Abstract
Mycoplasma haemocanis is a hemotrophic mycoplasma (hemoplasma), blood pathogen that may cause acute disease in immunosuppressed or splenectomized dogs. The genome of the strain Illinois, isolated from blood of a naturally infected dog, has been entirely sequenced and annotated to gain a better understanding of the biology of M. haemocanis. Its single circular chromosome has 919 992 bp and a low G + C content (35%), representing a typical mycoplasmal genome. A gene-by-gene comparison against its feline counterpart, M. haemofelis, reveals a very similar composition and architecture with most of the genes having conserved synteny extending over their entire chromosomes and differing only by a small set of unique protein coding sequences. As in M. haemofelis, M. haemocanis metabolic pathways are reduced and apparently rely heavily on the nutrients afforded by its host environment. The presence of a major percentage of its genome dedicated to paralogous genes (63.7%) suggests that this bacterium might use antigenic variation as a mechanism to evade the host’s immune system as also observed in M. haemofelis genome. Phylogenomic comparisons based on average nucleotide identity (ANI) and tetranucleotide signature suggest that these two pathogens are different species of mycoplasmas, with M. haemocanis infecting dogs and M. haemofelis infecting cats.
Introduction
Hemotrophic mycoplasmas (hemoplasmas) are uncultivable cell-wall less bacteria, formerly classified as Haemobartonella and Eperythrozoon species, that adhere to the surface of the erythrocytes of their vertebrate hosts. These bacteria form a new clade within the Mycoplasma genus (class Mollicutes) and are phylogenetically related to the pneumoniae group of the mycoplasmas [1-5].
Mycoplasma haemocanisHaemobartonella canis was first described in Germany in 1928 in a splenectomized dog [6]. The name Bartonella canis was proposed and remained until 1939 when Tyzzer and Weinman created the new genus Haemobartonella[7]. M. haemocanis, proposed species name since 2002 [5], is a pleomorphic bacterium with coccoid and ring forms that can be visualized in the host’s peripheral blood smear either singly or in chains that can resemble a “violin-bow” form [8]. It may cause overt, hemolytic anemia in immunosuppressed [5,9] or splenectomized dogs [5,10], and has a worldwide distribution with prevalence of infection varying from 0.5% to 40% [11-14].
Similarities with the feline hemoplasma M. haemofelisHaemobartonella felis, together with the fact that hemoplasmas are not species-specific as previously thought [15,16], led some research groups to hypothesize that these two bacteria could be the same species infecting different hosts [17,18]. Moreover, there are some reports in the old literature stating that M. haemocanis could infect cats; however M. haemofelis did not cause infection in dogs [19-21]. In 1961, Dr Lumb published the manuscript “Canine haemobartonellosis and its feline counterpart”, reporting cross-transmission experiments: it was shown that when blood from cats infected with M. haemocanis was injected into susceptible splenectomized dogs, organisms could be seen on their peripheral blood smears [8]. It was concluded based on this evidence that the feline might act as a reservoir for M. haemocanis. However, blood from dogs previously injected with M. haemofelis inoculated into susceptible cats failed to result in circulating organisms, leading to the conclusion that these two bacteria were different species [8,22]. Forty years later, the sequences of the 16S rRNA genes of these two bacteria were reported to have 99% identity [17] raising the same question again. In 2002, the sequences of the RNase P genes of these bacteria were reported having 94.3 to 95.5% identity [18]. While the results of the RNase P genes did not support the hypothesis that M. haemocanis and M. haemofelis were identical, this additional data was still considered insufficient to determine whether these organisms should be classified as different species, subspecies, or strains of the same species [18].
Recently, three species of hemoplasmas, including M. haemofelis, had their genomes completely sequenced and annotated [23-27]. The aim of this study was to sequence the whole genome of M. haemocanis in order to better understand its biology and to perform a complete genomic comparison with its counterpart, M. haemofelis.
Materials and methods
Bacterial strain and DNA isolation
M. haemocanis organisms were isolated from the blood of a naturally infected dog at peak of bacteremia [17]. Written informed consent was obtained from the client for publication of this report. Bacterial genomic DNA was extracted using Quick-gDNA MiniPrep kit according to the manufacturer’s instructions (Zymo Research, Irvine, CA, USA).
M. haemocanis strain Illinois sequencing and assembly
Whole genome was sequenced from paired-end libraries (TruSeq DNA sample preparation kit, Illumina, San Diego, CA, USA) using 20% of an Illumina® v3 chemistry lane (HiScanSQ). Sequencing resulted in 15.7 million high-quality filtered read pairs with an average read length of 2 × 100 nucleotides and a > 3400 X genome equivalent coverage. Reads were assembled using ABySS-PE v1.2.7 utilizing 20% of the reads with “kmer” set to 95 bases [28]. Predicted scaffolds with significant BLAST matches to canine DNA were excluded and the remaining mycoplasma scaffolds were then organized based on the orientation predicted in the assembly and on the genome sequence of M. haemofelis strain Ohio2. A total of 13 gaps were identified and closed using conventional PCR followed by Sanger sequencing.
Genome annotation and analyses
First pass annotation was achieved using the NCBI annotation pipeline. Manual annotation/curation of each gene was performed using the annotation tool Manatee, provided by the Institute for Genome Sciences (IGS) at the University of Maryland, School of Medicine. Comparative analyses with other bacterial genomes were performed based on genomic data deposited in the NCBI database (NCBI, Bethesda, MD, USA).
