Abstract
The silencer information regulator (Sir) family of proteins has attracted much attention during the past decade due to their prominent role in metabolic homeostasis in mammals. The Sir1-4 proteins were first discovered in yeast as nicotinamide adenine dinucleotide (NAD+)-dependent deacetylases, which through a gene silencing effect promoted longevity. The subsequent discovery of a homologous sirtuin (Sirt) family of proteins in the mammalian systems soon led to the realization that these molecules have beneficial effects in metabolism- and aging-related diseases. Through their concerted functions in the central nervous system, liver, pancreas, skeletal muscle, and adipose tissue, they regulate the body’s metabolism. Sirt1, -6, and -7 exert their functions, predominantly, through a direct effect on nuclear transcription of genes involved in metabolism, whereas Sirt3-5 reside in the mitochondrial matrix and regulate various enzymes involved in the tricarboxylic acid and urea cycles, oxidative phosphorylation, as well as reactive oxygen species production. An interesting aspect of sirtuin’s functionality involves their regulation by the circadian rhythm, which impacts their function via cyclically regulating systemic NAD+ availability, further establishing the link of these proteins to metabolism. In this review we will discuss the relation of sirtuins to NAD+ metabolism, their mechanism of function, and their role in metabolism and mitochondrial functions. In addition, we will describe their effects in the cardiovascular and central nervous systems.
Keywords: Aging
Introduction
Silent information regulator 1-4 (Sir1-4) proteins were initially discovered in Saccharomyces cerevisiae as transcriptional repressors of the silent hidden MAT left (HML) and hidden MAT right (HMR) mating type loci 1–3 - which are suppressed copies of mating loci MATa and MATα for changing mating type in haploid cells - and of telomeres 4. This effect is partly due to deacetylation of histones H3 and H4 and heterochromatin formation by Sir2 5. In addition, the Sir proteins are involved in double stranded DNA repair 6, as well as suppression of mitotic and meiotic intrachromosomal recombination of ribosomal DNA (rDNA) in the nucleolus 7, 8, which is responsible for extrachromosomal accumulation of the senescence-related rDNA circles 9. This family of protein is mainly recognized for its role in longevity, as the phenotypes of the Sir2/3/4 mutants were characterized by a shortened lifespan 10, 11. In the Sir3 and Sir4 mutants, this appears to be due to the concomitant expression of mating-type information a and α, while the Sir2 mutant failed to suppress rDNA recombination. Conversely, overexpression of Sir2 extended the lifespan of wild type yeast. It was later discovered that Sir2 is a nicotinamide adenine dinucleotide (NAD+)-dependent deacetylase enzyme 12–14, which is a unique feature among deacetylase, via which it deacetylates K9 and K14 of H3 and K16 of H4 12. The deacetylase activity is indeed responsible for Sir2-dependent gene silencing, suppression of rDNA recombination, and longevity in yeast 12. In fact, the Sir family appears to be the main NAD+-dependent histone deacetylase in yeast, as this activity is abrogated when Sir2 and its four homologues are deleted 14. NAD+ is not simply a regulator in this reaction but a substrate; it is hydrolyzed by Sir2 into nicotinamide and adenosine diphosphate (ADP)-ribose, which is an acceptor of the acetyl group hydrolyzed from an acetyl-lysine substrate, thus, forming 2’-O-acetyl-ADP-ribose 15, a reaction that is conserved from yeast to humans 16.
Yeast Sir2, in particular, is conserved from bacteria to humans 17. There are currently 7 known isoforms of Sir2-like proteins in mammals (sirtuins 1-7). Frye et al cloned human SirT1-7, which are ubiquitously expressed and exhibit NAD+-dependent deacetylase activity, and in the case of Sirt4 an ADP-ribosyltransferase activity, with Sirt1 being the most homologous to yeast Sir2 18, 19. Sirt1, Sirt6, and Sirt7 harbor a nuclear localization signal and, accordingly, are predominantly nuclear. However, Sirt1 is also present in the cytoplasm of various cell types under different developmental conditions 20. For example, in E12.5 cardiac myocytes, Sirt1 is exclusively nuclear, whereas in the adult heart it is also found in the cytoplasm. This could be explained by the fact that in addition to its nuclear localization signals, Sirt1 harbors 2 nuclear export signals that allow for its nuclear-cytoplasmic shuttling 20. On the other hand, Sirt2 is predominantly cytoplasmic 21, while Sirt3-5 are predominantly mitochondrial 22–23.
Like its yeast homolog, Sirt1 has also been implicated in the longevity of C. elegans and D. melanogaster by mimicking the effects of calorie restriction 24. Similarly, Sirt1 mediates anti-apoptotic effects of calorie restriction in mammalian cells via deacetylating Ku70, which sequesters Bax 25. Most significantly, it is required for the characteristic increase in physical activity 26 and enhanced sensitivity of muscle to insulin 27 that is associated with calorie restriction. Additionally, overexpression of Sirt1 was sufficient for preventing insulin resistance following a high-fat diet in mice 28. Thus, it is not surprising that sirtuin mimics are being therapeutically exploited in an attempt to improve the quality and duration of life. Some of the main functions of sirtuins discovered to-date will be discussed below and their role in calorie restriction will be highlighted throughout.
Pyridine nucleotides and the regulation of sirtuins’ activity
NAD+ metabolism
The pyridine nucleotides NAD+ and NADP are known for their roles in metabolism as hydride acceptors (forming NADH and NADPH), and subsequently donors, for oxidoreductases. On the other hand, NAD+ also serves as a substrate for 4 classes of enzymes, mono-ADP-ribose transferases (MART), poly-ADP-ribose polymerases (PARP), cyclic-ADP-ribose synthases (cADP), and Sirtuin deacetylases, which accordingly couples them to the metabolic status of the cell. These enzymes utilize NAD+ by hydrolyzing it into nicotinamide (NAM) and ADP-ribose that serves as the acceptor of the acetyl group, thus, forming 2’-O-acetyl-ADP-ribose 15. NAD+ levels are subsequently reconstituted through de novo or nicotinic acid/nicotinamide (vitamin D3 or niacin) salvage biosynthesis pathways (Fig.1). NAM, via a negative feedback loop, can inhibit the deacetylase activity 29, thus, underscoring the importance of the salvage pathways in eliminating this byproduct and restoring NAD+ levels and sirtuins’ activity.
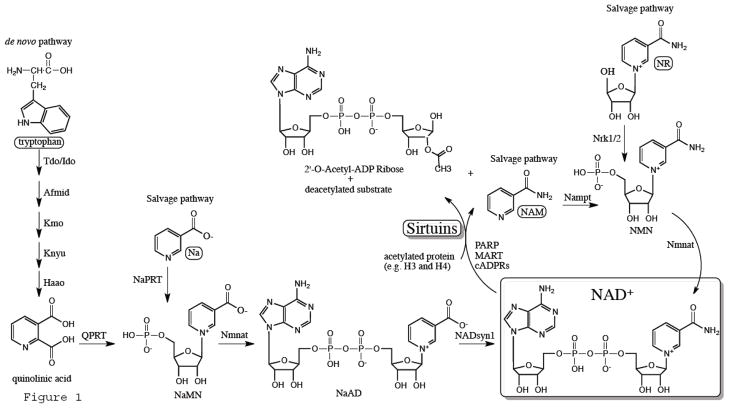
Figure 1.
An illustration of the reactions involved in NAD+ biosynthesis and its consumption by sirtuins. Abbreviation include: mono-ADP-ribose transferases (MART), poly-ADP-ribose polymerases (PARP), cyclic-ADP-ribose synthases (cADP), nicotinamide (NAM), indolamine 2,3-dioxygenase (Ido), kynurenine formamidase (Afmid), kynurenine 3-hydroxylase (Kmo), kynureninase (Knyu), 3-hydroxyanthranilate 3,4-dioxygenase (Haao), nicotinate mononucleotide (NaMN), nictoniate adenine dinucleotide (NaAD), nicotinamide mononucleotide adenylyl transferase (Nmnat), nicotinamide mononucleotide (NMN), phosphoribosyl transferase (NAMPT), nicotinic acid (Na), Na phosphoribosyltransferase (NaPRT), nicotinamide ribosde (NR), nicotinamide riboside kinase (Nrk).
NAD+ can be replenished by de novo synthesis from L-tryptophan, particularly when nicotinic acid or nicotinamide are deficient. First, L-tryptophan is converted to quinolinic acid in the liver via 4 steps catalyzed by indolamine 2,3-dioxygenase, kynurenine formamidase, kynurenine 3-hydroxylase, kynureninase, 3-hydroxyanthranilate 3,4-dioxygenase, respectively (reviewed in detail in ref. 30). Next, a phosphoribosyl group is added to quinolinic acid by quinolinate phosphoribosyltransferase to generate nicotinate mononucleotide (NaMN), which is then converted to nictoniate adenine dinucleotide (NaAD) via nicotinamide mononucleotide adenylyl transferase. The latter enzyme also catalyzes the conversion of nicotinamide mononucleotide (NMN) and NaMN into NAD+ and NaAD, respectively, in the salvage pathways. Finally, NAD+ synthase converts NaAD into NAD+ 30 (Fig. 1).
A major salvage pathway for NAD+ synthesis utilizes NAM, the byproduct of NAD+ consumption by the various glycohydrolases. It was thus suggested that it is the main pathway responsible for reconstituting NAD+ in mammalian cells 30. It involves 2 steps, the conversion of NAM into NMN by NAM phosphoribosyl transferase (NAMPT), which is then converted to NAD+ via NAM mononucleotide adenylyl transferases (NMNATs), NAMPT being the rate-limiting enzyme in this pathway 31. Intracellular Nampt (iNampt) is particularly enriched in brown adipose tissue, heart, muscle, liver, and kidney, but is undetectable in the brain and pancreas 32. It is worth noting that this enzyme is also secreted into the circulation by various tissues including visceral fat, liver, and leuckocytes where it is proposed to function as a pre-B-cell enhancing factor (PBEF) 33, also known as Visfatin, with mixed results regarding its role in diabetes and obesity 34–36. In addition, it exhibits pro-inflammatory functions 37–39, thus linking NAD+ metabolism to inflammation as well.
A second salvage pathway utilizes nicotinic acid (Na). In fungi and bacteria, but not in mammalian cells, nictoinamidase converts NAM into NA. On the other hand, NA maybe supplied to mammalian cells in the form of niacin/vitamin B3, which can be converted into NAD+ in 3 steps 40. First, Na is converted to NaMN by Na phosphoribosyltransferase (NaPRT). Second, NaMN is converted to Na adenine dinucleotide (NaAD), which at this juncture converges on the de novo pathway. Yet a third pathway is activated during H. influenzae infections in humans, in which the hydrolysis of NMN to nicotinamide ribosde (NR) is reversed by nicotinamide riboside kinase 41.
NAD World
“NAD world” is a term introduced by Dr. Imai that describes the functional relation between the circadian cycle, extracellular Nampt (eNampt) levels and systemic NAD+ biosynthesis, Sirt1, and metabolism 42. Intriguingly, it was discovered that Nampt regulates the biosynthesis of NAD+ in a circadian-dependent oscillatory fashion, which, accordingly, adjusts the activity of Sirt1 and the sensitivity of its response to nutritional stimuli 43, 44. Sirt1, through a negative feedback loop, inhibits the circadian cycle by counteracting the acetyltransferase function of CLOCK:BMAL1 - a positive transcriptional regulator of Nampt. This NAD+-driven CLOCK-Nampt-Sirt1 loop has been dubbed “NAD world” 45. Since eNampt is secreted from adipose tissue into the plasma 34–36, the effect of this circadian loop would have a systemic influence. NAD+ as well is found extracellularly, as it is released from cells through connexin43 hemi channels 46. As expected, plasma eNampt converts NAM into NMN, which can be uploaded by cells and converted to NAD+, thereby, regulating whole body NAD+ availability 32.
