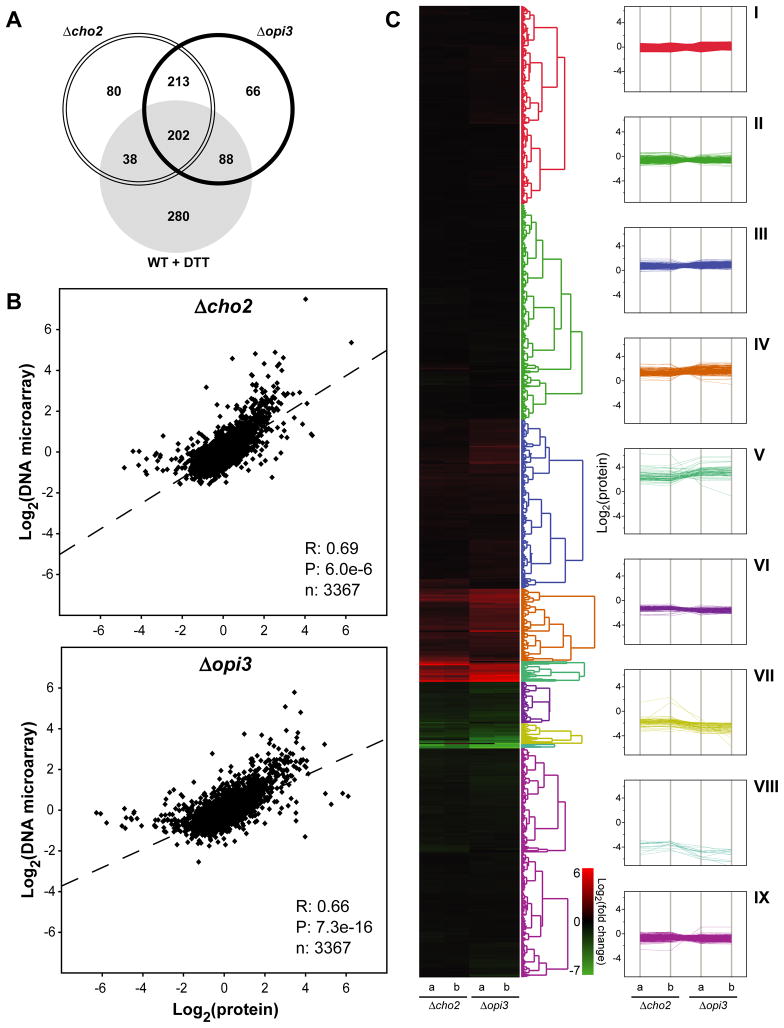
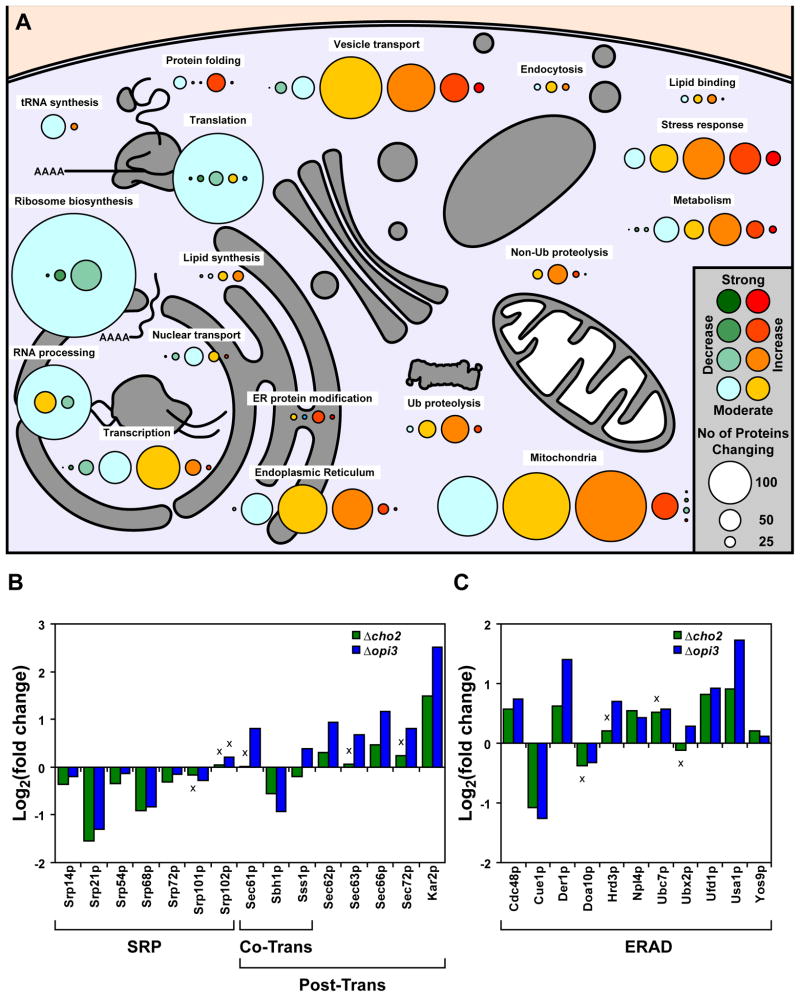
To understand how cells buffer homeostatic shocks, quantitative transcriptional and proteomic profiles were obtained for mutant and wild type cells. First, DNA microarray analysis was performed using RNA extracted from cells grown in synthetic media without added choline. To validate the quality of microarray data, qPCR was performed on a gene subset (Figure S2). The breadth and extent of changes were dramatic. Overall, 527 genes were upregulated at least 2-fold in Δcho2 and Δopi3 cells and 152 genes were downregulated (Figure 2A). For a detailed accounting of protein changes, global quantitative proteomic analysis was performed using isobaric Tandem Mass Tag (TMT) labeling (Ting et al., 2011). Log2 values of each protein were used for hierarchical clustering of protein expression levels in Δcho2 and Δopi3 (Huang da et al., 2009a, b). Except for a small number of outliers, comparison of the two data sets shows changes in steady state protein levels are in excellent agreement with microarray data (Figure 2B). Functional clustering analysis revealed strong elevation of major protein chaperones involved in protein folding, repair, and quality control (Figure 2C and 3A; Table S2 and S3). Along these lines, components required for posttranslational translocation into the ER are upregulated (Figure 3B). Notably, the major catabolic pathways of the ubiquitin-proteasome system (UPS) as well as the conventional and autophagic arms of the vacuolar (lysosomal) system are coordinately induced. Among UPS feeder pathways, components of the ER-associated degradation pathway (ERAD), which disposes aberrant proteins, are coordinately elevated (Figure 3C). Of major stress regulatory pathways, numerous targets of the heat shock response (HSR) and the unfolded protein response (UPR) are strongly activated (Figure 3A, 4A and 4B). The UPR monitors the state of the ER and regulates several hundred genes to maintain homeostasis (Travers et al., 2000). UPR activation is not surprising because PC is the most abundant lipid constituent of ER membranes (Zinser et al., 1991). On the other hand, the HSR is sometimes termed the “cytosolic UPR” because it can respond to unfolded cytosolic proteins independent of temperature change (Metzger and Michaelis, 2009). Their activation suggests that protein homeostasis is broadly disrupted by lipid disequilibrium. In agreement with microarray data, most enzymes of lipid biosynthetic pathways were unchanged or somewhat reduced. Although this finding doesn’t rule out remodeling of membrane lipid composition as part of the compensatory response, it is notable that proteins that stabilize and regulate the structure of membranes (Hsp12p and Rtn2p) were strongly elevated. A particularly intriguing pattern is the broad upregulation of proteins comprising the secretory pathway and the mitochondria. It is unclear whether their regulation compensates for reduced activities or to elevate their functions for increased loads.
Figure 2. Genomic and proteomic cellular response to lipid disequilibrium.
(A) Venn diagram representation of upregulated genes at a minimum of two folds compared to untreated WT cells. Total RNA was extracted from cells grown to early log phase at 30°C. Probes were prepared using Low Input Quick Amp Labeling System with 100 ng of total RNA as starting material following manufacturer instruction. (B) Pearson correlation of relative protein and RNA abundance. R, Pearson correlation; P, p-value. (C) Hierarchical clustering of Log2 levels for each protein in Δcho2 and Δopi3 (left panel) and parallel coordinate plot of the nine major clusters (right panel). Major GO terms for plots i to ix are listed in Table S2.