The assignment of paralogous gene families was performed using BLASTclust tool provided by Max-Planck Institute for Developmental Biology [29], with 70% covered length and 30% sequence identity thresholds. Subcellular localization and protein sorting signals were predicted for each unique protein coding sequence (CDS) of M. haemocanis and M. haemofelis using PSORTb v.3.0 [30,31]. Metabolic pathways were predicted based on the KEGG pathway database [32] and the study reported by Yus et al. [33]. Presence of lipoproteins was predicted by LipoP version 1.0 software [34]. In addition, the tandem repeats were identified using the Tandem repeats finder program [35]. Comparative analyses of the whole genome of M. haemocanis and M. haemofelis strain Ohio2 were performed using the same tools mentioned above and all the CDSs from both genomes were evaluated using BLASTp and/or BLASTn in order to obtain a complete detailed comparison. CDSs were assigned using BLASTp and considered unique to M. haemocanis or M. haemofelis when there were no matching sequences in the aligned sequences list with ≥ 90% coverage and ≥ 30% identity or ≥ 80% coverage and ≥ 40% identity to the query sequence. Extended similarity group method for automated protein function prediction (ESG software) [36] was applied for both sets of unique CDSs.
Species differentiation analyses
The average nucleotide identity (ANI; MUMmer algorithm) and tetranucleotide signature correlation index between genomes were calculated using JSpecies software as previously described [37]. In addition to the genome of M. haemocanis strain Illinois, the following genome sequences were used in the analyses: M. haemofelis strain Ohio2 (CP002808.1), M. haemofelis strain Langford (FR773153.2), M. suis strain Illinois (CP002525.1), and M. suis strain KI3806 (FQ790233.1). If two organisms had ANIm and tetranucleotide coefficients greater than 94% and 0.99, respectively, they were considered the same species [37].
Results
Mycoplasma haemocanis strain Illinois genome features
The complete singular circular chromosome of M. haemocanis strain Illinois has a size of 919 992 base pairs (bp) and G + C content of 35%; these genomic features are similar to other hemoplasmas species sequenced to date [23,24,26,27] and within the range reported for other members of the genus Mycoplasma (Table 1). As described for all sequenced mycoplasmas (24 species to date), M. haemocanis also uses the opal stop codon (UGA) for tryptophan. The 16S, 23S and 5S rRNA genes are represented as single copies and share the same operon. The manual genome annotation suggests the presence of 1173 CDSs and 31 tRNAs, covering all amino-acids. Putative functions of most of the CDSs are represented as hypothetical proteins (75.62%), which are mostly due to its large repertoire of paralogous genes (63.76%) (Additional file 1: Table S1). These and other genome features were compared with other hemoplasmas and mycoplasmas members of the pneumoniae group (Table 1). The total number of CDSs of M. haemocanis classified by role (according to TIGR microbial role categories) was compared to those found in the M. haemofelis genome (Table 2).
Table 1.
General genomic characteristics of Mycoplasma haemocanis strain Illinois compared to members of pneumoniae group of mycoplasmas
|
Characteristic |
Pneumoniae Group |
|||||||
|---|---|---|---|---|---|---|---|---|
| |
Hemoplasmas |
M. pneumoniae |
M. gallisepticum |
M. genitalium |
M. penetrans |
|||
| M. haemocanis strain Illinois | M. haemofelis strain Ohio2 | 'Candidatus M. haemominutum' strain Birmingham 1 | M. suis strain Illinois | |||||
| Genome size (base pairs) |
919 992 |
1 155 937 |
513 880 |
742 431 |
816 394 |
1 012 800 |
580 076 |
1 358 633 |
| % of G + C |
35.3 |
38.8 |
35.5 |
31.1 |
40 |
31 |
31.7 |
25.7 |
| Total of genes |
1207 |
1584 |
582 |
883 |
733 |
817 |
524 |
1069 |
| Coding sequences (CDSs) |
1173 |
1549 |
547 |
844 |
689 |
763 |
475 |
1037 |
| CDSs with predicted function |
286 (24.3%) |
299 (19.3%) |
219 (40%) |
293 (34.7%) |
333 (48.3%) |
469 (61.46%) |
323 (68%) |
585 (56.4%) |
| No. of tRNAs |
31 |
31 |
32 |
32 |
37 |
32 |
36 |
29 |
| No. of rRNAs |
|
|
|
|
|
|
|
|
| 16S |
1 |
1 |
1 |
1 |
1 |
2 |
1 |
1 |
| 23S |
1 |
1 |
1 |
1 |
1 |
2 |
1 |
1 |
| 5S |
1 |
1 |
1 |
1 |
1 |
3 |
1 |
1 |
| Genes in paralogous families | 748 (63.76%) | 1103 (71.2%) | 134 (24.5%) | 361 (42.8%) | 132 (19.1%) | 110 (14.4%) | 25 (5.2%) | 245 (23.6%) |
Data was obtained from GenBank database using the following accession numbers: M. haemocanis strain Illinois (CP003199.1), M. haemofelis strain Ohio2 (CP002808), ‘Candidatus Mycoplasma haemominutum’ strain Birmingham 1 (HE613254.1), M. suis strain Illinois (CP002525), M. pneumoniae (U00089), M. gallisepticum (AE015450), M. genitalium (L43967), M. penetrans (BA000026). Paralogous gene families were assigned using BLASTclust, with 70% coverage and 30% sequence identity thresholds.