Organs that are deficient in Nampt, such as neurons and the pancreas, rely on systemic NMN to replenish their NAD+ supply 32, these are known as the frailty points in the NAD world. It has indeed been shown that the pancreas is dependent on eNampt and extracellular NMN for glucose-induced insulin secretion, which may partly explain why dysregulation of the circadian cycle affects glucose metabolism 32. Indeed, metabolism 47–49, as well as aging 50, 51, are known to be under the influence of the circadian rhythm, which further supports their connection to NAD+ world and sirtuins. The brain, which centrally regulates metabolism via Sirt1, is also deficient in iNampt, however, there is no direct evidence yet of how essential circulating NMN is for it function.
Although the cardiovascular system has relatively high iNampt, it could potentially be indirectly impacted by eNampt and NAD+ levels through their effect on pancreatic insulin secretion, or alternatively through circadian fluctuations in iNampt and intracellular NAD+ and Sirt1 activity. Indeed, cardiovascular diseases including ischemia 52, infarction 53, stroke 53, and sudden cardiac arrest 54 have all been shown to have a circadian pattern in the frequency of their occurrence. Until it is verified, it is interesting to speculate that these events may coincide with the periods during which Nampt, NAD+, and Sirt1 levels are at their lowest during the circadian cycle.
PARP and the consumption of NAD+
Fluctuations in PARP activity alone can dramatically perturb the levels of intracellular NAD+ and impact cell viability, as shown in a type I diabetes model 55. In particular, that study shows that activation of PARP by streptozocin resulted in consumption of ~70% of intracellular NAD+ and apoptosis of pancreatic islet cells, both of which were reversed by the knockout of PARP. From those data, one can predict that activation of PARP would compromise the activity of other NAD+-consuming enzymes and vice versa. This was proven to be the case when Bai et al deleted PARP-1 in brown adipose tissue and found that NAD+ availability and Sirt1 activity were increased 56. A reciprocal relation between Sirt1 and PARP may also be true, as deletion of Sirt1 enhanced PARP activity 57.
Transcriptional and posttranscriptional regulation of Sirtuins’ expression
In addition to their regulation by NAD+, sirtuins’ levels are also regulated at the transcriptional and posttranscriptional levels. For example, in nutrient-deprived PC12 rat cells, forkhead box O3a (FoxO3a) interacts with p53 to induce the transcription of the mouse Sirt1 gene via two p53-binding sites in its promoter 58. Under normal conditions, however, p53 suppresses the mouse Sirt1 promoter. In addition to FoxO3, FoxO1 has also been reported to activate the Sirt1 promoter 59. Likewise, the human Sirt1 promoter harbors a p53-binding site that is also required for its activation via a p53-FoxO3a-dependent mechanism during calorie restriction, but differs from the mouse promoter in that p53 has no impact on its activity under normal nutrient availability 60, 61. Interestingly, the p53-binding site in the human Sirt1 promoter exhibits a C/T variation, wherein the C variant reduces p53 binding and, thereby, promoter activity during nutrient restriction 61. In accordance, obese human subjects carrying at least one T allele have higher Sirt1 levels in their skeletal muscle after 3–6 months of calorie restriction. In addition to p53, human and mouse Sirt1 are induced by cAMP response-element-binding protein (CREB) during calorie restriction, which otherwise remains suppressed by carbohydrate response-element-binding protein 27. Additionally, peroxisome proliferator-activated receptor (PPAR) beta/delta indirectly enhances transcription of human Sirt1 via an Sp1 binding-site in its promoter 62. In contrast, to these activating factors, Sirt1 transcription is inhibited by PARP-2 in mice 63.
Sirt1 has the capacity to auto-regulate itself through its interaction with hypermethylated in cancer 1 (HIC1), a complex which inhibits Sirt1’s transcription in both mouse and human fibroblast 64. This effect is reversed when glycolysis is inhibited (for example during calorie restriction), as the redox sensor C-terminal-binding protein (CtBP) interacts with HIC1, thus, releasing Sirt1’s promoter from its inhibitory effect and enhancing transcription 65. The human promoter contains two HIC1-binding sites in close proximity to the p53-binding site, to which HIC1 binding is also reduced during nutrient restriction and replaced by p53 61.
Sirt1 is also differentially regulated under conditions that are unrelated to nutrient availability and in a cell type dependent manner. For instance, E2F1 regulates Sirt1 expression during DNA damage in human non-small cell lung cancer cells 66, while early growth response (Egr-1) is responsible for stretch-induced Sirt1 in skeletal muscle 67. Overall, the data reveal the diversity of transcription factors and conditions that differentially regulate the transcription of Sirt1 and the high degree of their conservation between mice and humans.
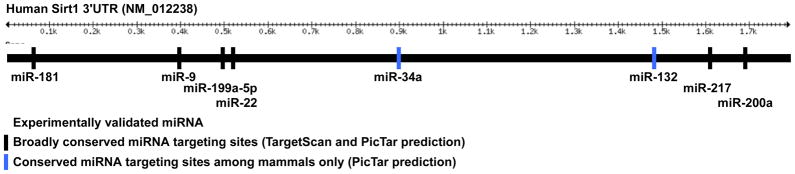
Sirt1 is also regulated at the posttranscriptional level, mainly via miRNAs (Fig. 2). In general, these posttranscriptional regulators have a profound effect on gene expression during development and disease (reviewed in 68). MiR-34a, which is a tumor suppressor and a transcriptional target of p53, directly binds the 3’UTR and suppresses translation of Sirt1 mRNA in human cell lines 69. As a result, overexpression of miR-34a in human colon cancer cells induces downregulation of Sirt1, upregulation of acetylated p53, and, thereby, apoptosis. In endothelial cells, both miR-217 70 and miR-34a 71 target and suppress Sirt1 and promote senescence of human umbilical vein, aortic, and coronary artery endothelial cells 70. In human fibroblast, an increase in miR-22 also induces senescence by targeting Sirt1 72. MiR-132 also targets Sirt1 in human adipocytes, resulting in an increase in acetylated p65 and activation of NFκB 73. In pancreatic β-cells, glucose-dependent insulin secretion is associated with an increase in miR-9, which target and suppresses Sirt1 74. In hippocampal cells, miR-9 and miR-181c target Sirt1 75. In mammary epithelial cells, miR-200a, whose downregulation is required for the upregulation zinc finger E-box binding homeobox proteins and the mediation of endothelial-to-mesenchymal transition 68, also targets Sirt1 76. In cardiac myocytes, miR-199a-5p is rapidly downregulated during hypoxia, which results in an increase both its targets, Sirt1 and Hif-1α 77. However, this increase is reversed upon prolonged deleterious periods of hypoxia. In addition, miR-195 mediates palmitate-induced apoptosis in cardiac myocytes by targeting Sirt1 78. All the miRNAs except for miR-195, are predicted by the one or more of the miRNA target-prediction engines, miRanda, Pictar, or TargetScan, to have a conserved binding site in the 3’UTR of Sirt1 (Fig. 2). Thus, in addition to transcriptional regulation, a wide array of miRNAs regulate the translation of Sirt1 during cell survival, senescence and metabolism, plausibly in a cell type- and condition-specific fashion.
Figure 2.
MicroRNAs that target the 3’UTR of Sirt1. The illustration shows the various miRNAs that are predicted by TargetSan and PicTar applications that target Sirt1 through broadly conserved (black bars) or conserved (excludes chicken, blue bars) sites in its 3’UTR. The ones listed are only those that have been experimentally validated.
Mechanisms of gene regulation by sirtuins
The Sir2 family of proteins was first discovered in yeast as a gene silencer of the silent mating loci and teleomeres 1–4. The underlying mechanism of its function was found to involve deacetylation of histones H3 and H4, which favors the formation of transcriptionally silent heterochromatin 5. In particular, Sir2 deacetylates K9 and K14 of H3 and K16 of H4, which proved to be responsible for its gene silencing and prolongation of lifespan effects in yeast 12. In mammalian cells, Sirt1 deacetylates K9 of H3 and K16 of H4, in addition to its interaction with, and deacetylation of, K26 of H1, also responsible for inducing gene silencing 79. Moreover, Sirt1 deacetylates and induces degradation of the histone H2A variant H2A.z, which was shown to selectively activate gene transcription in cardiac myocyte 80. In yeast, H2A.z prevents the spread of silent heterochromatin into euchromatin regions near telomeres 81. Heterochromatin formation also relies on an increase in histone methylation. It was soon discovered that Sirt1 can promote this function through an interaction with the methyl transferase enzyme SUV39H1, activating it through deacetylation of lysine 266 82. Accordingly, during starvation in mammalian cells, an increase in the NAD+/NADH ratio results in activating Sirt1, which then deacetylates H3 and SUV39H1 and induces silencing of rRNA transcription 83. As a result, energy would be conserved due to the inhibition of the high energy-consuming ribosome biosynthesis. This mechanism potentially links Sirt1 to energy conservation during caloric restriction conditions. Collectively, these findings prove that one of the main mechanisms that are employed by Sirt1 in regulating gene expression involves differential gene silencing via promoting heterochromatin formation (Fig. 3).
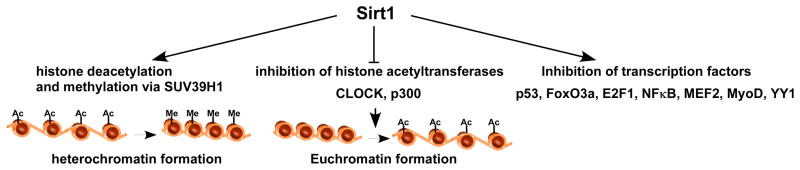
Figure 3.
Modes of transcriptional regulation by Sirt1. Sirt1 regulates transcription via 3 different mechanisms. The first involves histone deacetylation and heterochromatin formation, which results in gene silencing. This is complemented with a hypermethylation effect that is mediated by suppressor of variegation 3–9 homolog 1 (SUV39H1) methylase. The second involves inhibition of histone acetylases p300 and CLOCK, that also contributes to chromatin condensation. The third involves the deacetylation and inhibition or activation of specific transcription factors that include: p53, forkhead box O3a (FoxO3a), E2F transcription factor 1 (E2F), nuclear factor of kappa light polypeptide gene enhancer in B-cells (NFκB), myocyte enhancer factor 2 (MEF2), myogenic differentiation (MyoD), and ying yang 1 (YY1).
Additionally, sirtuins are known to indirectly impact transcription by negatively regulating the function of histone acetylases. Indeed, Sirt1 regulates the circadian clock genes by interacting with the CLOCK histone acetyltransferase and inhibiting its function through deacetylating its partner BMAL1 84. It also deacetylates the CLOCK/BMAL1 regulatory partner Period and induces its degradation 85. Likewise, Sirt1 interacts with deacetylates as it inhibits the histone acetyltransferase p300 86. Other mechanisms employed by Sirt1 in regulating transcription involve direct modulation of transcription factors’ activities via deacetylation. In fact, Sirt1 is involved in a negative feedback loop with some of the factors that regulate its own expression. For example, while the p53/FoxO3a complex induces the expression of Sirt1, Sirt1 binds to, deacetylates, and deactivates, p53 87. It also associates with FoxO3a and inhibits its pro-apoptotic capacity 88, 89. Similarly, Sirt1 deacetylates E2F1 in a negative feedback loop, and inhibits its activity 66, as well as its partner Rb protein 90. Moreover, it inhibits NF-κB 91, myocyte enhancer factor 2 92, myogenic differentiation 93, and ying yang 1 73. Thus, whether it is through epigenetic modifications and chromatin remodeling or through regulating the activity of transcription factors, Sirt1 appears to have a predominantly silencing effect on gene expression (Fig. 3).