Figure 3. Broad remodeling of protein homeostasis networks in response to lipid disequilibrium.
(A) Protein expression in Δcho2 and Δopi3 compared to WT are illustrated by color-coding according to their fold change. Circle sizes are proportional to the number of proteins that were found to have similar fold change under the same cellular function. The summarized data was based on the hierarchical clustering from Figure 2C. (B) Expression of proteins part of the signal recognition particle (SRP), co-translational (Co-Trans) and post-translational (Post-Trans) translocation machineries. Based 2 logarithmic fold changes in protein expression were normalized to untreated WT cells. (C) Expression of proteins part of the ER-associated protein degradation (ERAD) machinery and data was normalized as in (B). Non-significant changes are denoted by “x” with p > 0.05, Student’s t-test compared to WT.
Major systems were also downregulated. The most dramatic include broad classes related to protein synthesis and its regulation. These include the ribosomal proteins themselves, assembly and nuclear transport factors, translation initiation factors, ribosome associated chaperones, and components of the signal recognition particle (SRP) and its receptor (Figure 3A and 3B). Other functions important in protein synthesis, including various amino acid permeases, are also strongly reduced.
Based solely on transcriptome and proteome profiles, the data suggest that a major physiologic consequence of lipid disequilibrium is the general disruption of protein homeostasis, which increases the concentration of abnormal proteins (Lee, 2004). In response, the MSR output enhances protein folding and repair functions while attenuating translational activity to reduce further stressing on the system. Augmenting this, catabolic pathways are activated to rid irreversibly defective molecules.
We wondered whether the complex changes are specific to lipid disequilibrium or reflects a general stress response to perturbed membranes. To address this question, we analyzed cells lacking phosphatidic acid phosphatase (encoded by PAH1, Figure 1A), a key enzyme in phospholipid metabolism. It was previously reported that membrane biosynthesis is dysregulated in Δpah1 cells leading to the formation of extended and distorted membranes (Han et al., 2008). This phenotype is easily observed when compared to the near normal morphologies of Δcho2 and Δopi3 cells (Figures S1A, S3A and S3B). Despite these perturbations, we found the Δpah1 transcriptome nearly unchanged from wild type (Figures 4A and 4B). Lipidomic profiling shows a relatively normal lipid composition compared to Δcho2 and Δopi3 strains, apart from a sharp reduction in DAG and TAG (Figure S5B). These data show that proteome remodeling in Δcho2 and Δopi3 is a specific response to lipid disequilibrium.
The UPR program is essential to counteract the lethal effects of lipid disequilibrium
Of the prominent regulatory signatures revealed, broad activation of UPR targets was perhaps the most striking (Travers et al., 2000). Many UPR target genes were activated strongly in Δopi3 cells compared to wild type cells treated with DTT, a powerful UPR inducer (Figures 2A and 4B). The pattern for Δcho2 cells is similar but the extent of gene activation was generally lower. Although there is sizable overlap of induced genes, of equal interest is the large number of DTT-induced genes not activated in Δcho2 and Δopi3 strains (Figures 2A and 4A). This pattern follows other specific mutants causing ER stress, where differential target gene regulation was observed depending on the defect (Thibault et al., 2011).
