The exomer complex, consisting of ChAPs and Chs5p, exports specialized cargoes from the TGN. ChAPs bind to Chs5p through TPR repeats, whereas cargo specificity of the ChAPs is outside these interaction modules. Chs3p and Chs6p may require a complex interaction to form a complex.
Abstract
The exomer complex is a putative vesicle coat required for the direct transport of a subset of cargoes from the trans-Golgi network (TGN) to the plasma membrane. Exomer comprises Chs5p and the ChAPs family of proteins (Chs6p, Bud7p, Bch1p, and Bch2p), which are believed to act as cargo receptors. In particular, Chs6p is required for the transport of the chitin synthase Chs3p to the bud neck. However, how the ChAPs associate with Chs5p and recognize cargo is not well understood. Using domain-switch chimeras of Chs6p and Bch2p, we show that four tetratricopeptide repeats (TPRs) are involved in interaction with Chs5p. Because these roles are conserved among the ChAPs, the TPRs are interchangeable among different ChAP proteins. In contrast, the N-terminal and the central parts of the ChAPs contribute to cargo specificity. Although the entire N-terminal domain of Chs6p is required for Chs3p export at all cell cycle stages, the central part seems to predominantly favor Chs3p export in small-budded cells. The cargo Chs3p probably also uses a complex motif for the interaction with Chs6, as the C-terminus of Chs3p interacts with Chs6p and is necessary, but not sufficient, for TGN export.
INTRODUCTION
The trans-Golgi network (TGN) is the central sorting station for exocytic and endocytic cargoes. In the yeast Saccharomyces cerevisiae, several sorting machineries and vesicular carriers operate along at least two routes to the cell surface, marked by high-density or low-density secretory vesicles (Harsay and Bretscher, 1995; Harsay and Schekman, 2002; Bard and Malhotra, 2006). In addition, a subset of cargoes travels directly to the plasma membrane in low-density carriers, a subset of which require the exomer complex. This complex is a potential coat complex formed by the peripheral Golgi protein Chs5p and a protein family termed ChAPs, for Chs5p- and Arf1p-binding proteins. In budding yeast, this family includes the paralogues Chs6p, Bud7p, Bch1p, and Bch2p (Ziman et al., 1998; Sanchatjate and Schekman, 2006; Trautwein et al., 2006; Wang et al., 2006). Chs5p and the ChAPs are recruited from the cytosol to the TGN membrane by the small GTPase Arf1p. Together, they facilitate the incorporation of specific transmembrane cargoes into secretory vesicles (Trautwein et al., 2006; Wang et al., 2006).
Some specialized cargoes, such as chitin synthase III (Chs3p) or Fus1p, depend on exomer for their transport to the cell surface (Santos and Snyder, 1997; Ziman et al., 1998; Barfield et al., 2009). However, Chs3p and Fus1p do not share a common sorting motif (Barfield et al., 2009), suggesting that the exomer complex recognizes cargoes individually, perhaps in order to allow differential sorting. This provides an attractive model system for a protein trafficking pathway that is distinct from the major transport routes, allowing the cell to fine tune the surface expression of cargoes depending on the cell cycle stage or potentially also in response to the nutrient status and/or stress conditions.
The exomer components display a functional hierarchy. Whereas individual ChAP deletions—or combinations thereof—lead to certain cellular defects, a deletion of CHS5 collectively causes all ChAPs-associated defects (Trautwein et al., 2006). Given that these phenotypes are most likely due to the inability of specific cargoes to leave the TGN, this places Chs5p functionally upstream of the ChAPs. For example, Δchs6 cells cannot export Chs3p and thus have chitin synthesis defects, whereas Δbch1 cells are sensitive to ammonium (Trautwein et al., 2006). Accordingly, cells lacking CHS5 are both chitin deficient and ammonium sensitive. Interestingly, Chs3p export is also blocked when BCH1 and BUD7 are simultaneously deleted, suggesting that the ChAPs have partially overlapping functions. Alternatively, the ChAPs may also play a structural role in exomer complex assembly.
Chs5p requires activated Arf1p for TGN recruitment, whereas the ChAPs require both Chs5p and Arf1p, reflecting the functional hierarchy. The ChAPs do not coprecipitate in the absence of Chs5p, suggesting that they do not directly bind to each other (Sanchatjate and Schekman, 2006; Trautwein et al., 2006). How Chs5p and the ChAPs associate into a complex has not been investigated in detail. Because of their association with distinct cargoes, it is believed that the ChAPs act as soluble receptors for transmembrane cargoes. However, their mode of cargo recognition has not been characterized.
In this study, we performed a functional analysis of the ChAP Chs6p and found that the ChAP family members contain five essential tetratricopeptide repeats (TPRs), four of which are required for binding to Chs5p and other ChAPs. Export from the TGN and bud-neck localization of the Chs6p-dependent cargo Chs3p were dependent on extended Chs6p-specific sequences outside of the TPRs, suggesting an extensive interaction between Chs3p and Chs6p. The N-terminal 244 amino acids (aa) were required for Chs3p export early and late in the cell cycle, whereas the central part (aa 405–612) was specifically engaged in Chs3p transport early in the cell cycle. Similarly, we found that the C-terminal part of Chs3p bound to Chs6p. Although this interaction was necessary for Chs3p export from the TGN, it was not sufficient, as transplanting the signal onto another protein did not make this protein an exomer-dependent cargo.
RESULTS
The ChAPs contain tetratricopeptide repeats
The ChAPs appear to interact directly with exomer-dependent cargoes. To gain a better understanding of how cargo recognition and the interaction with other exomer components are achieved, we decided to examine the domain structure of the ChAPs. To this end, we performed a BLASTP search of the S. cerevisiae ChAP CHS6 against other fungal genomes. The resulting alignment showed that particular stretches of the protein were highly conserved across species, whereas other sequences were more variable (Supplemental Figure S1A). We expected the more conserved stretches to correspond to domains essential for function, whereas the sequences with a higher degree of variation might represent parts of the protein that are not involved in functions specific to the ChAPs family. Alternatively, those variable domains could be engaged in cargo recognition, because the cargoes studied thus far, Fus1p and Chs3p, do not share obvious motifs that are commonly recognized by all ChAPs (Barfield et al., 2009).