Sirtuins in metabolism
Sirt1 in glucose metabolism
Sirt1 has been reported to regulate glucose metabolism at multiple levels, including insulin secretion and sensitivity, gluconeogenesis, and glycolysis. It regulates insulin secretion from pancreatic β-cells via directly inhibiting transcription of uncoupling protein 2 (UCP2) 94, 95. The UCP family of proteins are transporters found in the inner membrane of mitochondria, where UCP2 is induced by dietary fat and reduces the production of mitochondrial ROS, which in-turn results in suppressing glucose-induced insulin secretion 96. Consequently, the β-cell-specific Sirt1-overexpressing mice (BESTO) exhibit enhanced glucose-induced insulin secretion and better glucose tolerance 94. Interestingly, though, these benefits of Sirt1 were lost in the aging mice 97. Since the pancreas does not have detectable intracellular Nampt it relies on the circulating eNampt and NMN for producing NAD+ (see above “NAD+ world”), and because plasma levels of NMN were reduced in older mice, it could explain why Sirt1 became ineffective. Indeed, replenishing NMN restored Sirt1 activity and function in older mice, emphasizing the role of NAD world in age-associated insulin resistance. These results were confirmed by a general Sirt1 transgenic mouse reported to have phenotype that recapitulates that of calorie restriction, which exhibits lower plasma insulin and superior glucose tolerance 98. In addition to regulating insulin secretion, Sirt1 regulates insulin sensitivity. It is reduced in insulin resistant cells and was found to be both necessary and sufficient for improving insulin sensitivity by directly repressing transcription of protein tyrosine phosphatase 1B (PTP1B) 99, which is responsible for dephosphorylation and deactivation of the insulin receptor 100. While all of the above studies were in rodents, in humans, it was shown that Sirt1 levels positively correlated with insulin sensitivity in adipose cells 101.
During fasting, Sirt1 and peroxisome proliferative activated receptor, gamma, coactivator 1 alpha (PGC-1α) are upregulated in hepatocytes, which leads to their direct association and the deacetylation and activation of PGC-1α by Sirt1 102. This enables PGC-1α to promote gluconeogenesis by enhancing the expression of the genes involved in this process, while inhibiting those involved in glycolysis 103. As a consequence, knockdown of Sirt1 in the mouse liver results in reduced gluconeogenesis, mild hypoglycemia and an increase in insulin sensitivity, particularly during fasting 104. In agreement, knockdown of Sirt1 in the liver of a rat model of type 2 diabetes inhibited gluconeogenesis, reduced fasting blood glucose levels, and increased insulin sensitivity, which was associated with increased acetylation of PGC-1α and reduced levels of phosphoenolpyruvate carboxylase kinase and glucose-6- phosphatase 105. These results, have been challenged by Liu et al, who report that during the early phase of fasting, gluconeogenesis is induced by CREB-regulated transcription coactivator 2 (CRTC2) not Sirt1 106. However, during the later stage, Sirt1 suppresses CRTC2, while FoxO1 activates gluconeogenesis. Thus, according to this model, knockdown of Sirt1 in the liver increases CRTC2 activity and, thereby, gluconeogenesis.
Yet another study shows that calorie restriction decreases, not increases, NAD+ and Sirt1 activity in the liver, which is contrary to what is observed in white adipose tissue and muscle 107. According to this study, mice with liver-specific knockout of Sirt1 are normal when maintained on a regular or calorie-restricted diet, and did not differ in any aspect, including gluconeogenesis, from their wild-type counterpart. On the other hand, when fed a high fat diet they had less body and liver fat, hypoglycemia, low insulin, and better glucose tolerance v. wild type. This contrasts with another study that shows that the same mice displayed an increase in hepatic lipids and a decrease in fatty acid oxidation 108.
Since glucose homeostasis is a concerted function of the whole body and not that of a single organ, it may be more relevant to examine the role of Sirt1 in a systemic fashion. There are two general Sirt1 transgenic models that were engineered for that purpose, one with a Sirt1 knock-in into the beta actin gene 98 and another Sirt1 bacterial artificial chromosome overexpressor (BACO) 28. The first model is characterized by a leaner mouse that has lower levels of plasma fatty acids, leptin, and adiponectin, and improved glucose tolerance. Although the reduced levels of adiponectin here is hard to reconcile with its role in increasing insulin sensitivity 109. The second model showed no difference in body weight, but conferred protection against type 2 diabetes during high fat intake and in the db/db mice due to increased hepatic insulin sensitivity, which may be an effect of hyperadiponectinemia secreted from white adipose tissue. Sirt1, plausibly via a decrease in acetyl-FoxO1 and an increase in adiponectin, also increased the levels of phospo-AMPK and PPARα. One major difference that could account for some of the discrepancies between the 2 models is the lack of an increase in hepatic Sirt1 in the first model. In agreement with these models, injecting mice with the Sirt1-specific activator SRT1720 reduced obesity and improved insulin sensitivity after being fed a high fat diet, with no change in gluconeogenesis 110.
Thus, while liver-specific knockout of Sirt1 improved insulin sensitivity in some reports, systemic overexpression of Sirt1 also had a similar effect, in seemingly inconsistent outcomes. Since glucose homeostasis is the net effect of its regulation by various organs, we may conclude that the systemic effects of Sirt1 may be a more faithful representation of its in vivo effect on glucose metabolism. In support, Sirt1 levels were lower than normal in humans exhibiting insulin resistance and metabolic syndrome, albeit that this study involved only 13 subjects 111. Moreover, incubating human monocytes with high doses of glucose reduced Sirt1 and Nampt, and increased p53 111. The results also confirm the conservation of some of Sirt1’s functionality between rodents and humans.
Sirt1 in lipid metabolism
Sirt1, like calorie restriction, functions to preserve body energy through increasing fatty acid oxidation. This effect is partly mediated through the deacetylation and activation of the transcriptional coactivator PGC-1α in skeletal muscle 112. In addition to enhancing the transcription of gluconeogenic genes, PGC-1α associates with PPARα and activates the transcription of genes involved in fatty acid oxidation 113. Both Sirt1 transgenic models described above support a role for Sirt1 in promoting fatty acid oxidation. One model, where the body weight was normal when maintained on a regular diet, responded to food restriction by increasing fatty acid oxidation 98. In the second model, the mice were leaner and exhibited lower circulating fatty acids, LDL, and HDL, in addition to higher O2 consumption, plausibly also reflecting an increase in fatty acid oxidation 28. More conclusively, specific activation of Sirt1 in vivo by SRT1720 results in an increase in fatty acid metabolism in the liver, muscle, and brown adipose tissue, with less accumulation of lipid in the liver and white adipose tissue 110. This, however, was not associated with an increase in mitochondrial density or oxidative phosphorylation genes, but was associated with an increase in enzymes involved in fatty acid oxidation in all 3 organs. Notably, this phenotype was associated with reduced acetylation of the Sirt1 targets, PGC-1α, FoxO1, and p53. In addition, chronic treatment with SRT1720 resulted in activation of AMP-activated protein kinase AMPK, possibly as a result of reduced ATP levels 110.
Conversely, mice with a liver-specific deletion of Sirt1 exhibited lower fatty acid oxidation after fasting, while accumulating more lipid in the liver and white adipose tissue when fed a Western diet 108. This was associated with reduced expression of genes involved in fatty acid oxidation. In addition, there was an increase in hepatic cholesterol due to a reduction in the enzymes that converts it into bile. In this model, PPARα, a known regulator of genes involved in fatty acid oxidation, was found to directly interact with Sirt1 in the liver, an association that was required for deacetylating its coactivator PGC-1α. Similarly, the Sirt1+/− mouse exhibited an increase in liver triglycerides, glycerol, cholesterol, and body fat, after a moderate fat diet, possibly as a result of increased lipogenesis 114. Fatty acid oxidation remained unchanged under these conditions, but was unexamined during calorie restriction. In contrast to the above studies, another shows that Sirt1−/− mice have reduced plasma HDL-cholesterol and triglycerides when fed a normal diet, which has been attributed to Sirt1’s requirement for the deacetylation and activation of the liver x receptor (LXR) that regulates both lipid and cholesterol homeostasis 115. Overall, however, the consensus derived from these studies is that Sirt1 enhances fatty acid oxidation and reduces body fat.
Sirt6 in metabolism
Sirt1 is not the only sirtuin to regulate metabolism; it appears that nuclear Sirt6 is involved in mediating some of Sirt1’s effects. Although Sirt6−/− mice have a phenotype that is mainly characterized by defects in the excision/repair system and genomic instability, it is also associated with extremely low levels of insulin-like growth factor-1 and serum glucose 116. It was found that Sirt1/FoxO3a/nuclear respiratory factor 1 complex activates the expression of Sirt6 during caloric restriction. Sirt6 then through deacetylating H3K9, reduces the expression of genes involved in glycolysis and triglyceride synthesis, while increasing those required for fatty acid oxidation, as evidenced by a liver-specific knockout of Sirt6 117. This model also suffers from fatty liver. On the other hand, a transgenic mouse overexpressing Sirt6 has a phenotype that is reminiscent of Sirt1 transgenics when fed a high fat diet 118. In specific, these mice accumulate less visceral fat, triglycerides, and LDL-cholesterol, in addition to improved glucose-induced insulin secretion and glucose tolerance. Thus, there appears to be redundancies in the functions of some sirtuin family members that may account for some of the variations observed between the different transgenic and knockout animal models.
Sirtuins in mitochondrial metabolism and biogenesis
From the discussion above it became evident that the nuclear sirtuins, Sirt1 and Sirt6, impact mitochondrial metabolism through upregulating the transcription of nuclear genes involved in fatty acid oxidation via deacetylating and activating PGC-1α 112. This protein has also been implicated in the biogenesis of mitochondria through inducing upregulation of mitochondrial transcription factor A 119. Notably, though, a complementary role for PGC-1β is required for this process 120. Although resveratrol 121, exercise 122, and calorie restriction 123 are associated with an increase in Sirt1 and mitochondrial biogenesis, it remains uncertain whether Sirt1 is sufficient or even required 124 for increasing mitochondrial biogenesis. On the other hand, Sirt1 may reduce mitochondrial numbers through a Bnip3-dependent protective mitophagy during calorie restriction, as seen in kidney cells 125.
In addition to the nuclear sirtuins, there are other family members that are localized to the mitochondria (Sirt3-5) that have been shown to directly regulate the activity of some of the genes involved in mitochondrial functions (Fig. 4). Sirt3 is characterized as a NAD+-dependent histone deacetylase that is localized to the mitochondrial matrix 22, 126. Mice deficient in Sirt3, but not Sirt4 or Sirt5, exhibited an increase in acetylated mitochondrial proteins, including glutamate dehydrogenase (GDH), but with no overt phenotype at basal or fasting conditions 23. However, embryonic fibroblasts isolated from Sirt3−/− mice had a lower ATP content, which was attributed to the requirement of Sirt3 for the activation of Complex I through its interaction with, and deacetylation of, the NDUFA9 subunit 127. Sirt3 also deacetylates and activates acyl-CoA synthetase 2, which catalyzes the synthesis of acetyl-CoA 128, 129.