Table 2.
Comparison of the total number of protein coding sequences (CDSs) of M. haemocanis strain Illinois and M. haemofelis strain Ohio2 genomes classified by role according to TIGR microbial role categories
|
Role Category |
Number of CDSs (%) |
|
|---|---|---|
| M. haemocanis str. Illinois | M. haemofelis str. Ohio2 | |
| Purines, pyrimidines, nucleosides, and nucleotides |
33 (2.81%) |
29 (1.85%) |
| Fatty acid and phospholipid metabolism |
6 (0.51%) |
6 (0.38%) |
| Biosynthesis of co-factors, prosthetic groups, and carriers |
8 (0.68%) |
7 (0.45%) |
| Central intermediary metabolism |
1 (0.09%) |
1 (0.06%) |
| Energy metabolism |
25 (2.13%) |
22 (1.41%) |
| Transport and binding proteins |
35 (2.98%) |
32 (2.04%) |
| DNA metabolism |
44 (3.75%) |
52 (3.32%) |
| Transcription |
21 (1.79%) |
18 (1.15%) |
| Protein synthesis |
96 (8.18%) |
97 (6.2%) |
| Protein fate |
21 (1.79%) |
19 (1.21%) |
| Regulatory functions |
4 (0.34%) |
3 (0.19%) |
| Signal transduction |
3 (0.26%) |
2 (0.13%) |
| Cell envelope |
3 (0.26%) |
7 (0.45%) |
| Cellular processes |
14 (1.19%) |
10 (0.64%) |
| Unknown functions |
8 (0.68%) |
8 (0.51%) |
| Hypothetical proteins |
887 (75.62%) |
1253 (80.06%) |
| Total* | 1209 | 1566 |
*Some of the CDSs are in more than one category.
Metabolic pathways predictions suggest similar growth requirements for M. haemocanis and M. haemofelis
Prediction of M. haemocanis metabolic pathways based on the KEGG pathway database [32] and Yus’s report [33] revealed that they are identical to those predicted for M. haemofelis[24]. As shown for M. haemofelis, metabolic pathways in M. haemocanis are reduced with many of the nutrients and metabolic precursors imported from the blood environment [24]. ATP and DNA/RNA biosynthesis depend on the transport from the environment of glucose and ribose/base derivates, respectively. Imported bases include: hypoxanthine, adenine, guanine, uracil and cytidine 5’-monophosphate (CMP). Furthermore, amino acids, nicotinamide and any vitamins required for growth must be acquired from blood environment.
Comparative analyses of M. haemocanis and M. haemofelis genomes
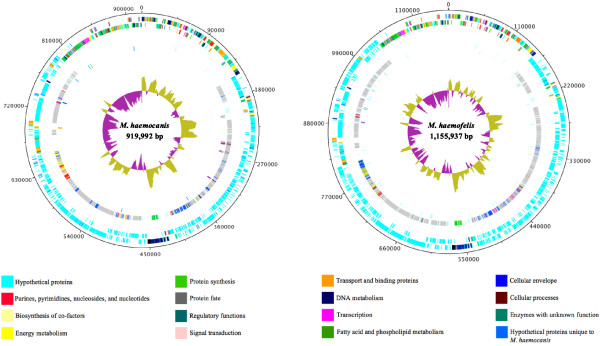
M. haemocanis genome was compared in its entirety to M. haemofelis strain Ohio2 (Figure 1): M. haemofelis has 376 CDSs more than M. haemocanis, however the majority of these CDSs are members of paralogous gene families also present in M. haemocanis; a set of only 67 CDSs was found to be different between these hemoplasmas. The canine hemoplasma possesses only 20 CDSs not identified in M. haemofelis genome, while 47 CDSs were unique to M. haemofelis. Most of these CDSs are hypothetical proteins, including one family of paralogous genes from M. haemocanis and four paralogous gene families from M. haemofelis (Additional file 2: Table S2). Predicted functions based on protein sequence similarity for these particular sets of CDSs were assigned using ESG software [36] (Additional file 2: Table S2). Analyses based on PSORTb parameters show that 35% of the unique CDSs of M. haemocanis are associated with the cytoplasmic membrane, while 17% of M. haemofelis CDSs are predicted to be associated with the membrane and 6.4% with extracellular (signal peptide detected) localization. For most of the unique CDSs, an unknown subcellular localization was predicted, corresponding to 60% and 51% in M. haemocanis and M. haemofelis genomes, respectively (Table 3). Thirteen out of the 20 (65%) unique CDSs of M. haemocanis and 35 out of 47 (74.5%) of M. haemofelis have at least one internal helix predicted. The number of predicted helices is also shown in the Additional file 2: Table S2. Fifteen lipoproteins were predicted in M. haemocanis compared with 17 for M. haemofelis genome, 13 of them are conserved between the two species. In addition, we identified 33 variable number tandem repeats (VNTRs) in the genome of M. haemocanis genome (Additional file 3: Table S3), while 61 were reported for M. haemofelis[24]. As in M. haemofelis genome, most of the VNTRs of M. haemocanis were localized within intergenic regions of hypothetical proteins. Five VNTRs were identified within the Type I restriction system operon; the presence of VNTRs in this operon was also described for M. haemofelis[24]. Other M. haemocanis VNTRs were identified within CDSs for SecD protein, efflux ABC transporter permease protein, PtsG protein, enolase, PotD protein, and for some of the hypothetical proteins.