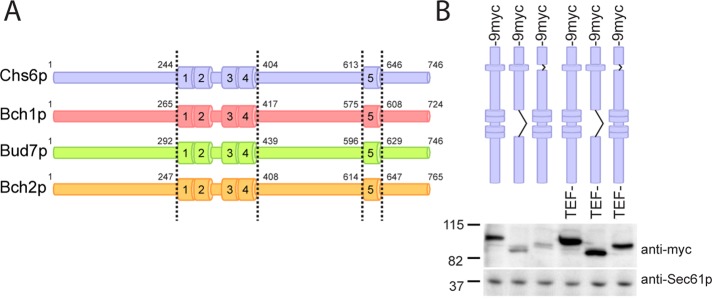
To analyze the conserved regions in more detail, we used a number of different algorithms of the Bioinformatics Toolkit (http://toolkit.tuebingen.mpg.de; Biegert et al., 2006). Interestingly, the conserved regions contained tetratricopeptide repeats (TPRs; Figure 1A), four of which were clustered in the central region of Chs6p, with a fifth one located toward the C-terminus. The TPRs were conserved among the different S. cerevisiae ChAPs, indicating that they may represent a common feature of this protein family (Figure 1A and Supplemental Figure S1B). This hypothesis is supported by the finding that automatic sequence annotation detected TPRs in ChAPs from Kluyveromyces lactis, Ashbya gossypii, and others (see, e.g., National Center for Biotechnology Information, www.ncbi.nlm.nih.gov/protein/CAG98421.1).
FIGURE 1:
Deletion of TPRs in ChAPs only mildly affects protein expression levels. (A) Domain structure of the ChAP family members. Numbers indicate the first and last amino acid of the TPR domains. The same coloring scheme is used in all subsequent figures. (B) Expression of 9myc-tagged TPR mutants of Chs6p, under the native and the TEF promoter. Immunoblot of yeast lysates; Sec61p serves as loading control. Note that all mutants were generated chromosomally.
TPRs are highly versatile protein–protein interaction domains. Each repeat consists of a degenerate 34–amino acid motif, which exhibits a conserved helix-turn-helix fold and the ability to form clusters of multiple repeats (Blatch and Lassle, 1999; Zhang et al., 2010). Interestingly, several cases of cargo recognition by TPRs have been described: peroxin 5, which harbors a six-TPR tunnel recognizing the C-terminal SKL motif for peroxisomal import (Gatto et al., 2000); Tom20, which facilitates mitochondrial import (Abe et al., 2000); and kinesin light chain, which binds multiple cargoes via its TPR domain (Kamal et al., 2000; Hammond et al., 2008). Alternatively, TPRs can also have more-structural roles, for example, in the assembly of multiprotein complexes such as the COPI vesicle coat (Hsia and Hoelz, 2010) or the anaphase-promoting complex (Zhang et al., 2010). Thus finding TPRs in the ChAPs family members raised the possibility that these repeats would be of functional importance for the exomer complex and could potentially provide protein–protein interaction surfaces.
The TPRs are essential for Chs6p function
The TPRs in the ChAPs might serve either as interaction modules for other exomer components or as cargo recognition sites. To distinguish between these possibilities, we created two internal truncations in Chs6p. The first truncation, Chs6(ΔTPR1-4), lacked the entire central cluster of TPRs. In the second construct, Chs6(ΔTPR5), the last and most conserved repeat in the protein was deleted (Figure 1A). The mutant proteins showed only a mild reduction in expression compared with wild type, indicating that removing one or more TPRs did not cause the protein to be largely unfolded and hence degraded (Figure 1B). The truncations did not massively shorten the proteins. Removing TPR1–4 reduced the molecular weight by ∼15 kDa, and eliminating TPR5 caused a 5-kDa reduction. To have consistently comparable expression levels, we decided to replace the endogenous promoter in all cases by the somewhat stronger TEF promoter (Figure 1B).
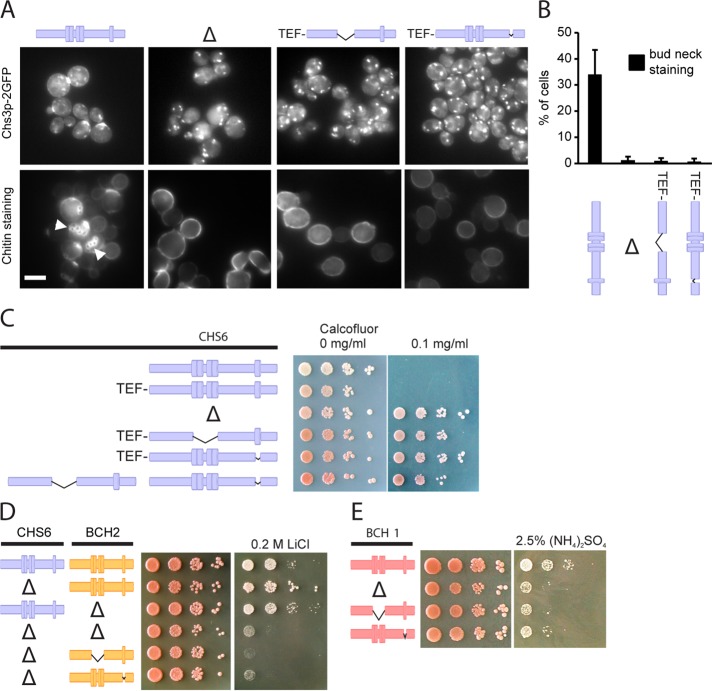
To assess the functionality of Chs6(ΔTPR1-4) and Chs6(ΔTPR5), we monitored the localization and activity of Chs3p, whose TGN export depends on functional Chs6p. Both truncation mutants were unable to export Chs3p–2 GFP from the TGN, as GFP staining was absent from the bud neck and Chs3p accumulated in intracellular structures, mimicking a CHS6 deletion (Figure 2, A and B). Chs3p synthesizes a chitin ring around the yeast bud neck, which can be visualized by calcofluor staining (Lord et al., 2002). The chitin ring was absent in Δchs6, Chs6(ΔTPR1-4), and Chs6(ΔTPR5) (Figure 2A and Table 1). All three mutants were calcofluor resistant, a hallmark of chitin synthesis–defective cells (Ziman et al., 1998), demonstrating a lack of chitin synthase III activity at the plasma membrane (Figure 2C).