To determine if the UPR signaling mechanism is directly stimulated, we applied an established UPR-specific reporter assay (Cox et al., 1997). Here, both mutants displayed a strong constitutive response, with Δopi3 cells measuring the highest for any physiological inducer analyzed to date (Figure 4C) (Thibault et al., 2011). These data are consistent with a large-scale screen for mutants activating a UPR-specific reporter (Jonikas et al., 2009). To determine if the UPR is stimulated as a direct consequence of lipid disequilibria, mutant cells were grown in media supplemented with 1 mM choline to restore PC/PE balance. As shown in Figure 4C, the addition of choline entirely abrogates UPR activation. By contrast, Δpah1 cells displayed no UPR induction using this assay, consistent with microarray data (Figure 4 and S3).
Interestingly, the CHO2 and OPI3 genes display synthetic lethality with the UPR signaling genes HAC1 and IRE1 (Costanzo et al., 2010 and our unpublished results). Because the UPR is normally quiescent, these interactions suggest that UPR activation buffers otherwise lethal deficiencies (Cox et al., 1997; Thibault et al., 2011). If so, we reasoned the relationship can be exploited to reveal the normally hidden adverse effects of lipid disequilibrium. For this, analysis of lipid dysregulated cells must be performed in the absence of UPR activation. To this end, we created “conditional synthetic lethality” mutants to overcome the inviability of double nulls. Applying the strategy outlined in Figure S4A, temperature sensitive (ts) alleles of CHO2 and OPI3 genes were created in combination with the wild type or null allele of IRE1 (Thibault et al., 2011). All strains grew equally well at the permissive temperature (Figure S4B, 25°C). At 37°C, the cho2-1Δire1 and opi3-1Δire1 strains were strongly impaired while Δire1, cho2-1, and opi3-1 strains grew like wild type (Figure S4B). Complete rescue of the cho2-1Δire1 and opi3-1Δire1 ts phenotype by choline supplementation shows that growth impairment is due to PC depletion specifically in the absence of the UPR (Figure S4B). The robust growth of UPR proficient opi3-1 and cho2-1 mutants at 37°C indicates that UPR induction compensates for an otherwise lethal condition (Figure S4B). Together, these experiments validate the use of conditional synthetic lethal mutants to examine the UPR’s role in alleviating lipid disequilibrium.
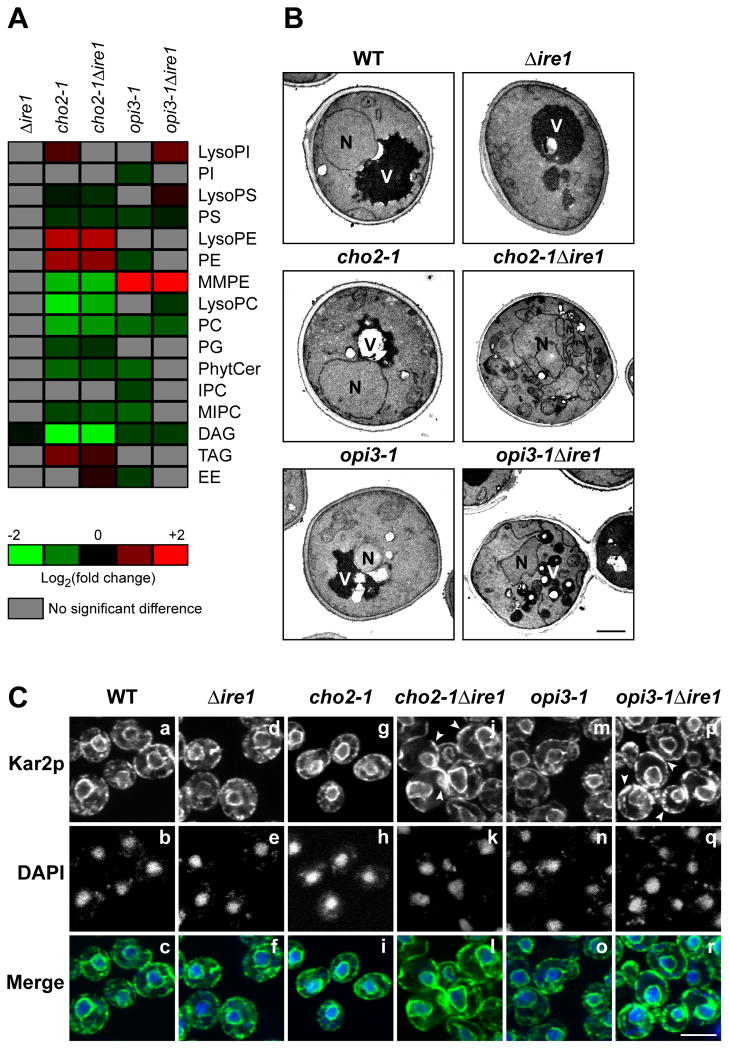
As part of the MSR, the UPR might function to adjust lipid composition because lipid metabolic genes are among its targets (Travers et al., 2000). Lipid profiles obtained for Δcho2 and Δopi3 cells reflect UPR-induced, adapted cells but do not by themselves inform on whether any changes are consequences of UPR induction (Figure 1B and Table S1). To determine if UPR activation adjusts membrane composition in response to stress, lipids were extracted from cho2-1, opi3-1, cho2-1Δire1, and opi3-1Δire1 strains following temperature shift and quantitative lipid profiles were obtained. The profiles measured for cho2-1 and cho2-1Δire1 cells showed no significant differences, indicating membrane remodeling is not part of the UPR compensatory program for diminished Cho2p (Figure 5A). Similarly, between opi3-1 and opi3-1Δire1 strains, only small differences were observed (Figure 5A). As a control, the profile of Δire1 cells is indistinguishable from wild type under the same conditions (Figure 5A). In addition, the lipid compositions for both ts mutants at permissive temperature were largely unchanged from wild type (Figure S5A). As expected, the presence or absence of the UPR has no impact on the lipid composition of cells lacking PAH1 (Figure S5B). Together, these data show that the ER stress response to lipid disequilibrium, though essential, does not include compensatory changes to lipid composition.