Figure 1.
Circular representation of the genomes of Mycoplasma haemocanis strain Illinois and Mycoplasma haemofelis strain Ohio2 showing a similar content and organization of the coding sequences. The dnaA gene is at position zero in both genome plots, and the rRNAs (16S, 23S and 5S) are represented in black on the outermost circle. Outer to inner circles correspond to: circle 1: predicted coding sequences (CDSs) on the positive strand; circle 2: predicted CDSs on the negative strand. Each CDS is classified by TIGR role category according to the color designation in the legend below the plots; circle 3: CDSs in paralogous gene families (larger than 5 CDSs) with each family represented by a different color in each genome and homologous families by the same corresponding color in both genomes; circle 4: unique CDSs of each genome with colors corresponding to their role or paralogous family if applicable; circle 5: GC skew. Paralogous families with less than 5 CDSs are represented in light blue. The diagrams were generated using Artemis 12.0 - DNAPlotter version 1.4, Sanger Institute. (M. haemofelis plot was modified from Santos et al. [24])
Table 3.
Subcellular localization of the unique protein coding sequences (CDSs) of M. haemocanis strain Illinois and M. haemofelis strain Ohio2 genomes
|
Subcellular Localizationa |
Number of unique CDSs* |
|||||
|---|---|---|---|---|---|---|
| |
M. haemocanis strain Illinois |
M. haemofelis strain Ohio2 |
||||
| CDSs in paralogous families | CDSs not in paralogous families | CDSs with predicted function | CDSs in paralogous families | CDSs not in paralogous families | CDSs with predicted function | |
| Cytoplasmic membrane |
3 |
3 |
1 |
0 |
8 |
0 |
| Cytoplasmic |
0 |
0 |
1 |
3 |
7 |
2 |
| Extracellular |
0 |
0 |
0 |
3 |
0 |
0 |
| Unknown |
5 |
6 |
1 |
18 |
5 |
1 |
| Total | 8 | 9 | 3 | 24 | 20 | 3 |
a prediction using PSORTb version 3 software.
* Unique CDSs were assigned using BLASTp when there were no matching sequences with ≥ 90% coverage and ≥ 30% identity or ≥ 80% coverage and ≥ 40% identity to the query sequence.
Only 3 CDSs with known function are exclusive to M. haemocanis genome when compared to M. haemofelis strain Ohio2; however phosphotransferase system glucose-specific IIBC component (MHC_04460) is only present in the genome of M. haemofelis strain Langford, while two ribosomal proteins (MHC_00995 and MHC_05355) are in neither of the feline hemoplasma strains (Additional file 2: Table S2). Another 3 CDSs with known function were identified only in the genome of M. haemofelis: two of these proteins are C-5 cytosine-specific DNA methylases (MHF_1273 and MHF_1319), and the other protein is a type II site-specific deoxyribonuclease (MHF_1274) (Additional file 2: Table S2). Small CDSs (corresponding to 30–100 amino acids) characterized as fragments of paralogous genes were excluded from these analyses since they presented a coverage and/or identity below the cutoff to be considered as a member of a paralogous gene family (70% coverage and 30% identity threshold).
Phylogenomic comparison of M. haemocanis to other hemoplasmas
ANI and tetranucleotide signature correlation indexes are shown in Table 4. As indicated, M. haemocanis had an ANI of approximately 85% in comparison to all other hemoplasma genomes, including M. haemofelis. This is below the cutoff value of 94% for species circumscription. The tetranucleotide correlation indexes of M. haemocanis with other genomes were also below the 0.99 cutoff limit, being approximately 0.95 for M. haemofelis strains and 0.45 for M. suis strains. Based on these analyses, M. haemocanis is indeed a distinct species infecting the dog.
Table 4.
Average nucleotide identity* (ANI) and tetranucleotides signature correlation indexes (Tetra) of selected hemotrophic mycoplasmas
| |
M. haemofelis Ohio2 |
M. haemofelis Langford |
M. suis illinois |
M. suis KI3806 |
||||
|---|---|---|---|---|---|---|---|---|
| ANI | Tetra | ANI | Tetra | ANI | Tetra | ANI | Tetra | |
|
M. haemocanis Illinois |
85.11 |
0.959 |
85.21 |
0.962 |
85.59 |
0.452 |
85.5 |
0.453 |
|
M. haemofelis Ohio2 |
|
|
97.3 |
0.999 |
85.41 |
0.365 |
85.3 |
0.372 |
|
M. haemofelis Langford |
|
|
|
|
84.83 |
0.366 |
87.74 |
0.372 |
| M. suis illinois | 97.74 | 0.997 | ||||||
* = ANI was calculated using MUMmer algorithm in JSpecies software.