FIGURE 2:
The TPRs are essential for the function of the ChAPs. (A) Chs3p-2GFP localized exclusively to internal structures in Δchs6, Chs6(ΔTPR1-4), and Chs6(ΔTPR5) strains. Accordingly, whereas calcofluor-stained wild-type cells showed bud scar chitin staining (arrowheads), this was absent in the mutants. Scale bar, 5 μm (B) Quantification of results in A. Graph shows an average of three experiments. Bud-neck staining was scored for the entire cell population in at least 100 cells per experiment. Bars, SD. (C) Chs6(ΔTPR1-4) and Chs6(ΔTPR5) strains were resistant to calcofluor. This defect was as pronounced as for a Δchs6 strain. The two mutant alleles showed no cross-complementation. Drop tests: plates were incubated at 30°C for 2–3 d. Blue, Chs6p alleles. Δ refers to Δchs6. (D) Bch2p requires TPRs for functionality. A CHS6 deletion in combination with a Δbch2, Bch2(ΔTPR1-4), or Bch2(ΔTPR5) allele led to lithium sensitivity. Drop tests were performed as described. Yellow, Bch2p alleles. Δ refers to Δchs6 and Δbch2, respectively. (D) Bch1p requires TPRs for functionality. Bch1(ΔTPR1-4) and Bch1(ΔTPR5) cells, like Δbch1, were sensitive to ammonium. Red, Bch1p alleles. Δ refers to Δbch1.
TABLE 1:
Summary of results for Chs6p mutants used in the study.
 |
The ChAPs form complexes with Chs5p in varying stoichiometries (Trautwein et al., 2006). We therefore wondered whether the truncations, when expressed together, could cross-complement and rescue the calcofluor sensitivity. However, this was not the case, indicating that each Chs6p molecule must contain the full set of TPR motifs (Figure 2C). In summary, these findings demonstrate that the TPRs of Chs6p are required for export of Chs3p from the TGN.
TPR function is conserved in the ChAPs
ChAPs share some degree of redundancy, indicated by the fact that some cellular phenotypes only arise upon deletion of multiple ChAPs (Trautwein et al., 2006; Barfield et al., 2009). For example, double deletion of CHS6 and BCH2 renders cells lithium sensitive, a phenotype that could not be observed for either single deletion (Figure 2D). This finding implicates Chs6p in the export of another, yet-unidentified, cargo involved in lithium homeostasis.
We used this paradigm to test whether the TPRs in other ChAPs might be of equal importance for function. Indeed, Bch2(ΔTPR1-4 or ΔTPR5), combined with a CHS6 deletion, also displayed the lithium-sensitivity phenotype (Figure 2D). Moreover, we constructed analogous truncation mutants in Bch1p and tested these for ammonium sensitivity, which is a characteristic phenotype of Δbch1 cells (Trautwein et al., 2006). Both TPR mutants behaved like the BCH1 deletion (Figure 2D), indicating that at least three (Chs6p, Bch2p, Bch1p) of the four ChAPs require their TPRs for functionality.
Chs6p requires its TPRs for efficient Golgi recruitment
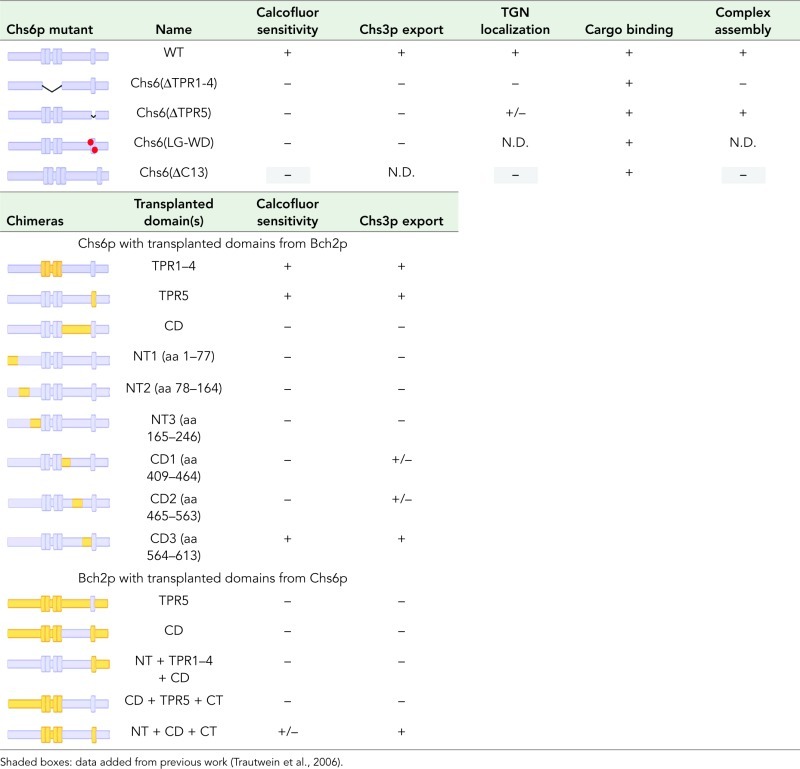
The strong defect of the TPR mutants in cargo export could be explained by impaired recruitment of the mutant proteins to the Golgi, failure to form a productive exomer–cargo complex, or a combination of both. We therefore tested first whether the TPRs were required for Golgi association and determined the subcellular localization of the TPR mutants using differential centrifugation. Chs6(ΔTPR1‑4)‑9myc and Chs6(ΔTPR5)‑9myc were depleted from the fractions containing Golgi membranes and were found almost exclusively in the cytosol (Figure 3A).