Figure 5. UPR activation maintains membrane morphologies under lipid disequilibrium.
(A) Heat maps reflecting all significant lipid changes to WT. Cells were grown to early log phase at 23°C and shifted to 37°C for 2 h in selective synthetic complete media before being harvested. Lipids were extracted with chloroform:methanol mixture from lyophilized cells. Organic extraction was concentrated and analyzed by HPLC-MS. p < 0.05, Student’s t-test. (B) Cells were grown at 23°C and shifted to 37°C for 2 h in selective synthetic complete media before being prepared for TEM analysis as described in experimental procedures. N, nucleus; V, vacuole. Scale bar, 1 μm. (C) Cells were grown to early log phase at 23°C and shifted to 37°C for 2 h in selective synthetic complete media before being fixed in formaldehyde and permeabilized. Staining was performed using anti-Kar2p primary antibody followed by Alexa Fluor 488 goat anti-rabbit secondary antibody. DAPI staining marks the position of nuclei. Scale bar, 5 μm.
UPR activation is required to maintain membrane morphology
In contrast to Δpah1 cells, indirect immunofluorescence and transmission electron microscopy (TEM) imaging revealed no morphological differences of internal membranes between Δcho2 and Δopi3 cells to wild type (Figure S1A and S1B). To determine if the UPR program plays a role in maintaining membrane integrity, imaging of cho2-1, opi3-1, cho2-1Δire1, and opi3-1Δire1 cells were performed before and after temperature shift. At the permissive temperature, internal membrane structures of all four strains were similar to wild type (Figure S5C and S5D). After the temperature shift, dramatic differences cho2-1Δire1 and opi3-1Δire1 cells were apparent. Both displayed distorted and enlarged ER/nuclear membranes (Figure 5B). Visualization of these cells by TEM revealed highly disorganized membranes and distorted or fragmented organelles (Figure 5B). By contrast, cho2-1 and opi3-1 cells appeared similar to wild type except for the appearance of lipid droplets (Figure 5C and 5D). These data show that the UPR plays a critical role in maintaining the internal architecture of membranes under phospholipid disequilibrium.
The UPR compensates for protein biogenesis and quality control deficiencies during lipid disequilibrium
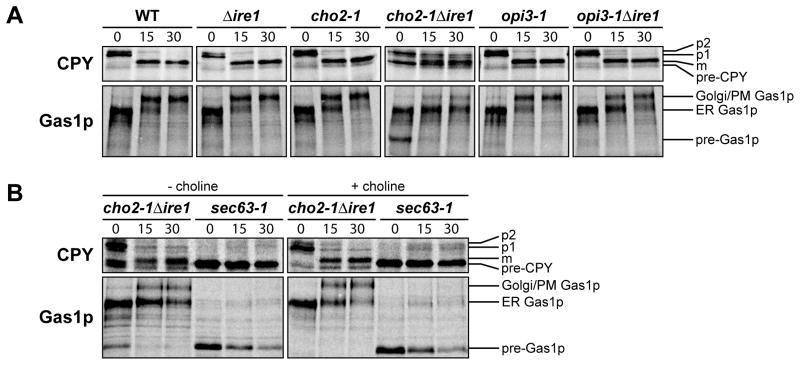
Among its functions, the UPR regulates ER protein synthesis and quality control (Casagrande et al., 2000; Ng et al., 2000; Travers et al., 2000). Because of the strong UPR response, we wondered whether these functions are compromised during lipid disequilibrium. For this, the synthesis and processing of endogenous secretory proteins were analyzed. We selected carboxypeptidase Y (CPY) and the glycosylphosphatidylinositol (GPI)-linked membrane protein Gas1p (Doering and Schekman, 1996; Hann and Walter, 1991; Ng et al., 2000; Robinson et al., 1988; Simons et al., 1995). CPY and Gas1p are translocated into the ER using a post-translational mechanism. Translocation efficiency in vivo can be monitored by protein gel mobility shifts reflecting the addition of N-linked glycans in the ER (Figure 6A). In the cho2-1Δire1 strain, translocation of CPY and Gas1p was delayed at 37°C as detected by the appearance of “pre” forms (Figure 6A and S6A). This strain also displayed a strong delay in the ER-to-Golgi/plasma membrane (PM) transport of Gas1p compared to controls (Figure 6A). This delay could be due to defect in the addition of the GPI-anchor, folding, transport, or a combination of these. A similar but less pronounced effect was observed for CPY maturation suggesting a general defect in protein maturation and/or transport. N-linked glycosylation was unaffected in cho2-1Δire1 cells because underglycosylated forms were not observed (Figure 6A). We did not observe translocation defects in the SRP-dependent substrate DPAP B suggesting that the co-translational pathway remains intact (data not shown) (Hann and Walter, 1991). The difference is intriguing because components dedicated to posttranslational protein translocation are coordinately upregulated while SRP components are downregulated (Figure 3B). No translocation defect was observed in opi3-1Δire1 for CPY and Gas1p. This could be due to a milder lost of function of opi3-1 allele compared to cho2-1 or the difference in lipid composition. Δcho2 and Δopi3 cells (UPR activated) did not display CPY defects indicating the robustness of the UPR in functional compensation (Figure S6B). Gas1p trafficking from the ER was delayed in these strains suggesting that the biogenesis of GPI-anchored proteins might be more sensitive to lipid imbalance. The Δpah1 and Δpah1Δire1 strains were unaffected in secretory protein processing compared to WT and Δire1 strains despite their altered ER/nuclear morphologies (Figure S6C). To confirm that the observed defects are the consequence of perturbed membrane lipid composition, we examined the biogenesis of CPY and Gas1p in the presence of exogenous choline. Choline supplementation rescued the translocation defect in cho2-1Δire1 but not in the control translocation mutant sec63-1 (Figure 6B).