As expected, strains of the same species (M. suis Illinois and KI3806; M. haemofelis Ohio2 and Langford1) showed high ANI and tetranucleotide correlation indexes, which were above the proposed thresholds for species definition. In contrast, ANI and tetranucleotide correlation indexes between M. suis and M. haemofelis were approximately 85% and 0.37, respectively, correctly separating these organisms as two different species of mycoplasmas.
Discussion
The complete genome sequence and annotation of M. haemocanis extends our understanding of the biology of hemoplasmas and provides clues about the growth requirements for in vitro cultivation of these bacteria. Based on the metabolic pathway predictions and specific metabolic deficiencies, a more comprehensive medium can be designed [33]. To date, only three other species of hemoplasmas have been entirely sequenced [23-27]. The genome features of M. haemocanis, including its small size, low G + C content and use of UGA codon to encode tryptophan, are similar to those of other hemoplasmas and are typical of members of the genus Mycoplasma. It is believed that the reduced metabolic pathways of hemoplasmas are probably a consequence of the adaptation to the nutrient-rich blood environment [23,24]. The predicted metabolic pathways of M. haemocanis are very similar to those of M. haemofelis having orthologs for all the CDSs identified in the genome of this feline hemoplasma [24]; this is not surprising since both species are obligate red cell pathogens that reside in the blood of their hosts. As suggested for other hemoplasmas, it is likely that M. haemocanis takes advantage of the erythrocyte’s metabolism, scavenging nutrients, which leads to diminished erythrocyte life-span and exacerbation of anemia during acute disease.
Additional primary virulence factors were not identified in the genome of M. haemocanis. The o-sialoglycoprotein endopeptidase, related to the cleavage of glycophorin A, is conserved among hemoplasmas; the superoxide dismutase (SOD), identified in M. haemofelis[24,26] is also present in M. haemocanis, but not found in any other sequenced mycoplasma. Although SOD may protect these bacteria from superoxide anion toxicity faced in the blood environment, it is unlikely that this enzyme plays a determinant role in the primary pathogenicity associated with M. haemofelis infection or in the opportunistic infection caused by M. haemocanis.
As with other hemoplasmas, M. haemocanis contains an abundance of paralogous gene families (63.7% of all its CDSs) and the presence of strategically located tandem repeats. Although there is evidence supporting the role of paralog genes and the presence of tandem repeats in the development of antigenic diversity in Mycoplasma species [38,39], additional studies are needed to verify the ability of hemoplasmas to undergo antigenic variation. The presence of irregular cyclic episodes of bacteremia in splenectomized dogs reported following experimental infection with M. haemocanis[40], and the possibility that such cycles are due to phase variation is also an area of active investigation in our laboratory.
Comparison of the genomes of M. haemocanis and M. haemofelis revealed remarkable genetic similarities. Most of the coding and non-coding sequences were conserved and topography of genes within their chromosomes was similar. Even the paralogous gene families were conserved between the two species; the only exceptions were one family with 8 members in M. haemocanis, and four small families of M. haemofelis with 8, 5, 4 and 3 members, and two with 2 members. The major difference in the paralogous families is the number of duplicate genes inside each of the common families. Thus, as with other bacteria that cannot survive without their host, it appears that maintaining paralogous gene families to generate antigenic variants is a high priority for the hemoplasmas too [41]. On the other hand, CDSs that are unique to M. haemocanis or M. haemofelis might represent a set of proteins related to differences in virulence and/or related to host specificity. Most of these unique proteins are hypothetical. Although we attempted to improve the function prediction accuracy using the ESG software [36], most of the probabilities assigned were less than 50% and results remained inconclusive (Additional file 2: Table S2). Regarding the subcellular localization of the unique CDSs, it is important to mention that the PSORTb software only predicts cytoplasmic membrane localizations when 3 or more transmembrane helices are present within the sequence, otherwise unknown localization is returned. Therefore, these predictions based on strict criteria might have underestimated the potential for membrane localization of these CDSs.
CDSs with known function that are unique to M. haemocanis do not appear to have a significant impact on its pathogenicity since they code for an enzyme involved in sugar transport and for ribosomal proteins. On the other hand, M. haemofelis possesses a type II restriction enzyme and two C-5 cytosine-specific DNA methylases (C5 Mtase); the restriction endonuclease is located in the same operon as one of the C5 Mtase, indicating that this operon is functional [42]. Moreover, this endonuclease/methyltranferase pair is not present in any of the other hemoplasmas and the restriction enzyme is absent in the strain Langford 1 of M. haemofelis. DNA methylation has been associated with virulence in other bacteria [43]; however, the function of these pair in M. haemofelis Ohio2 is unknown.
As mentioned previously, the hemoplasmas cannot be cultivated in vitro. This has resulted in a lack of detailed phenotypic and genotypic characterization, which has hampered our ability to correctly classify these organisms within the Mycoplasmataceae family. In addition, the 16S rRNA gene failed to provide sufficient resolution to separate M. haemocanis and M. haemofelis as different species of Mycoplasma[5,17]. To date, the genotypic evidence for species differentiation of these two hemoplasmas is solely based on phylogenetic studies using a 177 bp fragment of their RNase P genes [18,44]. Herein, we performed a phylogenomic comparison between M. haemocanis and strains of M. haemofelis to resolve this long lasting controversy. In recent years, the sequencing of entire genomes has allowed the in silico evaluation of genomic similarities between different organisms. ANI and tetranucleotide signatures have been used as surrogates to previous methods of species circumscription, such as 16S rRNA gene phylogeny and DNA-DNA hybridization [37]. With both ANI and tetranucleotide indexes below the proposed thresholds for species definition, our results show that the M. haemocanis strain Illinois and M. haemofelis (strains Langford and Ohio2) are different species of mycoplasmas infecting two distinct animal species. This conclusion is also supported by the transmission studies done more than 50 years ago [8].