FIGURE 3:
Chs6(ΔTPR1-4) and Chs6(ΔTPR5) cannot be efficiently recruited to the Golgi. (A) Chs6(ΔTPR1-4)-9myc and Chs6(ΔTPR5)-9ymc display a reduced membrane association in cell lysates. Ten OD600 of cells were spheroplasted, regenerated, and subsequently lysed in hypotonic buffer. Lysates were cleared of unbroken cells and subjected to differential centrifugation at 4°C. TCL, total cell lysate; P13, 13,000 × g pellet; S100, 100,000 × g supernatant; P100, 100,000 × g pellet; PM, plasma membrane. All constructs were chromosomally expressed under the native CHS6 promoter. (B) TPR mutants show inefficient Golgi localization in vivo. Chs6p‑3GFP and Chs6(ΔTPR1-4)-3GFP were chromosomally expressed under the native CHS6 promoter. Chs6(ΔTPR1-4)-3GFP was almost entirely cytoplasmic and showed no association with Golgi membranes. Chs6(ΔTPR5)-3GFP, expressed at a level similar to wild-type Chs6p using an inducible methionine promoter, was partially Golgi localized (arrowheads). Scale bar, 5 μm. (C) Quantification of results in B. Graph shows a total of three experiments. At least 95 cells were scored per experiment; only budded cells were used for scoring; only GFP dots overlapping with Sec7-dsRed were considered as TGN. Drawn with Origin software (OriginLab, Northampton, MA). Lower whisker represents 5th percentile; box represents 25th, 50th, and 75th percentiles; upper whisker represents 95th percentile.
To corroborate our findings, we also monitored the localization of the truncations by live imaging. A 3×GFP-tagged version of wild-type Chs6p mostly localized to punctate structures, which overlapped with the TGN marker Sec7p-dsRed (Figure 3B). As previously observed, some Chs6p-3GFP was also found in the cytoplasm (Ziman et al., 1998; Trautwein et al., 2006). Consistent with the in vitro fractionation, in vivo, both Chs6(ΔTPR1-4)-3GFP and Chs6(ΔTPR5)‑3GFP were not efficiently recruited to the TGN, as Chs6(ΔTPR1-4)-3GFP was found entirely in the cytoplasm, whereas a minor fraction of Chs6(ΔTPR5)-3GFP was present at the TGN (Figure 3, B and C). Thus all five TPRs contribute to efficient Golgi recruitment, whereby TPRs 1–4 seem to play a more predominant role.
TPR1–4 are required for interaction with Chs5p and other ChAPs
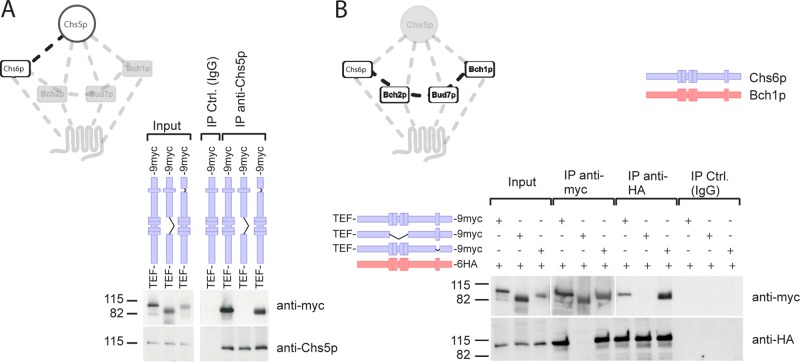
We showed previously that the ChAPs require Chs5p for steady-state Golgi localization (Trautwein et al., 2006). Therefore, we asked next whether Chs5p interaction was also impaired in the TPR mutants and whether this was the cause of the cytoplasmic localization of the mutants. Chs6(ΔTPR1-4) could not be coprecipitated with Chs5p (Figure 4A), indicating that the lack of this interaction might be the cause of the cytoplasmic localization of Chs6(∆TPR1-4). In contrast, Chs5p and Chs6(ΔTPR5) coprecipitated, suggesting that TPR5 is not involved in Chs5p binding.
FIGURE 4:
Chs6(ΔTPR1-4) fails to interact with other exomer components. (A) Chs6(ΔTPR1-4) failed to bind to Chs5p, whereas Chs6(ΔTPR5) still interacted. Coimmunoprecipitation experiments were performed using an anti-Chs5p antibody and lysates generated from cells expressing chromosomally tagged Chs6(ΔTPR1-4)-9myc or Chs6(ΔTPR5)-9myc. (B) Chs6(ΔTPR1-4) also failed to coprecipitate with other ChAPs, such as Bch1p. Blue, Chs6p alleles; red, Bch1p alleles. Two different exposures were cropped together because of the strong signal of the precipitated myc-tagged constructs.
Chs6p copurifies with Bch1p in a Chs5p-dependent manner (Sanchatjate and Schekman, 2006). Thus we expected that Chs6(ΔTPR5) would still bind to other ChAPs, whereas Chs6(ΔTPR1-4) would not. Alternatively, TRP5 of Chs6p could interact with the TPR5 of other ChAPs. Chs6(ΔTPR1-4), but not Chs6(ΔTPR5), specifically failed to interact with Bch1p (Figure 4B). Similarly, Bud7p bound to Chs6(∆TPR5) but not Chs6(∆TPR1-4) (Supplemental Figure S2). These results suggest that the ChAPs require their first four TPRs for the association with Chs5p and thus for assembly into a complex with other exomer components.