Figure 6. The UPR compensates protein biogenesis defect during lipid imbalance.
Cells were grown to early log phase at 23°C and shifted to 37°C for 30 min (sec63-1) or 2 h in selective synthetic complete media in the absence (A) or presence (B) of 1 mM choline before being pulse-labeled at 37°C with L-[35S]-methionine/cysteine for 5 min followed by a chase at the indicated times. Immunoprecipitated proteins using anti-CPY or anti-Gas1p were resolved by SDS-PAGE and visualized by phosphoimager analysis.
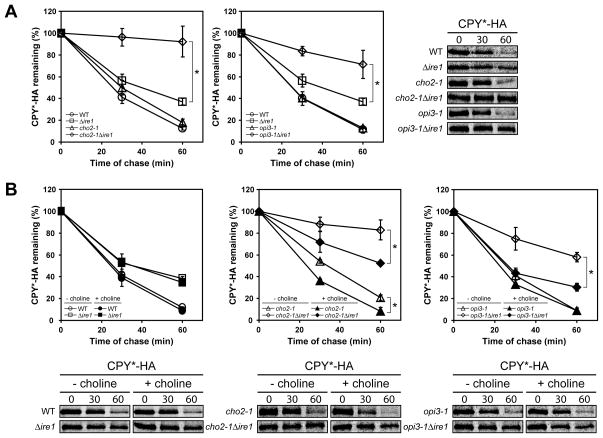
Misfolded proteins in the ER are degraded by ER-associated degradation (ERAD) (Romisch, 2005; Vembar and Brodsky, 2008). CPY*, a misfolded mutant of CPY, is a classical substrate of ERAD (Finger et al., 1993). To examine the functionality of the ERAD machinery during lipid disequilibrium, CPY* turnover was measured by metabolic pulse chase. CPY* expression causes mild ER stress and consequently a minor ERAD defect in control Δire1 cells as reported previously (Figure 7A) (Casagrande et al., 2000; Ng et al., 2000). In cho2-1Δire1 and opi3-1Δire1 cells, CPY* was strongly stabilized while in cho2-1 and opi3-1 cells, degradation was as efficient as wild type (Figure 7A and S7A). These results demonstrate that both forms of lipid disequilibrium severely disrupt ERAD function that is detected and alleviated by UPR activation. The exceptional sensitivity of ERAD to lipid disequilibrium is highlighted in Δcho2 and Δopi3 cells, which remains partially defective in ERAD (Figure S7B). In these strains, UPR activation fails to fully restore ERAD perhaps reflecting the limit of the response when disequilibrium is particularly severe. Although ERAD is highly sensitive to changes to lipid composition, not all forms of membrane dysfunction compromise its function. No significant defect in CPY* degradation was observed in Δpah1 and Δpah1Δire1 cells (Figure S7C). To confirm that the observed ERAD defects are due to lipid disequilibria, we monitored CPY* degradation in the presence of exogenous choline. In cho2-1Δire1 strain, the availability of choline strongly relieved the ERAD defect but is less efficient compared to the Δire1 control (Figure 7B, compare left and middle panels). For opi3-1Δire1, choline supplementation fully restored CPY* degradation to the rate observed for the Δire1 control (Figure 7B, compare left and right panels).
Figure 7. The UPR compensates protein quality control defect during lipid imbalance.
Cells were grown to early log phase at 23°C and shifted to 37°C for 2 h in selective synthetic complete media in the absence (A) or presence (B) of 1 mM choline before being pulse-labeled at 37°C with L-[35S]-methionine/cysteine for 10 min followed by a chase at the indicated times. Immunoprecipitated proteins using anti-HA were resolved by SDS-PAGE and quantified by phosphoimager analysis. Data plotted is the mean ± SD of three independent experiments. *p < 0.02, Student’s t-test.
DISCUSSION
Changes in cellular and circulating lipid composition are associated with numerous human pathologies (Sanyal, 2005; Wenk, 2005). It is therefore widely acknowledged that lipid homeostasis is critical for health. Despite many lipid metabolic pathways characterized in detail, the mechanisms used to monitor and maintain lipid homeostasis are not well understood. We sought to understand how cells respond and cope with lipid disequilibrium using PC/PE homeostasis as a focus. A yeast model based on the PEMT-deficient mouse system of steatohepatitis was created to analyze the global response to lipid disequilibrium.
Cells reacted to Cho2p or Opi3p deficiency by broadly altering transcription patterns resulting in a purposely-remodeled proteome. The MSR output, although broad in scope, provided insight into the consequences of lipid disequlibrium as well as the cellular strategy used to buffer them. The coordinate regulation of protein synthesis, folding, modification, quality control, transport and degradation functions mirrors the defined collection of pathways termed the proteostasis (protein homeostasis) network (Balch et al., 2008; Hartl et al.; Powers et al., 2009). Despite the dramatic changes, we were surprised that regulation of lipid metabolic pathways to remodel membrane composition is not part of the response. Regulation of the proteostasis network has featured prominently in various protein conformational diseases. It was therefore proposed that its therapeutic control to restore balance could be a key in disease intervention (Balch et al., 2008; Hartl et al.; Powers et al., 2009). The results of this study broaden this view to lipid disorders and provide a de facto proof-of-principle using a simple model organism.