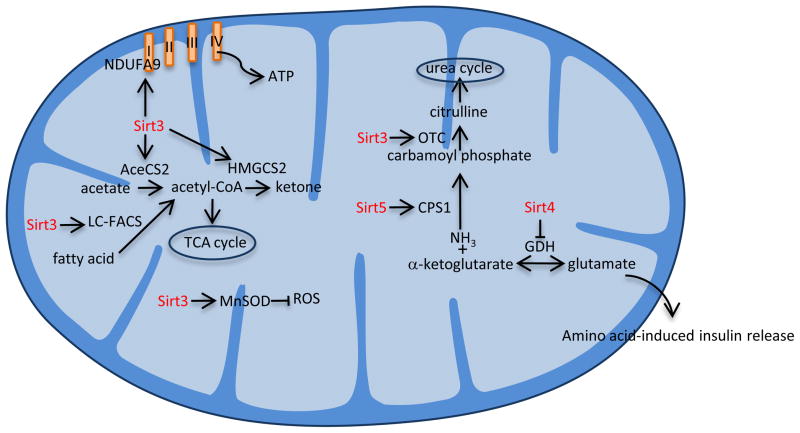
Figure 4.
Mitochondrial function of sirtuins. Sirt3 deacetylates and activates acetyl-CoA synthetase 2 (AceCS2) and long chain fatty acyl-CoA (LC-FACS), which convert acetate and fatty acid, respectively, into acetyl-CoA. It also activates 3-hydroxy-3methylglutaryl CoA synthase 2 (HMGCS2), which is involved in ketogenesis; ornithine transcarbamoylase (OTC), which is involved in the urea cycle; manganese superoxide dismutase (MnSOD), which inhibits reactive oxygen species (ROS) production; and NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9 (NDUFA9), which a subunit of complex I in the electron transport chain. Sirt4 inhibits glutamate dehydrogenase (GDH), which is involved in the synthesis of glutamate and amino acid-induced insulin release, while Sirt5 is also involved in the urea cycle through activating carbamoyl-phosphate synthase 1 (CPS1).
Upon a closer look, it was noted that Sirt3−/− mice exhibited lower levels of fatty acid oxidation in the liver that was associated with reduced ATP and higher acetylated long-chain acyl-CoA synthetase levels 130. In addition, the knockout mice exhibited a defect in the urea cycle due to hyperacetylation and deactivation of ornithine transcarbamoylase enzyme 131. Ketogenesis was also impacted due to hyperacetylation and deactivation of the rate limiting mitochondrial 3-hydroxy-3methylglutaryl CoA synthase 2 132. In addition, Sirt3 deacetylates and activates manganese superoxide dismutase (MnSOD), thus, reducing mitochondrial superoxide, as evidenced by studies in the liver of Sirt3−/− mice133, 134.
Calorie restriction induces Sirt3-dependent deacetylation of MnSOD as a means of reducing oxidative stress 134. In contrast, high-fat diet results in downregulation of Sirt3 and hyperacetylation of liver mitochondrial protein 135. Further reduction in Sirt3 levels by genomic deletion enhanced the development of obesity, insulin resistance, hyperlipidemia, and hepatic steatosis in these mice. This phenotype is partly attributed to an increase in stearoyl-CoA desaturase 1. There is evidence to suggest that Sirt3 may have a similar function in humans, where a single nucleotide polymorphism that reduced the activity of Sirt3 was found associated the metabolic syndrome 135.
While mitochondrial protein deacetylation is beneficial during calorie restriction and in metabolic syndrome, it should be noted that this effect might enhance hepatotoxicity by drugs that bind to lysine residues made accessible by deacetylation. Indeed, it was recently shown that fasting exacerbates, while Sirt3 knockout ameliorates, acetaminophine-induced liver injury in mice 136. One of the major targets of Sirt3 deacetylase activity that seems responsible for this function is aldehyde dehydrogenase 2 (ALDH2), which is known to oxidize and, thereby, detoxify aldehydes including the acetaminophine metabolite N-acetyl-p-benzoquinoneimine. This is an example of why precaution should be exercised when considering the benefits of calorie restriction if combined with any drugs, even common analgesics.
As Sirt3 takes center stage in mitochondrial metabolism, little is known about the functions of mitochondrial Sirt4 and Sirt5. Unlike the rest of the family members, Sirt4 is mainly an ADP-ribosyltransferase that is ubiquitously expressed. It ADP-ribosylates glutamate dehydrogenase (GDH), leading to its inhibition 137. A Sirt4−/− mouse does not have any overt defects, but does exhibit an increase in GHD activity, but not protein, that is a result of a reduction in its ADP-ribosylation 137. GDH is the enzyme involved in the reversible reaction involving synthesis of glutamate from ammonia and α-ketoglutarate. It was expected and subsequently proven that Sirt4 inhibits this function and, thus, indirectly inhibits amino acid-induced insulin release. Sirt5, on the other hand, regulates the urea cycle by deacetylating and activating carbamoyl phosphate synthetase 1 138, which is the enzyme involved converting ammonia into carbamoyl phosphate. As a result, Sirt5−/− mice develop hyperammonemia during fasting or a high protein diet 138. Notably, a recent finding shows that Sirt5 is also a NAD+-dependent protein lysine demalonylase and desuccinylase, which in addition to deacetylating can desuccinylate carbamoyl phosphate 139.
Sirtuins in the heart
Sirtuins in myocardial ischemia and infarction
Sirt1, Sirt3, and Sirt7 have beneficial roles in the heart that can retard the damaging effects of aging and disease (Fig. 5). It was first noted that NAM-induced inhibition of Sirt1 in cardiac myocytes leads to p53-dependent programmed cell death and, conversely, forced expression of Sirt1 rescues the cells from starvation-induced apoptosis 140. A role for Sirt1 in myocyte survival became evident during cardiac ischemia/reperfusion (I/R) when Sirt1 was downregulated, whereupon further reduction of the protein by cardiac-specific deletion led to aggravating the damage 141. This function was confirmed with replenishment of Sirt1 by a cardiac-specific transgene, which reduced the size of the infarct and enhanced functional recovery. The effects of Sirt1 here are at least partly mediated through activation of FoxO1 and its downstream target, manganese superoxide dismutase. Similarly, Sirt1 reduced paraquat-induced oxidative stress in the heart 142 and mediated the cardioprotective effect of mIGF 143. The role of Sirt1 in cardioprotection was further substantiated by data that showed that reconstituting Nampt levels in the heart during I/R restored NAD+ content and reduced injury 144.
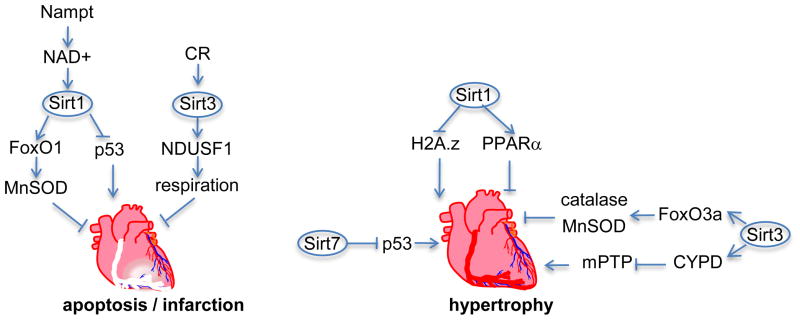
Figure 5.
Sirtuins inhibit cardiac ischemic injury and hypertrophy. Overexpression of phosphoribosyl transferase (NAMPT) in the heart increases NAD+ biosynthesis through the salvage pathway, which activates Sirt1. Sirt1 through inhibition of p53 and activation of forkhead O1 (FoxO1) and its downstream target manganese superoxide dismutase (MnSOD) reduces infarct size after ischemia/reperfusion. On the other hand, calories restriction (CR) activates Sirt3, which protects the heart through activation of the mitochondrial protein NADH dehydrogenase (ubiquinone) Fe-S protein 1 (NDUSF1), a subunit of complex I in the electron transport chain. Sirt1 inhibits hypertrophy by deacetylating and inducing degradation of histone 2A.z (H2A.z), as well as activating peroxisome proliferator-activated receptor alpha (PPARα). Sirt3 inhibits hypertrophy via activating forkhead O3a (FoxO3a) and cyclophilin D (CYPD), which inhibit the mitochondrial permeability transition pore (mPTP). Sirt 7 inhibits hypertrophy via deacetylating and inhibiting p53.
Calorie restriction and sirtuins in the heart
Several reports show that calorie restriction can confer cardioprotection during aging, myocardial ischemia, or hypertrophy 145–148, however, the exact mechanism has not been fully elucidated. Although, we have learned that sirtuins play a major role in mediating the beneficial metabolic effects of calorie restriction, there is limited direct evidence that they mediate its cardioprotective effects. For example, one study shows that long-term calorie restriction, which is associated with increased Sirt1 and reduced H3 acetylation, reduced infarct size and improved recovery after I/R in a Sirt1-dependent fashion 149. Another reports that caloric restriction protected the mitochondria from I/R-induced reduction in respiration, which was associated with increased deacetylation of mitochondrial proteins, including the Sirt3 substrate NDUFS1 150. More recently, it was reported that ischemia preconditioning shares a common feature with caloric restriction in the form of a reduction in acetylated proteins and enhanced Sirt1 activity/expression in the heart 77, 151.
Calorie restriction activates AMPK, which, like Sirt1, mediates some of its metabolic effects 152. On the other hand, AMPK activates Sirt1, which has also been shown to mediate its effects 153. Like calorie restriction, AMPK and Sirt1 have been independently shown to have cardioprotective effects. Thus, we may extrapolate from this that the cardioprotective effects of calorie restriction are at least in part mediated by AMPK via Sirt1.
Sirtuins in cardiac hypertrophy
Sirt1 exerts anti-hypertrophic effects in the heart, as it reduces aging-related cardiac hypertrophy, fibrosis, and contractile dysfunction, in cardiac-specific Sirt1 transgenic mice expressing 2.5–7.5 fold higher levels of Sirt1 142. Also, forced expression of Sirt1 in cardiac myocytes inhibits endothelin-1 80 and phenylephrine-induced cardiac hypertrophy 154. This is mediated in part by deacetylation and degradation of the histone variant H2A.z 80 and the deacetylation and activation of peroxisome proliferator-activated receptor-α (PPARα) 154. It should be noted, that Sirt1 increases during pressure overload-induced hypertrophy up to 4–8 fold, possibly as an adaptive mechanism, which, in addition to curbing hypertrophy, reduces myocyte apoptosis 80, 155. In contrast, however, 12.5-fold higher Sirt1 in the heart exacerbates age-induced cardiac hypertrophy and dysfunction 142. In concordance with this latter observation, Sirt1+/− mice reduced the extent of pressure overload-induced hypertrophy and normalized ejection fraction 156. This effect of Sirt1 was mediated through its interaction with PPARα, as the complex inhibited targets of estrogen-related receptor, which include genes regulating mitochondrial metabolism and cardiac contraction 156. Thus, it would appear that moderate levels of Sirt1 that accumulate during the early stage of hypertrophy exert an anti-hypertrophic effect, but a spike in Sirt1 concentration during the later stages of hypertrophy and failure contributes to cardiac dysfunction.