The TPRs are dispensable for cargo recognition
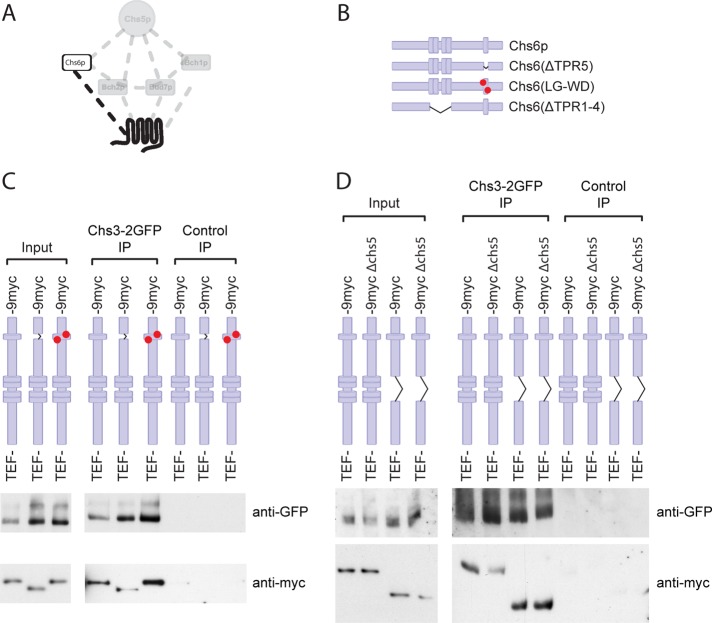
Chs6p interacts with both Chs5p and the cargo Chs3p, and Chs5p is required for Chs6p binding to Chs3p (Trautwein et al., 2006; Figure 5A). Because the cargo interaction site in the ChAPs is not known and TPRs mediate protein–protein interactions, we tested whether the TPRs would be involved in this process. The binding between a cargo and its cargo receptor is usually rather transient (Appenzeller et al., 1999; Muniz et al., 2000; Zhang et al., 2005). To “freeze” the interaction, we performed immunoprecipitations after chemical cross-link from yeast lysates. This approach has been used previously to detect exomer–cargo interactions (Sanchatjate and Schekman, 2006; Trautwein et al., 2006; Barfield et al., 2009). Interestingly, both TPR mutants were efficiently cross-linked to Chs3p (Figure 5, C and D), indicating that the potential to recognize cargo in vitro was not strongly impaired in Chs6(ΔTPR1‑4) or Chs6(ΔTPR5). This result was somewhat unexpected because in the wild-type situation Chs6p requires the presence of Chs5p to interact efficiently with Chs3p in vitro (Trautwein et al., 2006; Figure 5D). Yet, Chs6(∆TPR1-4) did not bind Chs5p and interacted with cargo independent of the presence of Chs5p. These findings would indicate that cargo binding and Chs5p interaction are separable in Chs6p but that Chs5p may negatively influence cargo binding by weakening the receptor–cargo interaction. To test this hypothesis, we used another truncation of Chs6p, one in which the C-terminal 13 amino acids were deleted (Chs6(∆C13)). This truncation fails to bind Chs5p and cannot be recruited to the Golgi apparatus (Trautwein et al., 2006). Again, like the TPR mutants, Chs6(ΔC13) still bound Chs3p (Supplemental Figure S3D). Taken together, our data suggest that Chs5p binding to Chs6p decreases the stability of Chs6p–cargo interaction.
FIGURE 5:
TPRs are not required for stable association with cargo. (A) Schematic representation of the interaction between the cargo, Chs3p, and the exomer–cargo recognition subunit, Chs6p. (B) Chromosomally generated Chs6p TPR mutants. (C) Chs6(ΔTPR5) and Chs6(LG-WD) interact with cargo. To assess cargo interaction, Chs3p-2GFP was precipitated from DSP cross-linked lysates with anti-GFP monoclonal antibodies, and the precipitates were probed for different Chs6p constructs. Control immunoprecipitations were performed using monoclonal HA antibody. Chs6(ΔTPR5) and a double TPR5 point mutant Chs6(LG-WD) retained association with Chs3p. (D) Chs6(ΔTPR1-4) interacts with Chs3p and does so independently of Chs5p, the core exomer subunit. The cross-linker immunoprecipitation was performed as described in B.
So far we used deletions of the different TPRs. Despite the small size of the deletions, they still may change the structure of the protein and hence influence the binding. To less disturb the overall structure, we constructed a point mutant, Chs6p-L619W/G620D (LG-WD), in which two critical residues of the TPR5 backbone were mutated (Magliery and Regan, 2004) but the protein was otherwise left intact. As expected, this mutant also caused Chs3p‑2GFP to accumulate in the TGN and was calcofluor resistant (Supplemental Figure S3, A–C). Again, this protein also interacted with Chs3p in vitro (Figure 5C). Therefore, the results presented so far indicate no major role of the TPRs in cargo recognition and specificity.