Remodeling of the proteostasis network suggested that lipid disequilibrium interfered with protein biogenesis and quality control functions. These include increased chaperone concentrations in all compartments, the enhancement of quality control pathways for aberrant proteins and the associated catabolic pathways of the proteasome and vacuole (lysosome). In addition, levels of the membrane stabilizing protein Hsp12p and curvature inducing protein Rtn2p are both strongly increased. Of great interest is the coordinate downregulation of protein biosynthetic machinery, particularly in the secretory pathway. The ribosome and associated proteins are repressed, as are components of the signal recognition particle, glycosylation factors, plasma membrane transporters, etc. Interestingly, factors dedicated to SRP-independent translocation are coordinately upregulated (Figure 3B). This could be due to the upregulation of ER chaperones, vacuolar proteases, and other soluble luminal proteins, which tend to use the SRP-independent pathway while membrane proteins tend to use the SRP pathway (Ng et al., 1996; Ulbrandt et al., 1997).
As part of a comprehensive membrane stress response, the yeast UPR is specifically activated. UPR activation was previously reported for several models of metabolic disease and lipid disequilibrium (see Introduction). Of particular interest, a recent study reported that the fatty liver tissue of obese mice led to changes in lipid composition and notably in a change of the PC/PE ratio (Fu et al., 2011). The disequilibrium causes sarco/endoplasmic reticulum calcium ATPase (SERCA) dysfunction, leading to chronic ER stress that was previously associated with the development of type II diabetes (Ozcan et al., 2004). Restoring the PC/PE ratio or exogenous supplementation of SERCA function reduced ER stress and improved glucose homeostasis.
In this study, the data show that broad deployment of the UPR program is essential for viability under stress. Evidence came from the use of conditional synthetic lethal mutants that allow controlled initiation of lipid perturbation, with and without the UPR. These analyses show that the UPR can alleviate some forms of stress resulting from lipid disequilibrium without remodeling membrane composition. Instead, the translocation, trafficking, and ERAD defects observed when PC/PE ratios are modestly altered are entirely alleviated by UPR activation. It should be noted that lipid biosynthetic genes are part of the UPR program (Travers et al., 2000). It is therefore likely that their activation may be required for other forms of lipid perturbation. In a recent report, mutants of ARV1, encoding a protein involved in sterol transport, leads to ER stress (Shechtman et al., 2011). Thus, it is plausible that UPR activation under a distinct form of membrane stress might involve membrane remodeling.
UPR synthetic lethality has been particularly useful in revealing physiologically linked functions to the stress pathway (Ng et al., 2000). Indeed, near genome-wide SGA analysis suggests that well over a hundred client functions are monitored by the UPR (Costanzo et al., 2010). This demonstrates the remarkable breadth of the UPR in maintaining functionality of the early secretory pathway. Despite this, the UPR is not generally deployed. For example, it is not activated by heat stress in yeast as one might expect intuitively and UPR mutants are not temperature sensitive (Cox et al., 1993). Similarly, Δpah1 cells do not activate the UPR despite disruption of a major lipid metabolic pathway that causes broad changes in membrane morphology. However, detailed analyses may explain the basis for the apparent paradox. We detected few changes in lipid composition besides major decreases in diacylglycerol and triacylglycerol, which are not major components of membrane bilayers (Figure 1B). The relative balance in lipid composition likely involves phosphatidic acid metabolic redistribution by alternative synthetic routes (Gaspar et al., 2007). In addition, no defects in ER protein translocation, trafficking, or ERAD were detected. Viability of Δpah1 cells is not based on a prototypical stress response because few genes were found to be significantly upregulated compared with wild type (Figure 4) (Han et al., 2008).
Upon ER stress, ER membrane expansion can occur in concert with the UPR to relieve stress (Schuck et al., 2009). However, conditions that lead to membrane expansion in cho2-1 and opi3-1 cells (UPR off) are insufficient to relieve stress. The reduction in PC of CHO2 and OPI3 null and ts strains dramatically changes the chemical composition of the membranes. The thickness, fluidity, and spontaneous curvature of the bilayer are a product of the collective physical properties of the bilayer (Lee, 2004). As such, changing the zwitterionic lipid from PC to PE was shown to dramatically reduce the activity of the mechanosensitive channel MscL in E. coli (Powl et al., 2008). This effect likely explains the observed defects in ER protein translocation and ERAD, activities that are entirely dependent on ER membrane protein complexes (Vembar and Brodsky, 2008; Zimmermann et al., 2011). Remarkably, UPR activation completely alleviates these defects caused by the cho2-1 and opi3-1 mutations despite having altered membranes.
Although the importance of the proteostasis network in alleviating the consequences of PC/PE disequilibrium is apparent, DNA microarray and proteomic analysis revealed more extensive physiological remodeling. These include the INO1 gene, which encodes the inositol-3-phosphate synthase that catalyzes the rate-limiting step in the synthesis of the essential phospholipid precursor inositol (Carman and Henry, 1999; Donahue and Henry, 1981). Accordingly, the transcriptional activator genes for INO1, INO2, and INO4, are simultaneously activated. INO2 displays synthetic lethality with CHO2 and OPI3 indicating that its targets are an essential part of the overall compensatory mechanism (Costanzo et al., 2010). In addition, the heat shock genes are strongly upregulated in the Δcho2 and Δopi3 cells. This is not unexpected because PC/PE disequilibrium affects both bilayers of cellular membranes and the consequences could equally impact the ER lumen (UPR) and cytosol (heat shock response). The breadth and magnitude of transcriptional remodeling in these cells likely reflect the broad defects expected when PC is sharply reduced or eliminated. Microarray analysis of the UPR synthetic lethality mutants Δalg5, Δscj1, and Δlhs1 revealed relatively modest changes. Only selective parts of the UPR program were upregulated and, except for Δlhs1, other known stress pathways were not activated (Thibault et al., 2011). Although the UPR plays an essential compensatory role, the overall buffering effect is undoubtedly the coordinated effort of multiple regulatory pathways comprising the MSR.