In contrast to nuclear Sirt1 and Sirt6, little is known about the third nuclear family member Sirt7, apart from the fact that it associates with rDNA and RNA polymerase I and enhances transcription of ribosomal RNA 157. In the heart, deletion of Sirt7 in mice induced cardiac hypertrophy and inflammation, and reduced life span 158. Similar to Sirt1, Sirt7 deacetylates and inactivates p53 as a plausible mechanism for the observed phenotype. Mitochondrial Sirt3 also plays a role in the development of cardiac hypertrophy. Sirt3−/− mice exhibit exaggerated cardiac hypertrophy after transverse aortic banding 159, 160, as well as accelerated age-related cardiac hypertrophy and fibrosis that is manifest at 13 months of age 160. Conversely, overexpression of Sirt3 attenuated angiotensin II- and isoproterenol-induced hypertrophy 160. The mechanisms underlying its function involves the inhibition of mitochondrial permeability transition pore (mPTP) opening, since it was noted that the Sirt3−/− mouse is associated with hyperacetylation of cyclophilin D and mitochondrial swelling 160, in addition to deacetylation and activation of FoxO3a and its downstream targets, manganese superoxide dismutase and catalase 159. Thus, both nuclear Sirt1 and Sirt7, and mitochondrial Sirt3 all have anti-hypertrophic functions, albeit through different mechanisms. Additionally, Sirt1 also exhibits pro-hypertrophic functions that may be dependent on a higher dosage.
Situins in the vascular system
Steady laminar flow, such as that observed in the thoracic aorta, is responsible for higher levels of Sirt1 in the endothelial lining, which account for lower levels of acetylated eNos through a direct association and deacetylation reaction 161. This leads to activation of eNos, which promotes vasodilatation 162 and alleviates vasoconstriction after a high fat diet 163. In contrast, oscillatory flow such as that seen in the atherosclerotic prone aortic arch, does not induce an increase in Sirt1, but does increase the expression of PARP-1 161. As discussed above, PARP-1 is a NAD+ consumer that could deplete the substrate pool and reduce Sirt1’s activity, which could promote atherogenesis, as described below. It should be noted though, that the effect of Sirt1 on vascular tone was challenged by another study, in which no significant differences were detected in vascular relaxation between ApoE−/−/SIRT1+/− and ApoE−/−/SIRT1+/+ mice 164.
Sirt1 is also required for the budding and migration of endothelial cells, as well as, ischemia-induced neovascularization after hind limb ischemia 165. This function appears to be mediated through its interaction with FoxO1 and the regulation of an array of genes that are involved in vascular growth including fms-related tyrosine kinase 1, chemokine (C-X-C motif) receptor 4, platelet-derived growth factor receptor β, angiopoietin- like 2, matrix metallopeptidase 14, and EPH receptor B2. More recently, it was discovered that Sirt1 regulates endothelial cell budding via deacetylating Notch’s intracellular domain 166. Sirt1 also promotes the longevity of endothelial cells, as Nampt, which is the rate-limiting enzyme in the conversion of NAM to NAD+, was shown to retard senescence that is precipitated by high glucose levels via activation of Sirt1 167. Conversely, in senescent endothelial cells, miR-34a 168 and miR-217 70 are upregulated, both of which directly target and suppress the expression of Sirt1 mRNA. Moreover, overexpression of either miRNA accelerated senescence and reduced angiogenesis via suppressing Sirt1 and increasing acetylated FoxO1.
There have been conflicting reports on the role of Sirt1 in atherogenesis. Some studies suggest that Sirt1 has an anti-atherogenic effect since it is reduced in human 70 and in ApoE−/− mice 163 atherosclerotic plaques. Its anti-atherogenic effect may be partly mediated via inhibiting NFκB signaling in endothelial cells and reducing inflammation 164. Additionally, Sirt1 has been shown to reduce neointimal hyperplasia via inhibiting vascular smooth muscle proliferation and migration 169. A contrasting study, however, shows that Sirt1-overexpressing mice have higher levels of triglycerides and cholesterol, and accordingly larger atherosclerotic plaques 170. It is plausible that the extent of Sirt1 overexpression dictates the nature of its effects, as was observed in the heart, where higher levels of the overexpressed Sirt1 had an adverse effect. Thus, it remains to be seen if precise normalization of endogenous Sirt1 levels during atherogenesis would affect the outcome.
Sirtuins in the central nervous system
As discussed earlier, sirtuins regulate metabolism through their local effects in the liver, pancreas, skeletal muscle, and adipose tissue. However, they also have the capacity to centrally regulate metabolism via regulating functions in the pituitary gland and hypothalamus. It was found that deletion of Sirt1 in neurons, astrocytes and glial cells resulted in dwarfism, associated with smaller pituitary gland / body weight ratio that produced lower levels of growth hormone, in addition to glucose intolerance that was manifest in older mice only 171. More significantly, calorie restriction-enhanced insulin sensitivity and physical activity were abolished in these mice, suggesting that the beneficial effects of calorie restriction are mainly regulated by the central activity of Sirt1 in the brain. In specific, it was discovered that chronic diet restriction induces upregulation of Sirt1 in the dorsomedial and lateral hypothalamic nuclei and increases their activity, as well as increase thermogenesis and physical activity 172. Gain- and loss-of-Sirt1 function in vivo proved that under these conditions Sirt1 induces the expression of orexin type 2 receptor in the dorsomedial and lateral hypothalamic nuclei, which in-turn increases their sensitivity to the stomach-released hormone gherlin. However, it isn’t clear how this phenotype reconciles with the known role of gherlin in increasing food intake. Gherlin, which is involved in various eating disorders, also regulates appetite and food intake via inhibiting the activity of proopiomelanocortin (POMC) neurons in the hypothalamic arcuate nuclei (reviewed in 173). These neurons secret α-melanocyte stimulating hormone, which suppresses appetite and increases thermogenesis. Sirt1 is also expressed in POMC neurons, where its deletion results in a significant increase in body weight (females more than males) versus wild type when the mice are maintained on high fat diet 174. The main cause of obesity in these Sirt1 mutant mice appears to be a result of reduced O2 consumption and thermogenesis, which ensued before weight gain. It is important to note that food intake, ambulation, and energy source, were unchanged in these mice. The phenotype may be attributed to noted defects in leptin signaling or reduced sympathetic nerve activity in the perigonadal white adipose tissue. This contrasts with the phenotype of a mouse in which Sirt1 was deleted in Agrp neurons 175. In this model, the Agrp neurons, which stimulate PMOC, were less sensitive to gherlin, resulting in reduced food intake and reduced body fat. Thus, Sirt1 has neuron-specific functions, however, the outcome of its combined effects in the brain may be best gauged in the model in which Sirt1 was deleted in all neurons 171.
Sirt1’s function in the brain extends beyond the regulation of metabolism through the hypothalamus. Recent studies suggest that it plays a role in cognition 176, 177 via regulating miR-134 177. MiR-134 is a brain-restricted miRNA, increasing gradually in the hippocampus where it plateaus at P13 after synaptic maturation and then becomes localized to dendrites near synaptic sites 178. Overexpressing it in hippocampal cells decreases the volume of dendritic spines and vice versa, an effect that is mediated by Lim domain-containing kinase-1 (LIMK-1). In agreement, a knockout model of this protein exhibits a similar phenotype 179. The dendritic spines are known to increase in size during synaptic excitation and, thereby, establish sites of synaptic contact. It was recently found that a Sirt1-YY1 complex suppresses miR-134 177. In this study, miR-134 was shown to suppress the expression of cAMP responsive element binding protein (CREB) and brain-derived neurotrophic factor (BDNF). In accordance, knockout of Sirt1 in the brain reduced synaptic plasticity and impaired memory in mice 176, 177. Another characteristic of advanced age that is regulated by Sirt1 includes reduced wakefulness 180. Accordingly, Sirt1-deficient mice exhibit reduced spontaneous wake time that is associated with reduced wake-active neuron neurotransmitters or the rate limiting enzymes that are involved in their synthesis.
A protective effect of Sirt1 against neurodegeneration was first recognized upon discovering that the protein Wlds, which is a fusion between ubiquitin assembly protein Ufd2a and Nmnat1, confers a delay of axonal degeneration in the Wallerian degeneration slow mice 181. In addition, Sirt1-induced α-secretase activity and reduction of Aβ peptides in cultured neurons from a mouse model of amyloid neuropathy, suggested that it might have a beneficial effect in Alzheimer’s disease (AD) 182. This was indeed confirmed in vivo in a mouse model of AD, in which injection of a Sirt1-expressing vector into the hippocampus, ameliorated neuronal degeneration 183. Likewise, overexpressing of Sirt1 in the brain reduced β-amyloid plaque formation in an AD mouse model by enhancing the expression of α-secretase gene (ADAM10) via deacetylating and activating its regulator, the retinoic acid receptor 184. Thus, increasing Sirt1 in the brain can potentially retard some of the aging-related pathologies.
Concluding remarks
Overall, the experimental evidence is quite compelling regarding the beneficial effects of sirtuins on metabolic homeostasis in both rodents and humans. This is particularly evident during calorie restriction, which elicits upregulation of Sirt1 and Sirt3. Sirt1 exerts its effects on metabolism both locally in the liver, pancreas, skeletal muscle, and adipose tissue, and centrally through regulating functions of the pituitary gland and hypothalamus. The results of Sirt1 knockout in the mouse brain, would argue that the effects of calorie restriction are mainly governed by neuronal Sirt1. Since neurons are deficient in Nampt, neuronal Sirt1 activity is expected to be sensitive to circulating levels of eNampt and NMN, and thereby, the general metabolic status of the whole body. The findings to-date show that the mechanisms underlying the metabolic effects of Sirt1 are mainly mediated through regulating the activity of several key nuclear transcription factors involved in the transcription of metabolic genes via its deacetylase activity. An equally critical and complementary role for Sirt3 in metabolism has also been described, which involves direct regulation of mitochondrial enzymes via its deacetylase activity. One should recall, though, that sirtuins are also histone deacetylases, with the potential for inducing heterochromatin formation and gene silencing. Until know we have little if any knowledge of whether this mechanism plays a major role are during calorie restriction in mammalian cells. Current state-of-the-art technologies, including high resolution profiling of histone modifications and chromatin remodeling, can be expected to help address this question.
The effects of Sirt1, in particular, extend beyond metabolism in various organs and cell types. For example, in the heart it protects it against ischemia/reperfusion damage, potentially by preventing p53-mediated apoptosis, although an equally beneficial metabolic effect as well has not been ruled out. Additionally, during cardiac hypertrophy and failure, Sirt1 regulates the transcription of contractile proteins, as well as, transcriptional regulators of metabolism. Sirt1 also impacts the vasculature, as it regulates vascular tone via deacetylating eNos and enhances endothelial cell migration and budding, while inhibiting smooth muscle cell proliferation. These are just a few examples that were described in this review, but the array of its functionality in other organs and cell types is vast and includes both metabolic and non-metabolic roles. However, since the activity of sirtuins is closely linked to the metabolic status of the body through the availability of NAD+, the function of this NAD+-dependent deacetylase family during metabolism remains a central focus.
Acknowledgments
The author thanks all the present and past lab members for their hard work, dedication, and contribution to research. The author also thanks Dr. Junichi Sadoshima, Chairman of the Department of Cell Biology and Molecular Medicine, for his support.
Sources of funding
The research in the author’s laboratory is supported by NIH grants HL057970 and HL104115.
List of non-standard abbreviations
- NAD+
nicotinamide adenine dinucleotide
- Sirt
sirtuin
- PARP
poly-ADP-ribose polymerases
- NAM
nictinamide
- ADP
adenosine diphosphate
- MNM
nicotinamide mononucleotide
- Nampt
phosphoribosyl transferase
- eNampt
extracellular Nampt
- FoxO
forkhead box O
- PPAR
peroxisome proliferator-activated receptor
- PGC-1α
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha
- I/R
ischemia/reperfusion
Footnotes
Disclosures
None.