The TPRs are interchangeable among the ChAPs
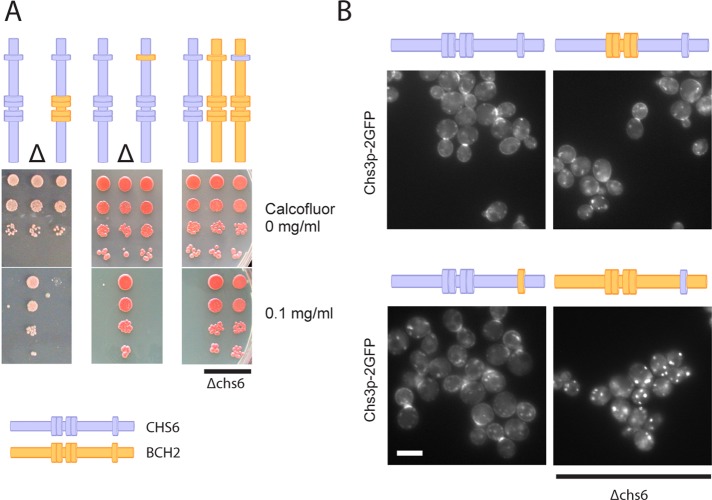
We have shown thus far that TPRs 1–4 are required for interaction with Chs5p and other ChAPs and that neither TPR1–4 nor TPR5 appeared to play a major role in cargo recognition. To corroborate our results, we aimed to least disturb the structure of the protein and constructed chromosomally chimeric mutants of the ChAPs. If the TPRs perform functions that are conserved among the ChAPs, such as Chs5p binding, they should be interchangeable between two different ChAPs. If the TPRs perform a specific function, such as cargo recognition, the TPR chimera should be nonfunctional. We chose CHS6 and BCH2 for these experiments because the functionality of Chs6p could be monitored easily by both Chs3p localization and chitin synthesis. Bch2p, on the other hand, is entirely dispensable for Chs3p traffic. As expected, transplantation of TPR1–4 or TPR5 from BCH2 to CHS6 had no effect on calcofluor sensitivity or Chs3p localization (Figure 6), demonstrating that Chs6p chimera carrying the alien TPR1–4 or TPR5 were indeed functional. Thus the TPRs in Chs6p are most likely not required for cargo recognition.
FIGURE 6:
The TPRs are interchangeable among ChAPs. (A) Chs6p bearing either TPR1–4 or TPR5 from Bch2p is fully functional. Chimeras in which TPRs from CHS6 were grafted into BCH2 (or vice versa) were created by delitto perfetto. Drop tests for calcofluor sensitivity were performed as in Figure 2. Transplanting TPR5 from Chs6p to Bch2p did not restore calcofluor sensitivity in a Δchs6 background. Blue, Chs6p domains; yellow, Bch2p domains. Δ refers to Δchs6. (B) Live fluorescence imaging of Chs3p-2GFP in the chimeras shown in A confirmed that the TPRs do not contribute to cargo specificity. Scale bar, 5 μm.
Cargo specificity of the ChAPs is not conveyed by a simple linear sequence
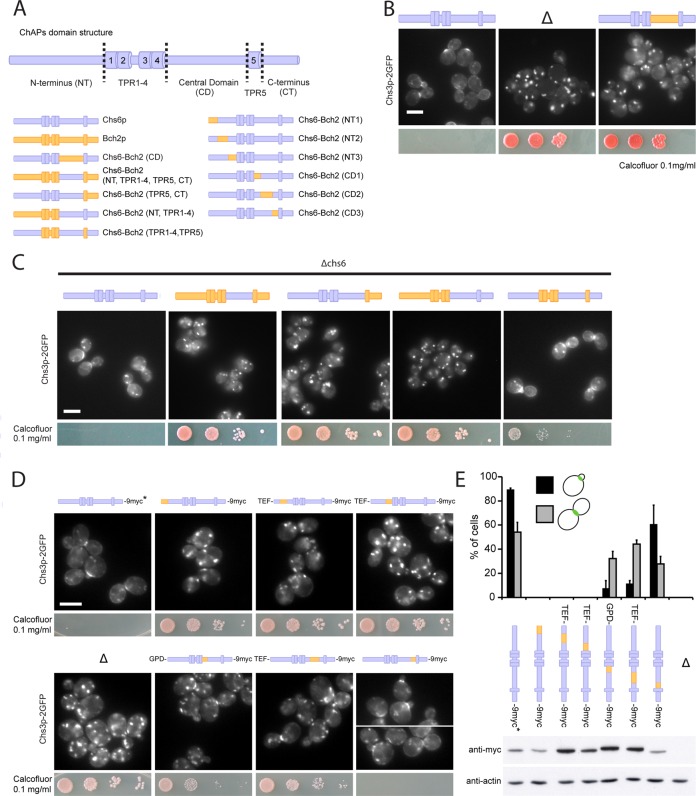
Because the TPRs were not involved in cargo specificity, we asked next where the cargo recognition site was located in Chs6p and how cargo specificity was achieved. We again used our chimera approach to address these questions and concentrated on the regions outside the TPRs (Figure 7A and Supplemental Figure S4). First, we exchanged the central domain (CD, located between TPR4 and TPR5) of Chs6p for the CD of Bch2p. This strain did not export Chs3p from the TGN and was calcofluor resistant, suggesting that this chimeric Chs6p was unable to recognize Chs3p as a cargo (Figure 7B). However, the inverse experiment—transplantation of the corresponding region from CHS6 to BCH2—did not change the cargo specificity of Bch2p and failed to rescue Δchs6 defects, indicating that the central domain of the ChAPs is necessary but not sufficient to convey cargo specificity (Figure 7C). Strikingly, similar results were obtained when we individually exchanged longer sequences, like the C-terminal half (aa 409–765) of Bch2p or even the N-terminus, TPR1–4, and the central domain together (aa 1–613) for the homologous sequences in Chs6p. These results were not due to a positioning effect in the genome, because insertion of the full-length CHS6 ORF into the BCH2 locus restored Chs3p export and calcofluor sensitivity (Figure 7C). Moreover, the chimeric constructs were expressed and stable (unpublished data). In summary, these results suggest that the N-terminal, central, and C-terminal domains were necessary for cargo specificity, but none was sufficient by itself. In fact, only transplanting all corresponding sequences except for TPR1–4 and TPR5 restored Chs3p bud-neck localization and reduced calcofluor resistance to close to wild-type levels (Figure 7C). These data suggest a model in which the TPRs provide the interaction surface for Chs5p, whereas the sequences outside of these repeats may be involved in cargo recognition.