In this report, we show that remodeling of the protein homeostasis network is a signature response against lipid disequilibrium, particularly during loss of PC/PE homeostasis. One facet of the response, the UPR, sustains protein biogenesis and ERAD functions. In addition, through a mechanism that remains unclear, UPR activation maintains membrane integrity and organization without the need to adjust lipid composition. Taken together, the data demonstrate the remarkable ability of cells to remodel their physiology to counteract perturbations to internal membranes. How each of the regulatory pathways contributes to stress alleviation will be the challenge of future studies.
EXPERIMENTAL PROCEDURES
Assays
Protocols and reagents used for proteomic analysis, lipidomic analysis, DNA microarray, β-galactosidase reporter assay, transmission electron microscopy, and real-time PCR are described in Supplemental Information.
Cell labeling and immunoprecipitation
Cell labeling and immunoprecipitation was carried out as previously described (Thibault et al., 2011). Typically, 3.0 A600 units of early log phase cells were labeled with 80 μCi of L-[35S]-methionine/cysteine mix (Perkin Elmer). Immunoprecipitated proteins were separated on SDS-PAGE and exposed to phosphor screens. Exposed screens were visualized using a Typhoon 8600 scanner (GE Healthcare) and quantified using ImageQuant TL software (GE Healthcare). All data plots reflect three independent experiments with mean ± SD indicated.
Indirect Immunofluorescence
Indirect Immunofluorescence was carried out as previously described (Wang and Ng, 2010). Typically, cells were grown to early log phase at 23°C, 30°C, or 37°C in selective synthetic complete media, fixed in 10% formaldehyde and permeabilized. After blocking with 3% BSA, staining was performed using anti-Kar2p primary antibody (1:1000 dilution) followed by Alexa Fluor 488 goat anti-rabbit secondary antibody (1:1000 dilution). The DNA dye DAPI was applied as a mounting medium mixture. Samples were visualized using a Zeiss Axiovert 200M microscope with a 100x 1.4 NA oil Plan-Apochromat objective (Carl Zeiss MicroImaging).
Supplementary Material
HIGHLIGHTS.
Membrane lipid imbalance activates the membrane stress response
The MSR broadly reprograms the protein homeostasis network
The MSR buffers the lethal effects of lipid disequilibrium
The UPR arm maintains membrane functionality without remodeling lipid composition
Acknowledgments
We are grateful to the TLL/NUS core facilities especially Xuezhi Ouyang for EM sample preparation. We thank Songyu Wang and Chengchao Xu for critical reading of the manuscript and Ron Kopito (Stanford University) for insightful discussion. This work was supported by funds from the Temasek Trust (G.T. and D.T.W.N.), the Singapore National Research Foundation under CRP Award No. 2007-04 (G.S. and M.R.W.), the National Institutes of Health GM67945 (S. P. G.), and the Singapore Millennium Foundation to G.T. (Postdoctoral fellowship).
Footnotes
Accession Numbers
The DNA microarray data discussed in this publication was deposited in NCBI’s Gene Expression Onmibus (GEO) under series number SE39419.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Carman GM, Henry SA. Phospholipid biosynthesis in yeast. Annu Rev Biochem. 1989;58:635–669. doi: 10.1146/annurev.bi.58.070189.003223. [DOI] [PubMed] [Google Scholar]
- Carman GM, Henry SA. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog Lipid Res. 1999;38:361–399. doi: 10.1016/s0163-7827(99)00010-7. [DOI] [PubMed] [Google Scholar]
- Casagrande R, Stern P, Diehn M, Shamu C, Osario M, Zuniga M, Brown PO, Ploegh H. Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol Cell. 2000;5:729–735. doi: 10.1016/s1097-2765(00)80251-8. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Chapman RE, Walter P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell. 1997;8:1805–1814. doi: 10.1091/mbc.8.9.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Daum G, Tuller G, Nemec T, Hrastnik C, Balliano G, Cattel L, Milla P, Rocco F, Conzelmann A, Vionnet C, et al. Systematic analysis of yeast strains with possible defects in lipid metabolism. Yeast. 1999;15:601–614. doi: 10.1002/(SICI)1097-0061(199905)15:7<601::AID-YEA390>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Doering TL, Schekman R. GPI anchor attachment is required for Gas1p transport from the endoplasmic reticulum in COP II vesicles. EMBO J. 1996;15:182–191. [PMC free article] [PubMed] [Google Scholar]
- Donahue TF, Henry SA. myo-Inositol-1-phosphate synthase. Characteristics of the enzyme and identification of its structural gene in yeast. J Biol Chem. 1981;256:7077–7085. [PubMed] [Google Scholar]
- Fei W, Shui G, Zhang Y, Krahmer N, Ferguson C, Kapterian TS, Lin RC, Dawes IW, Brown AJ, Li P, et al. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 2011;7:e1002201. doi: 10.1371/journal.pgen.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger A, Knop M, Wolf DH. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur J Biochem. 1993;218:565–574. doi: 10.1111/j.1432-1033.1993.tb18410.x. [DOI] [PubMed] [Google Scholar]