Taken together our results suggest that, although sharing very similar genomes, M. haemocanis and M. haemofelis are different mycoplasmal species infecting dogs and cats, respectively. The set of unique proteins may be a target for vaccine development against these hemoplasmas, especially for the feline hemoplasmosis that can cause acute disease in immunocompetent hosts.
Nucleotide sequence accession number
The genome of M. haemocanis strain Illinois was deposited in GenBank under the accession number CP003199.1.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed experiments: NCN, APS, AMSG, JBM. Performed experiments: NCN, APS, AMSG, JBM. Analyzed the data: NCN, APS, AMSG, PJS, JBM. Wrote and reviewed the paper: NCN, APS, AMSG, PJS, JBM. All authors read and approved the final manuscript.
Supplementary Material
Table S1. List of paralogous gene families* in the genome of Mycoplasma haemocanis strain Illinois. Complete list of paralogous gene families in Mycoplasma haemocanis strain Illinois genome assigned by BLASTclust tool provided by Max-Planck Institute for Developmental Biology, with 70% covered length and 30% sequence identity thresholds. GenBank accession numbers are provided.
Table S2. Analyses of the unique protein coding sequences (CDSs) of Mycoplasma haemocanis strain Illinois and M. haemofelis strain Ohio2. Analyses of all the unique CDSs of M. haemocanis strain Illinois and M. haemofelis strain Ohio2 were performed using ESG software, which predicts protein functions based on sequence similarity, and PSORTb version 3 software, which predicts subcellular localization. GenBank accession numbers of proteins corresponding to each CDS are also provided.
Table S3. Tandem repeats* identified in the genome of Mycoplasma haemocanis strain Illinois. Complete list of variable tandem repeats, identified using Tandem repeats finder program, indicating their sequences and position in the genome of M. haemocanis.
Contributor Information
Naíla C do Nascimento, Email: ncannes@purdue.edu.
Andrea P Santos, Email: santos1@purdue.edu.
Ana MS Guimaraes, Email: anamsg@purdue.edu.
Phillip J SanMiguel, Email: psanmiguel@purdue.edu.
Joanne B Messick, Email: jmessic@purdue.edu.
Acknowledgments
We are extremely grateful to The Genomic Core Facility at Purdue University and to Samuel Martin for constructing the library, Ann Feil for running the HiScanSQ, and Richard Westerman for performing the ABySS assembly. We also thank the University of Maryland’s Institute of Genomic Sciences, which provided us with automatic annotation data and the manual annotation tool Manatee. This work was kindly supported by the Morris Animal Foundation (grant D10FE-0042), and the Brazilian government through CNPq Fellowship Program for Postdoctoral Research.
References
- Rikihisa Y, Kawahara MWB, Kociba G, Fuerst P, Kawamori F, Suto C, Shibata S, Futohashi M. Western immunoblot analysis of Haemobartonella muris and comparison of 16S rRNA gene sequences of H. muris,H. felis, and Eperythrozoon suis. J Clin Microbiol. 1997;35:823–829. doi: 10.1128/jcm.35.4.823-829.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neimark H, Kocan KM. The cell wall-less rickettsia Eperythrozoon wenyonii is a Mycoplasma. FEMS Microbiol Lett. 1997;156:287–291. doi: 10.1111/j.1574-6968.1997.tb12742.x. [DOI] [PubMed] [Google Scholar]
- Neimark H, Johansson KE, Rikihisa Y, Tully JG. Proposal to transfer some members of the genera Haemobartonella and Eperythrozoon to the genus Mycoplasma with descriptions of 'Candidatus Mycoplasma haemofelis', 'Candidatus Mycoplasma haemomuris', 'Candidatus Mycoplasma haemosuis' and 'Candidatus Mycoplasma wenyonii'. Int J Syst Evol Microbiol. 2001;51:891–899. doi: 10.1099/00207713-51-3-891. [DOI] [PubMed] [Google Scholar]
- Neimark H, Johansson KE, Rikihisa Y, Tully JG. Revision of haemotrophic Mycoplasma species names. Int J Syst Evol Microbiol. 2002;52:683. doi: 10.1099/00207713-52-2-683. [DOI] [PubMed] [Google Scholar]
- Messick JB, Walker PG, Raphael W, Berent LM, Shi X. Candidatus Mycoplasma haemodidelphidis’ sp. nov., ‘Candidatus Mycoplasma haemolamae’ sp. nov. and Mycoplasma haemocanis comb. nov., haemotrophic parasites from a naturally infected opossum (Didelphis virginiana ), alpaca (Lama pacos ) and dog (Canis familiaris ): phylogenetic and secondary structural relatedness of their 16S rRNA genes to other mycoplasmas. Int J Syst Evol Microbiol. 2002;52:693–698. doi: 10.1099/ijs.0.01861-0. [DOI] [PubMed] [Google Scholar]