FIGURE 7:
The N-terminus, central domain, and C-terminus of the ChAPs are individually necessary and only together sufficient to convey cargo specificity. (A) Schematic representation of ChAPs domain structure and Chs6p-Bch2p chromosomal chimera constructs. (B) The central domain (CD) of the ChAPs is required for cargo specificity. Chimeric Chs6p bearing the CD of Bch2p was unable to export Chs3p-2GFP and rendered cells calcofluor resistant, like a Δchs6 strain. Chimeras were created by delitto perfetto. Blue, Chs6p domains; yellow, Bch2p domains. Δ refers to Δchs6. Drop assays were performed as in Figure 2. Scale bar, 5 μm. (C) The N-terminus (NT), CD, and C-terminus (CT) are necessary and together sufficient to determine cargo specificity. In a Δchs6 background, calcofluor sensitivity was restored by reintroduction of the CHS6 full-length open reading frame into the BCH2 locus but not by transplantation of the following domains from Chs6p to Bch2p: CD, NT + TPR1–4 + CD, or CD + TPR5 + CT. Transplantation of NT, CD, and CT together restored Chs3p export to the bud neck (by ∼82% compared with wild-type cells). Scale bar, 5 μm. (D) Chs3p bud-neck export requires the entire Chs6p N-terminal domain and the majority of the central domain, the latter only early in the cell cycle. Transplantation of short Bch2p NT fragments into Chs6p resulted in exclusive localization of Chs3-2GFP to internal structures and calcofluor resistance. Transplantation of two short Bch2p CD fragments (aa 409–464 and 465–563), but not the fragment containing aa 564–613, proximal to TPR5 resulted in severely compromised Chs3p cargo export in small-budded cells. Several chimera constructs were expressed chromosomally under the TEF or GPD promoter to achieve protein levels comparable to that of wild-type Chs6p. Drop assays were performed as in Figure 2. Scale bar, 5 μm. (E) Quantification of results in D and expression levels of particular chimera constructs. Graph shows a total of three experiments. Bud-neck staining was scored in 100 small-budded cells (G1/S phase) and 100 large-budded cells (M phase) in each experiment. Bars, SD. Actin serves as a loading control in immunoblot of yeast lysates. Chs6p in wild-type control used in microscopy studies in D and E is untagged, as indicated by the asterisk.
To test this hypothesis, we divided each of the N-terminus and the central domain again into three smaller regions and replaced them individually by the corresponding sequences from Bch2p (Figure 7A). All substitutions in the N-terminal region caused Chs3p to be in internal structures and conferred resistance to calcofluor (Figure7, D and E), indicating that indeed large parts of the N-terminal region of Chs6p are involved in Chs3p export. In contrast, we could narrow down the region in the central domain necessary for Chs3p export. The truncation in which aa 557–612 (closest to TPR5) had been swapped showed Chs3p localization similar to the wild-type control, and the strain was sensitive to calcofluor (Figure 7, D and E). Of importance, the other two chimera of the central domain mislocalized Chs3p only early in the cell cycle (Figure 7, D and E). The bud-neck localization of Chs3p in large-budded cells (late in the cell cycle) was mostly achieved in these strains. Consistently, the calcofluor resistance was reduced. These data imply a general role for the Chs6p N-terminus in cargo transport, whereas parts of the central domain would be required only early in the cell cycle and dispensable for transport late in the cell cycle.
Chs6p interacts with the C-terminus of Chs3p
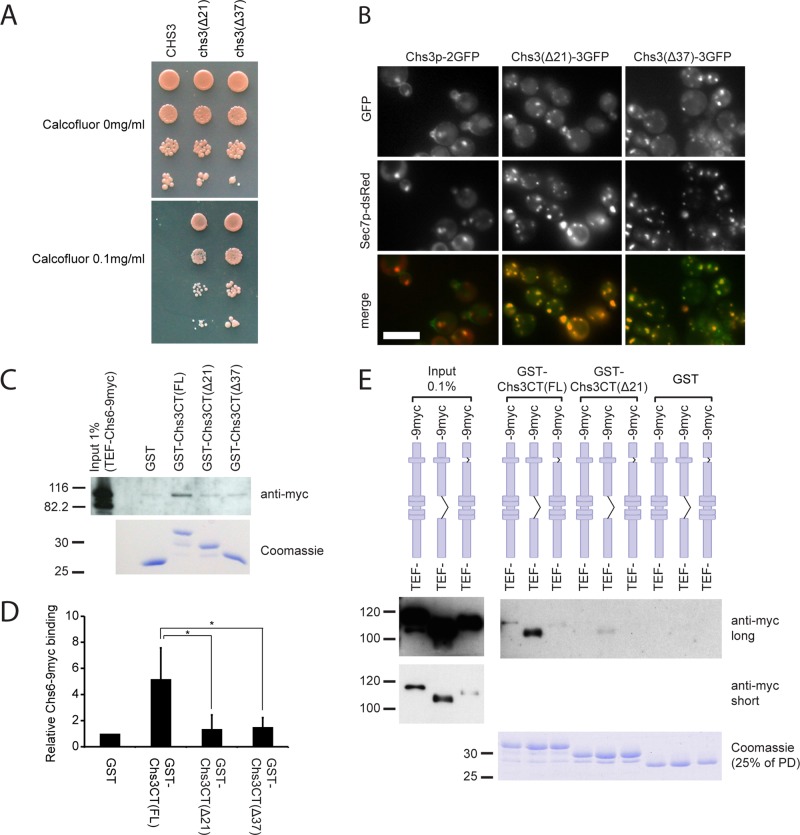
Although we could assign parts in Chs6 that were involved in cargo recognition, the size of the area—especially the N-terminal region—seemed to make it unlikely to identify a small motif that would provide the interaction site with Chs3p. On the other hand, we might be able to identify individual parts of Chs3p required for the interaction with Chs6p, similar to the short, linear motif in Fus1p that binds to exomer (Barfield et al., 2009). Because the topology of Chs3p is still disputed (Cos et al., 1998; Meissner et al., 2010), and even the number of transmembrane (TM) domains is debated—varying between four and eight—we decided to focus on the C‑terminal part of Chs3p. Cos et al. (1998) generated two C-terminal truncations that rendered the cells calcofluor resistant (Figure 8A), suggesting a defect in either Chs3p function or localization. Interestingly, we found that GFP-tagged versions of these mutant proteins failed to reach the cell surface and were retained at the TGN, indicated by colocalization with Sec7p-dsRed (Figure 8B). These results suggest that the C‑terminal 21 amino acids of Chs3p might be important for binding of the exomer complex and thus for incorporation into secretory vesicles.