References
- 1.Ivy JM, Klar AJ, Hicks JB. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rine J, Strathern JN, Hicks JB, Herskowitz I. A suppressor of mating-type locus mutations in Saccharomyces cerevisiae: evidence for and identification of cryptic mating-type loci. Genetics. 1979;93:877–901. doi: 10.1093/genetics/93.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shore D, Squire M, Nasmyth KA. Characterization of two genes required for the position-effect control of yeast mating-type genes. Embo J. 1984;3:2817–2823. doi: 10.1002/j.1460-2075.1984.tb02214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 5.Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 6.Tsukamoto Y, Kato J, Ikeda H. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature. 1997;388:900–903. doi: 10.1038/42288. [DOI] [PubMed] [Google Scholar]
- 7.Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 8.Gotta M, Strahl-Bolsinger S, Renauld H, Laroche T, Kennedy BK, Grunstein M, Gasser SM. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. Embo J. 1997;16:3243–3255. doi: 10.1093/emboj/16.11.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 10.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kennedy D, Wood SA, Ramsdale T, Tam PP, Steiner KA, Mattick JS. Identification of a mouse orthologue of the human ras-GAP-SH3-domain binding protein and structural confirmation that these proteins contain an RNA recognition motif. Biomed Pept Proteins Nucleic Acids. 1996;2:93–99. [PubMed] [Google Scholar]
- 12.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 13.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci U S A. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borra MT, O'Neill FJ, Jackson MD, Marshall B, Verdin E, Foltz KR, Denu JM. Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases. J Biol Chem. 2002;277:12632–12641. doi: 10.1074/jbc.M111830200. [DOI] [PubMed] [Google Scholar]
- 17.Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 18.Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 19.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 20.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 21.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 22.Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci U S A. 2002;99:13653–13658. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 25.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, deCabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. Epub 2004 Jun 2017. [DOI] [PubMed] [Google Scholar]
- 26.Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005:310. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- 27.Noriega LG, Feige JN, Canto C, Yamamoto H, Yu J, Herman MA, Mataki C, Kahn BB, Auwerx J. CREB and ChREBP oppositely regulate SIRT1 expression in response to energy availability. EMBO Rep. 2011;12:1069–1076. doi: 10.1038/embor.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banks AS, Kon N, Knight C, Matsumoto M, Gutierrez-Juarez R, Rossetti L, Gu W, Accili D. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 30.Magni G, Amici A, Emanuelli M, Orsomando G, Raffaelli N, Ruggieri S. Enzymology of NAD+ homeostasis in man. Cell Mol Life Sci. 2004;61:19–34. doi: 10.1007/s00018-003-3161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 32.Revollo JR, Korner A, Mills KF, Satoh A, Wang T, Garten A, Dasgupta B, Sasaki Y, Wolberger C, Townsend RR, Milbrandt J, Kiess W, Imai S. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab. 2007;6:363–375. doi: 10.1016/j.cmet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samal B, Sun Y, Stearns G, Xie C, Suggs S, McNiece I. Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol Cell Biol. 1994;14:1431–1437. doi: 10.1128/mcb.14.2.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haider DG, Schaller G, Kapiotis S, Maier C, Luger A, Wolzt M. The release of the adipocytokine visfatin is regulated by glucose and insulin. Diabetologia. 2006;49:1909–1914. doi: 10.1007/s00125-006-0303-7. [DOI] [PubMed] [Google Scholar]
- 35.Lopez-Bermejo A, Chico-Julia B, Fernandez-Balsells M, Recasens M, Esteve E, Casamitjana R, Ricart W, Fernandez-Real JM. Serum visfatin increases with progressive beta-cell deterioration. Diabetes. 2006;55:2871–2875. doi: 10.2337/db06-0259. [DOI] [PubMed] [Google Scholar]
- 36.Pagano C, Pilon C, Olivieri M, Mason P, Fabris R, Serra R, Milan G, Rossato M, Federspil G, Vettor R. Reduced plasma visfatin/pre-B cell colony-enhancing factor in obesity is not related to insulin resistance in humans. J Clin Endocrinol Metab. 2006;91:3165–3170. doi: 10.1210/jc.2006-0361. [DOI] [PubMed] [Google Scholar]
- 37.Brentano F, Schorr O, Ospelt C, Stanczyk J, Gay RE, Gay S, Kyburz D. Pre-B cell colony-enhancing factor/visfatin, a new marker of inflammation in rheumatoid arthritis with proinflammatory and matrix-degrading activities. Arthritis Rheum. 2007;56:2829–2839. doi: 10.1002/art.22833. [DOI] [PubMed] [Google Scholar]
- 38.Jia SH, Li Y, Parodo J, Kapus A, Fan L, Rotstein OD, Marshall JC. Pre-B cell colony-enhancing factor inhibits neutrophil apoptosis in experimental inflammation and clinical sepsis. J Clin Invest. 2004;113:1318–1327. doi: 10.1172/JCI19930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oki K, Yamane K, Kamei N, Nojima H, Kohno N. Circulating visfatin level is correlated with inflammation, but not with insulin resistance. Clin Endocrinol. 2007;67:796–800. doi: 10.1111/j.1365-2265.2007.02966.x. [DOI] [PubMed] [Google Scholar]
- 40.Magni G, Amici A, Emanuelli M, Raffaelli N, Ruggieri S. Enzymology of NAD+ synthesis. Adv Enzymol Relat Areas Mol Biol. 1999;73:135–182. doi: 10.1002/9780470123195.ch5. [DOI] [PubMed] [Google Scholar]
- 41.Sasiak K, Saunders PP. Purification and properties of a human nicotinamide ribonucleoside kinase. Arch Biochem Biophys. 1996;333:414–418. doi: 10.1006/abbi.1996.0409. [DOI] [PubMed] [Google Scholar]
- 42.Imai S. “Clocks” in the NAD World: NAD as a metabolic oscillator for the regulation of metabolism and aging. Biochim Biophys Acta. 2010;2010:1584–1590. doi: 10.1016/j.bbapap.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imai S. Dissecting systemic control of metabolism and aging in the NAD World: The importance of SIRT1 and NAMPT-mediated NAD biosynthesis. FEBS Lett. 2011;585:1657–1662. doi: 10.1016/j.febslet.2011.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruzzone S, Guida L, Zocchi E, Franco L, De Flora A. Connexin 43 hemi channels mediate Ca2+-regulated transmembrane NAD+ fluxes in intact cells. Faseb J. 2001;15:10–12. doi: 10.1096/fj.00-0566fje. [DOI] [PubMed] [Google Scholar]
- 47.Peret J, Chanez M, Pascal G. Schedule of protein ingestion and circadian rhythm of certain hepatic enzyme activities involved in glucose metabolism in the rat. Nutr Metab. 1976;20:143–157. doi: 10.1159/000175698. [DOI] [PubMed] [Google Scholar]
- 48.Pessacq MT, Gagliardino JJ. Glycogen metabolism in muscle: its circadian and seasonal variations. Metabolism. 1975;24:737–743. doi: 10.1016/0026-0495(75)90041-4. [DOI] [PubMed] [Google Scholar]
- 49.Wagner E, Deitzer GF, Fischer S, Frosh S, Kempf O, Stroebele L. Endogenous oscillations in pathways of energy transduction as related to circadian rhythmicity and photoperiodic control. Biosystems. 1975;7:68–76. doi: 10.1016/0303-2647(75)90044-1. [DOI] [PubMed] [Google Scholar]
- 50.Dubrovsky YV, Samsa WE, Kondratov RV. Deficiency of circadian protein CLOCK reduces lifespan and increases age-related cataract development in mice. Aging. 2010;2:936–944. doi: 10.18632/aging.100241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pepine CJ. Circadian variations in myocardial ischemia. Implications for management. Jama. 1991;265:386–390. [PubMed] [Google Scholar]
- 53.Muller JE, Stone PH, Turi ZG, Rutherford JD, Czeisler CA, Parker C, Poole WK, Passamani E, Roberts R, Robertson T, et al. Circadian variation in the frequency of onset of acute myocardial infarction. N Engl J Med. 1985;313:1315–1322. doi: 10.1056/NEJM198511213132103. [DOI] [PubMed] [Google Scholar]
- 54.Levine RL, Pepe PE, Fromm RE, Jr, Curka PA, Clark PA. Prospective evidence of a circadian rhythm for out-of-hospital cardiac arrests. Jama. 1992;267:2935–2937. [PubMed] [Google Scholar]
- 55.Burkart V, Wang ZQ, Radons J, Heller B, Herceg Z, Stingl L, Wagner EF, Kolb H. Mice lacking the poly(ADP-ribose) polymerase gene are resistant to pancreatic beta-cell destruction and diabetes development induced by streptozocin. Nat Med. 1999;5:314–319. doi: 10.1038/6535. [DOI] [PubMed] [Google Scholar]
- 56.Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, Schoonjans K, Schreiber V, Sauve AA, Menissier-de Murcia J, Auwerx J. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kolthur-Seetharam U, Dantzer F, McBurney MW, de Murcia G, Sassone-Corsi P. Control of AIF-mediated cell death by the functional interplay of SIRT1 and PARP-1 in response to DNA damage. Cell Cycle. 2006;5:873–877. doi: 10.4161/cc.5.8.2690. [DOI] [PubMed] [Google Scholar]
- 58.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 59.Xiong S, Salazar G, Patrushev N, Alexander RW. FoxO1 mediates an autofeedback loop regulating SIRT1 expression. J Biol Chem. 2011;286:5289–5299. doi: 10.1074/jbc.M110.163667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shang L, Zhou H, Xia Y, Wang H, Gao G, Chen B, Liu Q, Shao C, Gong Y. Serum withdrawal up-regulates human SIRT1 gene expression in a p53-dependent manner. J Cell Mol Med. 2009;13:4176–4184. doi: 10.1111/j.1582-4934.2008.00468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Naqvi A, Hoffman TA, DeRicco J, Kumar A, Kim CS, Jung SB, Yamamori T, Kim YR, Mehdi F, Kumar S, Rankinen T, Ravussin E, Irani K. A single-nucleotide variation in a p53-binding site affects nutrient-sensitive human SIRT1 expression. Hum Mol Genet. 2010;19:4123–4133. doi: 10.1093/hmg/ddq331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okazaki M, Iwasaki Y, Nishiyama M, Taguchi T, Tsugita M, Nakayama S, Kambayashi M, Hashimoto K, Terada Y. PPARbeta/delta regulates the human SIRT1 gene transcription via Sp1. Endocr J. 2010;57:403–413. doi: 10.1507/endocrj.k10e-004. [DOI] [PubMed] [Google Scholar]
- 63.Bai P, Canto C, Brunyanszki A, Huber A, Szanto M, Cen Y, Yamamoto H, Houten SM, Kiss B, Oudart H, Gergely P, Menissier-de Murcia J, Schreiber V, Sauve AA, Auwerx J. PARP-2 regulates SIRT1 expression and whole-body energy expenditure. Cell Metab. 2011;13:450–460. doi: 10.1016/j.cmet.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 65.Zhang Q, Wang SY, Fleuriel C, Leprince D, Rocheleau JV, Piston DW, Goodman RH. Metabolic regulation of SIRT1 transcription via a HIC1:CtBP corepressor complex. Proc Natl Acad Sci U S A. 2007;104:829–833. doi: 10.1073/pnas.0610590104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66.Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, Nemoto S, Finkel T, Gu W, Cress WD, Chen J. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8:1025–1031. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- 67.Pardo PS, Mohamed JS, Lopez MA, Boriek AM. Induction of Sirt1 by mechanical stretch of skeletal muscle through the early response factor EGR1 triggers an antioxidative response. J Biol Chem. 2011;286:2559–2566. doi: 10.1074/jbc.M110.149153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sayed D, Abdellatif M. MicroRNAs in Development and Disease. Physiol Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 69.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;28:28. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Menghini R, Casagrande V, Cardellini M, Martelli E, Terrinoni A, Amati F, Vasa-Nicotera M, Ippoliti A, Novelli G, Melino G, Lauro R, Federici M. MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation. 2009;120:1524–1532. doi: 10.1161/CIRCULATIONAHA.109.864629. [DOI] [PubMed] [Google Scholar]