- Kikuth W. Uber einen neuen anamieerreger, Bartonella canis nov. sp. Klin Wchnschr. 1928;7:1729–1730. doi: 10.1007/BF01738861. (in German) [DOI] [Google Scholar]
- Tyzzer EE, Weinman D. Haemobartonella n.g. (Bartonella olim pro parte), H. microti n. sp. of the field vole, Microtus pennsylvanicus. Am J Hyg. 1939;30:141–157. [Google Scholar]
- Lumb WV. Canine Haemobartonellosis and Its Feline Counterpart. Calif Vet. 1961;14:24–25. [Google Scholar]
- Bellamy JE, Macwillams PS, Searcy GP. Cold-agglutinin hemolytic anaemia and Haemobartonella canis infection in a dog. J Am Vet Med Assoc. 1978;173:397–401. [PubMed] [Google Scholar]
- Hulme-Moir KL, Barker EN, Stonelake A, Helps CR, Tasker S. Use of real-time polymerase chain reaction to monitor antibiotic therapy in a dog with naturally acquired Mycoplasma haemocanis infection. J Vet Diag Invest. 2010;22:582–587. doi: 10.1177/104063871002200413. [DOI] [PubMed] [Google Scholar]
- Biondo AW, Dos Santos AP, Guimaraes AM, Vieira RF, Vidotto O, Macieira Dde B, Almosny NR, Molento MB, Timenetsky J, de Morais HA, González FH, Messick JB. A review of the occurrence of hemoplasmas (hemotrophic mycoplasmas) in Brazil. Rev Bras Parasitol Vet. 2009;18:1–7. doi: 10.4322/rbpv.01803001. [DOI] [PubMed] [Google Scholar]
- Novacco M, Meli ML, Gentilini F, Marsilio F, Ceci C, Pennisi MG, Lombardo G, Lloret A, Santos L, Carrapiço T, Willi B, Wolf G, Lutz H, Hofmann-Lehmann R. Prevalence and geographical distribution of canine hemotropic mycoplasma infections in Mediterranean countries and analysis of risk factors for infection. Vet Microbiol. 2010;142:276–284. doi: 10.1016/j.vetmic.2009.09.069. [DOI] [PubMed] [Google Scholar]
- Roura X, Peters IR, Altet L, Tabar MD, Barker EN, Planellas M, Helps CR, Francino O, Shaw SE, Tasker S. Prevalence of hemotropic mycoplasmas in healthy and unhealthy cats and dogs in Spain. J Vet Diagn Invest. 2010;22:270–274. doi: 10.1177/104063871002200219. [DOI] [PubMed] [Google Scholar]
- Tennant KV, Barker EN, Polizopoulou Z, Helps CR, Tasker S. Real-time quantitative polymerase chain reaction detection of haemoplasmas in healthy and unhealthy dogs from Central Macedonia, Greece. J Small Anim Pract. 2011;52:645–649. doi: 10.1111/j.1748-5827.2011.01126.x. [DOI] [PubMed] [Google Scholar]
- Luria BJ, Levy JK, Lappin MR, Breitschwerdt EB, Legendre AM, Hernandez JA, Gorman SP, Lee IT. Prevalence of infectious diseases in feral cats in Northern Florida. J Feline Med Surg. 2004;6:287–296. doi: 10.1016/j.jfms.2003.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neimark H, Hoff B, Ganter M. Mycoplasma ovis comb. nov. (formerly Eperythrozoon ovis), an epierythrocytic agent of haemolytic anaemia in sheep and goats. Int J Syst Evol Microbiol. 2004;54:365–371. doi: 10.1099/ijs.0.02858-0. [DOI] [PubMed] [Google Scholar]
- Brinson JJ, Messick JB. Use of a polymerase chain reaction assay for detection of Haemobartonella canis in a dog. J Am Vet Med Assoc. 2001;218:1943–1945. doi: 10.2460/javma.2001.218.1943. [DOI] [PubMed] [Google Scholar]
- Birkenheuer AJ, Breitschwerdt EB, Alleman AR, Pitulle C. Differentiation of Haemobartonella canis and Mycoplasma haemofelis on the basis of comparative analysis of gene sequences. Am J Vet Res. 2002;63:1385–1388. doi: 10.2460/ajvr.2002.63.1385. [DOI] [PubMed] [Google Scholar]
- Regendanz P, Reichenow E. Beitrag zur kenntnis von Bartonella canis. Arch Schiffs-u Tropen-Hyg. 1932;36:305–322. [Google Scholar]
- Flint JC. PhD thesis. St. Paul, Minnesota, USA: University of Minnesota, School of Veterinary Medicine; 1955. Feline infectious anemia. [Google Scholar]
- Griesemer RA. Bartonellosis. J Natl Cancer Inst. 1958;20:949–954. [PubMed] [Google Scholar]
- Lumb WV. More information on haemobartonellosis in dogs. J Am Vet Med Assoc. 2001;219:732–733. doi: 10.2460/javma.2001.219.732. [DOI] [PubMed] [Google Scholar]
- Guimaraes AM, Santos AP, SanMiguel PJ, Walter T, Timenetsky J, Messick JB. Complete genome sequence of Mycoplasma suis and insights into its biology and adaption to an erythrocyte niche. PLoS One. 2011;6:e19574. doi: 10.1371/journal.pone.0019574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AP, Guimaraes AM, do Nascimento NC, SanMiguel PJ, Martin SW, Messick JB. Genome of Mycoplasma haemofelis, unraveling its strategies for survival and persistence. Vet Res. 2011;42:102. doi: 10.1186/1297-9716-42-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehlerking J, Kube M, Felder KM, Matter D, Wittenbrink MM, Schwarzenbach S, Kramer MM, Hoelzle K, Hoelzle LE. Complete genome sequence of the hemotrophic Mycoplasma suis strain KI3806. J Bacteriol. 