- Fu S, Yang L, Li P, Hofmann O, Dicker L, Hide W, Lin X, Watkins SM, Ivanov AR, Hotamisligil GS. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473:528–531. doi: 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar ML, Aregullin MA, Jesch SA, Nunez LR, Villa-Garcia M, Henry SA. The emergence of yeast lipidomics. Biochim Biophys Acta. 2007;1771:241–254. doi: 10.1016/j.bbalip.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Han GS, O’Hara L, Carman GM, Siniossoglou S. An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth. J Biol Chem. 2008;283:20433–20442. doi: 10.1074/jbc.M802903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann BC, Walter P. The Signal Recognition Particle in S. cerevisiae. Cell. 1991;67:131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Henry SA, Kohlwein SD, Carman GM. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 2012;190:317–349. doi: 10.1534/genetics.111.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermansson M, Hokynar K, Somerharju P. Mechanisms of glycerophospholipid homeostasis in mammalian cells. Prog Lipid Res. 2011;50:240–257. doi: 10.1016/j.plipres.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, Schuldiner M. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanipes MI, Henry SA. The phospholipid methyltransferases in yeast. Biochim Biophys Acta. 1997;1348:134–141. doi: 10.1016/s0005-2760(97)00121-5. [DOI] [PubMed] [Google Scholar]
- Kodaki T, Yamashita S. Yeast phosphatidylethanolamine methylation pathway. Cloning and characterization of two distinct methyltransferase genes. J Biol Chem. 1987;262:15428–15435. [PubMed] [Google Scholar]
- Kodaki T, Yamashita S. Characterization of the methyltransferases in the yeast phosphatidylethanolamine methylation pathway by selective gene disruption. Eur J Biochem. 1989;185:243–251. doi: 10.1111/j.1432-1033.1989.tb15109.x. [DOI] [PubMed] [Google Scholar]
- Lee AG. How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Lee AH, Glimcher LH. Intersection of the unfolded protein response and hepatic lipid metabolism. Cell Mol Life Sci. 2009;66:2835–2850. doi: 10.1007/s00018-009-0049-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Agellon LB, Allen TM, Umeda M, Jewell L, Mason A, Vance DE. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab. 2006;3:321–331. doi: 10.1016/j.cmet.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Li Z, Vance DE. Phosphatidylcholine and choline homeostasis. J Lipid Res. 2008;49:1187–1194. doi: 10.1194/jlr.R700019-JLR200. [DOI] [PubMed] [Google Scholar]
- McGraw P, Henry SA. Mutations in the Saccharomyces cerevisiae opi3 gene: effects on phospholipid methylation, growth and cross-pathway regulation of inositol synthesis. Genetics. 1989;122:317–330. doi: 10.1093/genetics/122.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger MB, Michaelis S. Analysis of quality control substrates in distinct cellular compartments reveals a unique role for Rpn4p in tolerating misfolded membrane proteins. Mol Biol Cell. 2009;20:1006–1019. doi: 10.1091/mbc.E08-02-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau P, Cassagne C. Phospholipid trafficking and membrane biogenesis. Biochim Biophys Acta. 1994;1197:257–290. doi: 10.1016/0304-4157(94)90010-8. [DOI] [PubMed] [Google Scholar]
- Ng DT, Brown JD, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DT, Spear ED, Walter P. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J Cell Biol. 2000;150:77–88. doi: 10.1083/jcb.150.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7:520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- Powl AM, East JM, Lee AG. Importance of direct interactions with lipids for the function of the mechanosensitive channel MscL. Biochemistry. 2008;47:12175–12184. doi: 10.1021/bi801352a. [DOI] [PubMed] [Google Scholar]
- Preitschopf W, Luckl H, Summers E, Henry SA, Paltauf F, Kohlwein SD. Molecular cloning of the yeast OPI3 gene as a high copy number suppressor of the cho2 mutation. Curr Genet. 1993;23:95–101. doi: 10.1007/BF00352006. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romisch K. Endoplasmic reticulum-associated degradation. Annu Rev Cell Dev Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, Fornek J, et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal AJ. Mechanisms of Disease: pathogenesis of nonalcoholic fatty liver disease. Nat Clin Pract Gastroenterol Hepatol. 2005;2:46–53. doi: 10.1038/ncpgasthep0084. [DOI] [PubMed] [Google Scholar]
- Schuck S, Prinz WA, Thorn KS, Voss C, Walter P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol. 2009;187:525–536. doi: 10.1083/jcb.200907074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechtman CF, Henneberry AL, Seimon TA, Tinkelenberg AH, Wilcox LJ, Lee E, Fazlollahi M, Munkacsi AB, Bussemaker HJ, Tabas I, Sturley SL. Loss of subcellular lipid transport due to ARV1 deficiency disrupts organelle homeostasis and activates the unfolded protein response. J Biol Chem. 2011;286:11951–11959. doi: 10.1074/jbc.M110.215038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons JF, Ferro-Novick S, Rose MD, Helenius A. BiP/Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J Cell Biol. 1995;130:41–49. doi: 10.1083/jcb.130.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault G, Ismail N, Ng DT. The unfolded protein response supports cellular robustness as a broad-spectrum compensatory pathway. Proc Natl Acad Sci U S A. 2011;108:20597–20602. doi: 10.1073/pnas.1117184109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting L, Rad R, Gygi SP, Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods. 2011;8:937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Ulbrandt ND, Newitt JA, Bernstein HD. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkey CJ, Yu L, Agellon LB, Vance DE. Biochemical and evolutionary significance of phospholipid methylation. J Biol Chem. 1998;273:27043–27046. doi: 10.1074/jbc.273.42.27043. [DOI] [PubMed] [Google Scholar]
- Wang S, Ng DT. Evasion of endoplasmic reticulum surveillance makes Wsc1p an obligate substrate of Golgi quality control. Mol Biol Cell. 2010;21:1153–1165. doi: 10.1091/mbc.E09-10-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- Zimmermann R, Eyrisch S, Ahmad M, Helms V. Protein translocation across the ER membrane. Biochim Biophys Acta. 2011;1808:912–924. doi: 10.1016/j.bbamem.2010.06.015. [DOI] [PubMed] [Google Scholar]
- Zinser E, Sperka-Gottlieb CD, Fasch EV, Kohlwein SD, Paltauf F, Daum G. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.