- 71.Ito T, Yagi S, Yamakuchi M. MicroRNA-34a regulation of endothelial senescence. Biochem Biophys Res Commun. 2010;398:735–740. doi: 10.1016/j.bbrc.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 72.Xu D, Takeshita F, Hino Y, Fukunaga S, Kudo Y, Tamaki A, Matsunaga J, Takahashi RU, Takata T, Shimamoto A, Ochiya T, Tahara H. miR-22 represses cancer progression by inducing cellular senescence. J Cell Biol. 2011;193:409–424. doi: 10.1083/jcb.201010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Strum JC, Johnson JH, Ward J, Xie H, Feild J, Hester A, Alford A, Waters KM. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol Endocrinol. 2009;23:1876–1884. doi: 10.1210/me.2009-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ramachandran D, Roy U, Garg S, Ghosh S, Pathak S, Kolthur-Seetharam U. Sirt1 and mir-9 expression is regulated during glucose-stimulated insulin secretion in pancreatic beta-islets. Febs J. 2011;278:1167–1174. doi: 10.1111/j.1742-4658.2011.08042.x. [DOI] [PubMed] [Google Scholar]
- 75.Schonrock N, Humphreys DT, Preiss T, Gotz J. Target Gene Repression Mediated by miRNAs miR-181c and miR-9 Both of Which Are Down-regulated by Amyloid-beta. J Mol Neurosci. 2011;2011:1. doi: 10.1007/s12031-011-9587-2. [DOI] [PubMed] [Google Scholar]
- 76.Eades G, Yao Y, Yang M, Zhang Y, Chumsri S, Zhou Q. MiR-200a regulates SIRT1 and EMT-like transformation in mammary epithelial cells. J Biol Chem. 2011;2011:19. doi: 10.1074/jbc.M111.229401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, Vatner DE, Vatner SF, Abdellatif M. Downregulation of MiR-199a Derepresses Hypoxia-Inducible Factor-1{alpha} and Sirtuin 1 and Recapitulates Hypoxia Preconditioning in Cardiac Myocytes. Circ Res. 2009;104:879–886. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu H, Yang Y, Wang Y, Li J, Schiller PW, Peng T. MicroRNA-195 promotes palmitate-induced apoptosis in cardiomyocytes by down-regulating Sirt1. Cardiovasc Res. 2011;2011:21. doi: 10.1093/cvr/cvr145. [DOI] [PubMed] [Google Scholar]
- 79.Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 80.Chen IY, Lypowy J, Pain J, Sayed D, Grinberg S, Alcendor RR, Sadoshima J, Abdellatif M. Histone H2A.z is essential for cardiac myocyte hypertrophy but opposed by silent information regulator 2alpha. J Biol Chem. 2006;281:19369–19377. doi: 10.1074/jbc.M601443200. [DOI] [PubMed] [Google Scholar]
- 81.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 82.Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 83.Murayama A, Ohmori K, Fujimura A, Minami H, Yasuzawa-Tanaka K, Kuroda T, Oie S, Daitoku H, Okuwaki M, Nagata K, Fukamizu A, Kimura K, Shimizu T, Yanagisawa J. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 84.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 86.Bouras T, Fu M, Sauve AA, Wang F, Quong AA, Perkins ND, Hay RT, Gu W, Pestell RG. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J Biol Chem. 2005;280:10264–10276. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- 87.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 88.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 89.Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 90.Wong S, Weber JD. Deacetylation of the retinoblastoma tumour suppressor protein by SIRT1. Biochem J. 2007;407:451–460. doi: 10.1042/BJ20070151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. Embo J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol Cell Biol. 2005;25:8456–8464. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 94.Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, Permutt MA, Imai S. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 95.Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, Easlon EJ, Lin SJ, Guarente L. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4:27. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pi J, Collins S. Reactive oxygen species and uncoupling protein 2 in pancreatic beta-cell function. Diabetes Obes Metab. 2010;2:141–148. doi: 10.1111/j.1463-1326.2010.01269.x. [DOI] [PubMed] [Google Scholar]
- 97.Ramsey KM, Mills KF, Satoh A, Imai S. Age-associated loss of Sirt1-mediated enhancement of glucose-stimulated insulin secretion in beta cell-specific Sirt1-overexpressing (BESTO) mice. Aging Cell. 2008;7:78–88. doi: 10.1111/j.1474-9726.2007.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, Gu W, Guarente L. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 99.Sun C, Zhang F, Ge X, Yan T, Chen X, Shi X, Zhai Q. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metab. 2007;6:307–319. doi: 10.1016/j.cmet.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 100.Maegawa H, Ide R, Hasegawa M, Ugi S, Egawa K, Iwanishi M, Kikkawa R, Shigeta Y, Kashiwagi A. Thiazolidine derivatives ameliorate high glucose-induced insulin resistance via the normalization of protein-tyrosine phosphatase activities. J Biol Chem. 1995;270:7724–7730. doi: 10.1074/jbc.270.13.7724. [DOI] [PubMed] [Google Scholar]
- 101.Rutanen J, Yaluri N, Modi S, Pihlajamaki J, Vanttinen M, Itkonen P, Kainulainen S, Yamamoto H, Lagouge M, Sinclair DA, Elliott P, Westphal C, Auwerx J, Laakso M. SIRT1 mRNA expression may be associated with energy expenditure and insulin sensitivity. Diabetes. 2010;59:829–835. doi: 10.2337/db09-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 103.Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 104.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Erion DM, Yonemitsu S, Nie Y, Nagai Y, Gillum MP, Hsiao JJ, Iwasaki T, Stark R, Weismann D, Yu XX, Murray SF, Bhanot S, Monia BP, Horvath TL, Gao Q, Samuel VT, Shulman GI. SirT1 knockdown in liver decreases basal hepatic glucose production and increases hepatic insulin responsiveness in diabetic rats. Proc Natl Acad Sci U S A. 2009;106:11288–11293. doi: 10.1073/pnas.0812931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, 3rd, Olefsky J, Guarente L, Montminy M. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tschritter O, Fritsche A, Thamer C, Haap M, Shirkavand F, Rahe S, Staiger H, Maerker E, Haring H, Stumvoll M. Plasma adiponectin concentrations predict insulin sensitivity of both glucose and lipid metabolism. Diabetes. 2003;52:239–243. doi: 10.2337/diabetes.52.2.239. [DOI] [PubMed] [Google Scholar]
- 110.Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 111.de Kreutzenberg SV, Ceolotto G, Papparella I, Bortoluzzi A, Semplicini A, Dalla Man C, Cobelli C, Fadini GP, Avogaro A. Downregulation of the longevity-associated protein sirtuin 1 in insulin resistance and metabolic syndrome: potential biochemical mechanisms. Diabetes. 2010;59:1006–1015. doi: 10.2337/db09-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. Embo J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu F, Gao Z, Zhang J, Rivera CA, Yin J, Weng J, Ye J. Lack of SIRT1 (Mammalian Sirtuin 1) activity leads to liver steatosis in the SIRT1+/− mice: a role of lipid mobilization and inflammation. Endocrinology. 2010;151:2504–2514. doi: 10.1210/en.2009-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kalaany NY, Gauthier KC, Zavacki AM, Mammen PP, Kitazume T, Peterson JA, Horton JD, Garry DJ, Bianco AC, Mangelsdorf DJ. LXRs regulate the balance between fat storage and oxidation. Cell Metab. 2005;1:231–244. doi: 10.1016/j.cmet.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 116.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 117.Kim HS, Xiao C, Wang RH, Lahusen T, Xu X, Vassilopoulos A, Vazquez-Ortiz G, Jeong WI, Park O, Ki SH, Gao B, Deng CX. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010;12:224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kanfi Y, Peshti V, Gil R, Naiman S, Nahum L, Levin E, Kronfeld-Schor N, Cohen HY. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell. 2009;9:162–173. doi: 10.1111/j.1474-9726.2009.00544.x. [DOI] [PubMed] [Google Scholar]
- 119.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 120.Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 121.Csiszar A, Labinskyy N, Pinto JT, Ballabh P, Zhang H, Losonczy G, Pearson K, de Cabo R, Pacher P, Zhang C, Ungvari Z. Resveratrol induces mitochondrial biogenesis in endothelial cells. Am J Physiol Heart Circ Physiol. 2009;297:8. doi: 10.1152/ajpheart.00368.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Little JP, Safdar A, Wilkin GP, Tarnopolsky MA, Gibala MJ. A practical model of low-volume high-intensity interval training induces mitochondrial biogenesis in human skeletal muscle: potential mechanisms. J Physiol. 2010;588:1011–1022. doi: 10.1113/jphysiol.2009.181743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, Moncada S, Carruba MO. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 124.Philp A, Chen A, Lan D, Meyer GA, Murphy AN, Knapp AE, Olfert IM, McCurdy CE, Marcotte GR, Hogan MC, Baar K, Schenk S. Sirtuin 1 (SIRT1) deacetylase activity is not required for mitochondrial biogenesis or peroxisome proliferator activated receptor-{gamma} coactivator-1{alpha} (PGC-1{alpha}) deacetylation following endurance exercise. J Biol Chem. 2011;2011:11. doi: 10.1074/jbc.M111.261685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kume S, Uzu T, Horiike K, Chin-Kanasaki M, Isshiki K, Araki S, Sugimoto T, Haneda M, Kashiwagi A, Koya D. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest. 2010;120:1043–1055. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr, Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hallows WC, Yu W, Smith BC, Devries MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM, Zhao S, Guan KL, Denu JM. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 2011;41:139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, Jacobson MP, Verdin E. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, Olivier AK, Spitz DR, Gius D. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 135.Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, Stancakova A, Goetzman E, Lam MM, Schwer B, Stevens RD, Muehlbauer MJ, Kakar S, Bass NM, Kuusisto J, Laakso M, Alt FW, Newgard CB, Farese RV, Jr, Kahn CR, Verdin E. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell. 2011;44:177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lu Z, Bourdi M, Li JH, Aponte AM, Chen Y, Lombard DB, Gucek M, Pohl LR, Sack MN. SIRT3-dependent deacetylation exacerbates acetaminophen hepatotoxicity. EMBO Rep. 1038;2011:121. doi: 10.1038/embor.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 138.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Alcendor RR, Kirshenbaum LA, Imai S, Vatner SF, Sadoshima J. Silent information regulator 2alpha, a longevity factor and class III histone deacetylase, is an essential endogenous apoptosis inhibitor in cardiac myocytes. Circ Res. 2004;95:971–980. doi: 10.1161/01.RES.0000147557.75257.ff. [DOI] [PubMed] [Google Scholar]