2011;193:2369–2370. doi: 10.1128/JB.00187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker EN, Darby AC, Helps CR, Peters IR, Heesom KJ, Arthur CJ, Crossett B, Hughes MA, Radford AD, Tasker S. Molecular characterization of the uncultivatable hemotropic bacterium Mycoplasma haemofelis. Vet Res. 2011;42:83. doi: 10.1186/1297-9716-42-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker EN, Darby AC, Helps CR, Peters IR, Hughes MA, Radford AD, Novacco M, Boretti FS, Hofmann-Lehmann R, Tasker S. Genome sequence for "Candidatus Mycoplasma haemominutum," a low-pathogenicity hemoplasma species. J Bacteriol. 2012;194:905–906. doi: 10.1128/JB.06560-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biegert A, Mayer C, Remmert M, Söding J, Lupas AN. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 2006;34:W335–339. doi: 10.1093/nar/gkl217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/S0968-0004(98)01336-X. [DOI] [PubMed] [Google Scholar]
- Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yus E, Maier T, Michalodimitrakis K, van Noort V, Yamada T, Chen WH, Wodke JA, Güell M, Martínez S, Bourgeois R, Kühner S, Raineri E, Letunic I, Kalinina OV, Rode M, Herrmann R, Gutiérrez-Gallego R, Russell RB, Gavin AC, Bork P, Serrano L. Impact of genome reduction on bacterial metabolism and its regulation. Science. 2009;326:1263–1268. doi: 10.1126/science.1177263. [DOI] [PubMed] [Google Scholar]
- Rahman O, Cummings SP, Harrington DJ, Sutcliffe IC. Methods for the bioinformatic identification of bacterial lipoproteins encoded in the genomes of Gram-positive bacteria. World J Microbiol Biotech. 2008;24:2377–2382. doi: 10.1007/s11274-008-9795-2. [DOI] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acid Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitale M, Hawkins T, Park C, Kihara D. ESG: extended similarity group method for automated protein function prediction. Bioinformatics. 2009;25:1739–1745. doi: 10.1093/bioinformatics/btp309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A, Scherer S, van Alphen L, Verbrugh H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1998;62:275–293. doi: 10.1128/mmbr.62.2.275-293.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citti C, Nouvel LX, Baranowski E. Phase and antigenic variation in mycoplasmas. Future Microbiol. 2010;5:1073–1085. doi: 10.2217/fmb.10.71. [DOI] [PubMed] [Google Scholar]
- Lumb WV. PhD thesis. St. Paul, Minnesota, USA: University of Minnesota, School of Veterinary Medicine; 1957. A study of canine haemobartonellosis. [Google Scholar]
- Palmer GH. The highest priority: what microbial genomes are telling us about immunity. Vet Immunol Immunopathol. 2002;85:1–8. doi: 10.1016/S0165-2427(01)00415-9. [DOI] [PubMed] [Google Scholar]
- Bickle TA, Krüger DH. Biology of DNA restriction. Microbiol Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue JP, Peek RM. In: Restriction and Modification Systems. Mobley HLT, Mendz GL, Hazell SL, editor. Washington (DC): ASM Press; 2001. Helicobacter pylori: Physiology and Genetics. Chapter 24. [Google Scholar]
- Tasker S, Helps CR, Day MJ, Harbour DA, Shaw SE, Harrus S, Baneth G, Lobetti RG, Malik R, Beaufils JP, Belford CR, Gruffydd-Jones TJ. Phylogenetic analysis of hemoplasma species: an international study. J Clin Microbiol. 2003;41:3877–3880. doi: 10.1128/JCM.41.8.3877-3880.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of paralogous gene families* in the genome of Mycoplasma haemocanis strain Illinois. Complete list of paralogous gene families in Mycoplasma haemocanis strain Illinois genome assigned by BLASTclust tool provided by Max-Planck Institute for Developmental Biology, with 70% covered length and 30% sequence identity thresholds. GenBank accession numbers are provided.
Table S2. Analyses of the unique protein coding sequences (CDSs) of Mycoplasma haemocanis strain Illinois and M. haemofelis strain Ohio2. Analyses of all the unique CDSs of M. haemocanis strain Illinois and M. haemofelis strain Ohio2 were performed using ESG software, which predicts protein functions based on sequence similarity, and PSORTb version 3 software, which predicts subcellular localization. GenBank accession numbers of proteins corresponding to each CDS are also provided.
Table S3. Tandem repeats* identified in the genome of Mycoplasma haemocanis strain Illinois. Complete list of variable tandem repeats, identified using Tandem repeats finder program, indicating their sequences and position in the genome of M. haemocanis.