- 141.Hsu CP, Zhai P, Yamamoto T, Maejima Y, Matsushima S, Hariharan N, Shao D, Takagi H, Oka S, Sadoshima J. Silent information regulator 1 protects the heart from ischemia/reperfusion. Circulation. 2010;122:2170–2182. doi: 10.1161/CIRCULATIONAHA.110.958033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 143.Vinciguerra M, Santini MP, Martinez C, Pazienza V, Claycomb WC, Giuliani A, Rosenthal N. mIGF-1/JNK1/SirT1 signaling confers protection against oxidative stress in the heart. Aging Cell. 2012;11:139–149. doi: 10.1111/j.1474-9726.2011.00766.x. [DOI] [PubMed] [Google Scholar]
- 144.Hsu CP, Oka S, Shao D, Hariharan N, Sadoshima J. Nicotinamide phosphoribosyltransferase regulates cell survival through NAD+ synthesis in cardiac myocytes. Circ Res. 2009;105:481–491. doi: 10.1161/CIRCRESAHA.109.203703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ahmet I, Tae HJ, de Cabo R, Lakatta EG, Talan MI. Effects of calorie restriction on cardioprotection and cardiovascular health. J Mol Cell Cardiol. 2011;51:263–271. doi: 10.1016/j.yjmcc.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dolinsky VW, Morton JS, Oka T, Robillard-Frayne I, Bagdan M, Lopaschuk GD, Des Rosiers C, Walsh K, Davidge ST, Dyck JR. Calorie restriction prevents hypertension and cardiac hypertrophy in the spontaneously hypertensive rat. Hypertension. 2010;56:412–421. doi: 10.1161/HYPERTENSIONAHA.110.154732. [DOI] [PubMed] [Google Scholar]
- 147.Niemann B, Chen Y, Issa H, Silber RE, Rohrbach S. Caloric restriction delays cardiac ageing in rats: role of mitochondria. Cardiovasc Res. 2010;88:267–276. doi: 10.1093/cvr/cvq273. [DOI] [PubMed] [Google Scholar]
- 148.Shinmura K, Tamaki K, Sano M, Murata M, Yamakawa H, Ishida H, Fukuda K. Impact of long-term caloric restriction on cardiac senescence: caloric restriction ameliorates cardiac diastolic dysfunction associated with aging. J Mol Cell Cardiol. 2011;50:117–127. doi: 10.1016/j.yjmcc.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 149.Shinmura K, Tamaki K, Bolli R. Impact of 6-mo caloric restriction on myocardial ischemic tolerance: possible involvement of nitric oxide-dependent increase in nuclear Sirt1. Am J Physiol Heart Circ Physiol. 2008;295:17. doi: 10.1152/ajpheart.00602.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shinmura K, Tamaki K, Sano M, Nakashima-Kamimura N, Wolf AM, Amo T, Ohta S, Katsumata Y, Fukuda K, Ishiwata K, Suematsu M, Adachi T. Caloric restriction primes mitochondria for ischemic stress by deacetylating specific mitochondrial proteins of the electron transport chain. Circ Res. 2011;109:396–406. doi: 10.1161/CIRCRESAHA.111.243097. [DOI] [PubMed] [Google Scholar]
- 151.Nadtochiy SM, Redman E, Rahman I, Brookes PS. Lysine deacetylation in ischaemic preconditioning: the role of SIRT1. Cardiovasc Res. 2011;89:643–649. doi: 10.1093/cvr/cvq287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Edwards AG, Donato AJ, Lesniewski LA, Gioscia RA, Seals DR, Moore RL. Life-long caloric restriction elicits pronounced protection of the aged myocardium: a role for AMPK. Mech Ageing Dev. 2010;131:739–742. doi: 10.1016/j.mad.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Planavila A, Iglesias R, Giralt M, Villarroya F. Sirt1 acts in association with PPARalpha to protect the heart from hypertrophy, metabolic dysregulation, and inflammation. Cardiovasc Res. 2011;90:276–284. doi: 10.1093/cvr/cvq376. [DOI] [PubMed] [Google Scholar]
- 155.Passariello CL, Zini M, Nassi PA, Pignatti C, Stefanelli C. Upregulation of SIRT1 deacetylase in phenylephrine-treated cardiomyoblasts. Biochem Biophys Res Commun. 2011;407:512–516. doi: 10.1016/j.bbrc.2011.03.049. [DOI] [PubMed] [Google Scholar]
- 156.Oka S, Alcendor R, Zhai P, Park JY, Shao D, Cho J, Yamamoto T, Tian B, Sadoshima J. PPARalpha-Sirt1 complex mediates cardiac hypertrophy and failure through suppression of the ERR transcriptional pathway. Cell Metab. 2011;14:598–611. doi: 10.1016/j.cmet.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. 2008;102:703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- 159.Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. 2009;119:2758–2771. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hafner AV, Dai J, Gomes AP, Xiao CY, Palmeira CM, Rosenzweig A, Sinclair DA. Regulation of the mPTP by SIRT3-mediated deacetylation of CypD at lysine 166 suppresses age-related cardiac hypertrophy. Aging. 2010;2:914–923. doi: 10.18632/aging.100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chen Z, Peng IC, Cui X, Li YS, Chien S, Shyy JY. Shear stress, SIRT1, and vascular homeostasis. Proc Natl Acad Sci U S A. 107:10268–10273. doi: 10.1073/pnas.1003833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mattagajasingh I, Kim CS, Naqvi A, Yamamori T, Hoffman TA, Jung SB, DeRicco J, Kasuno K, Irani K. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc Natl Acad Sci U S A. 2007;104:14855–14860. doi: 10.1073/pnas.0704329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhang QJ, Wang Z, Chen HZ, Zhou S, Zheng W, Liu G, Wei YS, Cai H, Liu DP, Liang CC. Endothelium-specific overexpression of class III deacetylase SIRT1 decreases atherosclerosis in apolipoprotein E-deficient mice. Cardiovasc Res. 2008;80:191–199. doi: 10.1093/cvr/cvn224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Stein S, Schafer N, Breitenstein A, Besler C, Winnik S, Lohmann C, Heinrich K, Brokopp CE, Handschin C, Landmesser U, Tanner FC, Luscher TF, Matter CM. SIRT1 reduces endothelial activation without affecting vascular function in ApoE−/− mice. Aging. 2010;2:353–360. doi: 10.18632/aging.100162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Potente M, Ghaeni L, Baldessari D, Mostoslavsky R, Rossig L, Dequiedt F, Haendeler J, Mione M, Dejana E, Alt FW, Zeiher AM, Dimmeler S. SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev. 2007;21:2644–2658. doi: 10.1101/gad.435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Guarani V, Deflorian G, Franco CA, Kruger M, Phng LK, Bentley K, Toussaint L, Dequiedt F, Mostoslavsky R, Schmidt MH, Zimmermann B, Brandes RP, Mione M, Westphal CH, Braun T, Zeiher AM, Gerhardt H, Dimmeler S, Potente M. Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature. 2011;473:234–238. doi: 10.1038/nature09917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Borradaile NM, Pickering JG. Nicotinamide phosphoribosyltransferase imparts human endothelial cells with extended replicative lifespan and enhanced angiogenic capacity in a high glucose environment. Aging Cell. 2009;8:100–112. doi: 10.1111/j.1474-9726.2009.00453.x. [DOI] [PubMed] [Google Scholar]
- 168.Zhao T, Li J, Chen AF. MicroRNA-34a induces endothelial progenitor cell senescence and impedes its angiogenesis via suppressing silent information regulator 1. Am J Physiol Endocrinol Metab. 2010;299:27. doi: 10.1152/ajpendo.00192.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Li L, Zhang HN, Chen HZ, Gao P, Zhu LH, Li HL, Lv X, Zhang QJ, Zhang R, Wang Z, She ZG, Wei YS, Du GH, Liu DP, Liang CC. SIRT1 acts as a modulator of neointima formation following vascular injury in mice. Circ Res. 2011;108:1180–1189. doi: 10.1161/CIRCRESAHA.110.237875. [DOI] [PubMed] [Google Scholar]
- 170.Qiang L, Lin HV, Kim-Muller JY, Welch CL, Gu W, Accili D. Proatherogenic abnormalities of lipid metabolism in SirT1 transgenic mice are mediated through Creb deacetylation. Cell Metab. 2011;14:758–767. doi: 10.1016/j.cmet.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cohen DE, Supinski AM, Bonkowski MS, Donmez G, Guarente LP. Neuronal SIRT1 regulates endocrine and behavioral responses to calorie restriction. Genes Dev. 2009;23:2812–2817. doi: 10.1101/gad.1839209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Satoh A, Brace CS, Ben-Josef G, West T, Wozniak DF, Holtzman DM, Herzog ED, Imai S. SIRT1 promotes the central adaptive response to diet restriction through activation of the dorsomedial and lateral nuclei of the hypothalamus. J Neurosci. 2010;30:10220–10232. doi: 10.1523/JNEUROSCI.1385-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hillman JB, Tong J, Tschop M. Ghrelin biology and its role in weight-related disorders. Discov Med. 11:521–528. [PubMed] [Google Scholar]
- 174.Ramadori G, Fujikawa T, Fukuda M, Anderson J, Morgan DA, Mostoslavsky R, Stuart RC, Perello M, Vianna CR, Nillni EA, Rahmouni K, Coppari R. SIRT1 deacetylase in POMC neurons is required for homeostatic defenses against diet-induced obesity. Cell Metab. 2010;12:78–87. doi: 10.1016/j.cmet.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Dietrich MO, Antunes C, Geliang G, Liu ZW, Borok E, Nie Y, Xu AW, Souza DO, Gao Q, Diano S, Gao XB, Horvath TL. Agrp neurons mediate Sirt1's action on the melanocortin system and energy balance: roles for Sirt1 in neuronal firing and synaptic plasticity. J Neurosci. 2010;30:11815–11825. doi: 10.1523/JNEUROSCI.2234-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Michan S, Li Y, Chou MM, Parrella E, Ge H, Long JM, Allard JS, Lewis K, Miller M, Xu W, Mervis RF, Chen J, Guerin KI, Smith LE, McBurney MW, Sinclair DA, Baudry M, de Cabo R, Longo VD. SIRT1 is essential for normal cognitive function and synaptic plasticity. J Neurosci. 2010;30:9695–9707. doi: 10.1523/JNEUROSCI.0027-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 179.Meng Y, Zhang Y, Tregoubov V, Janus C, Cruz L, Jackson M, Lu WY, MacDonald JF, Wang JY, Falls DL, Jia Z. Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron. 2002;35:121–133. doi: 10.1016/s0896-6273(02)00758-4. [DOI] [PubMed] [Google Scholar]
- 180.Panossian L, Fenik P, Zhu Y, Zhan G, McBurney MW, Veasey S. SIRT1 regulation of wakefulness and senescence-like phenotype in wake neurons. J Neurosci. 2011;31:4025–4036. doi: 10.1523/JNEUROSCI.5166-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 182.Qin W, Yang T, Ho L, Zhao Z, Wang J, Chen L, Zhao W, Thiyagarajan M, MacGrogan D, Rodgers JT, Puigserver P, Sadoshima J, Deng H, Pedrini S, Gandy S, Sauve AA, Pasinetti GM. Neuronal SIRT1 activation as a novel mechanism underlying the prevention of Alzheimer disease amyloid neuropathology by calorie restriction. J Biol Chem. 2006;281:21745–21754. doi: 10.1074/jbc.M602909200. [DOI] [PubMed] [Google Scholar]
- 183.Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, Puigserver P, Sinclair DA, Tsai LH. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. Embo J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell. 2010;142:320–332. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]