FIGURE 8:
The C-terminus of Chs3p contains an exomer-binding site required for Golgi export. (A) The last 21 amino acids of Chs3p are essential for chitin synthesis. Cells expressing Chs3p lacking the C‑terminal 21 or 37 amino acids were calcofluor resistant. (B) The C-terminus of Chs3p is required for Golgi export. Chromosomally generated Chs3(Δ21)-3GFP or Chs3(Δ37)‑3GFP was trapped in internal membranes and colocalized with the TGN marker Sec7p‑dsRed. Scale bar, 5 μm. (C) Chs6p binds to the C-terminus of Chs3p. Lysates from cells expressing Chs6p‑9myc were incubated with immobilized GST, GST fused to the C-terminus of Chs3p (FL), or truncated C-terminal constructs (Δ21 and Δ37). Chs6p-9myc bound to the full C-terminus, but binding to the truncations was abolished. (D) Quantification of results in C. Graph shows an average of three experiments. The integrated density of Chs6-9myc bands in GST-Chs3CT pull-downs was measured using ImageJ software and normalized to that in the GST pull-down. Bars, SD. *p < 0.05. (E) Chs6(ΔTPR1‑4)-9myc TPR mutant efficiently binds to the Chs3p C-terminus. GST pull-downs were performed as in C with lysates from cells expressing Chs6(ΔTPR1‑4)-9myc or Chs6(ΔTPR5)-9myc.
We therefore performed GST pull-down experiments using the C-terminal cytoplasmic tail of Chs3p, which has a total length of 55 amino acids following the last predicted TM domain. The corresponding truncation constructs lacked the final 21 and 37 amino acids, respectively. Immobilized GST fusion proteins were then incubated with whole-cell lysate and analyzed for binding of Chs6p. Chs6p-9myc bound to full-length glutathione S-transferase (GST)–Chs3CT but not to GST alone, GST-Chs3CT(Δ21), or GST-Chs3CT(Δ37) (Figure 8, C and D). Consistently, the Chs3p tail truncations did also not bind to Chs6(TPR1-4) and Chs6(TPR5) (Figure 8E). This result suggests that the Chs3p C-terminus contains an exomer recognition site, which is necessary for Chs6p binding in vitro and for Chs3p export in vivo. This site is likely to be located within the last 21 amino acids, as the Δ21 mutation was sufficient to abolish Chs3p transport to the cell surface and abrogate Chs6 binding.
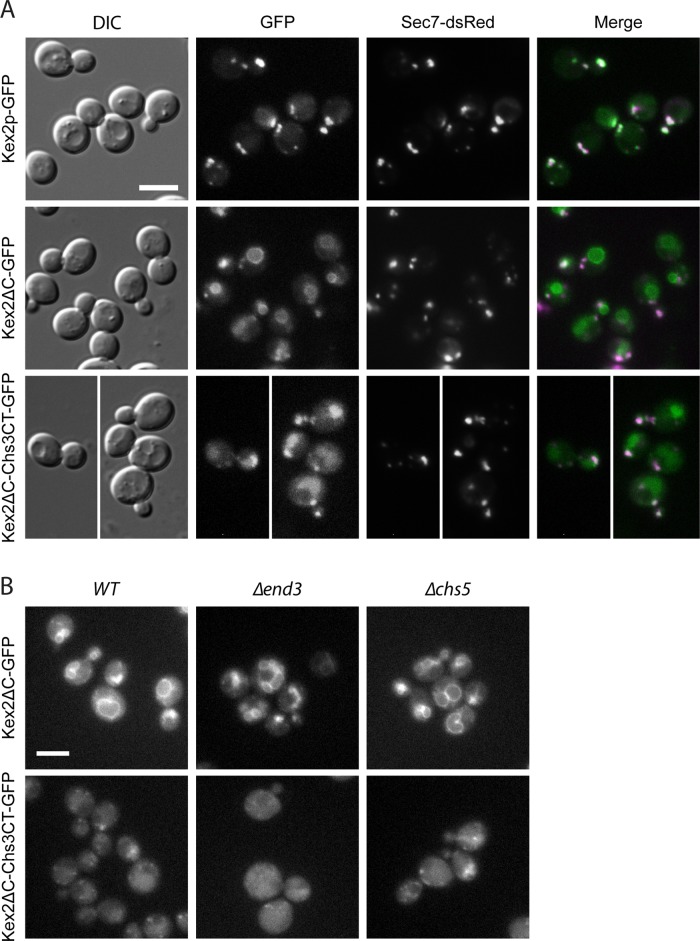
We next tested whether the C-terminal tail would be sufficient to drive TGN export and bud-neck localization of another, unrelated protein. For this, we replaced the C-terminus of the TGN/endosome–localized Kex2p protease with the one of Chs3p (Figure 9A), similar to the approach used for Fus1p (Barfield et al., 2009). The Chs3p C-terminal tail was not sufficient to cause export of Kex2∆C-Chs3CT-GFP to the plasma membrane in either the presence or absence of Chs5p (Figure 9). Yet Kex2p localization was partially altered, as some of it accumulated in the vacuolar lumen, an effect that was not entirely due to the removal of the Kex2p endogenous C-terminus, as Kex2∆C-GFP was most predominantly found on the vacuolar rim. One possible explanation of the difference in localization of both constructs is that Kex2∆C-Chs3CT-GFP could be exported to the plasma membrane and was then rapidly endocytosed. However, inhibiting endocytosis by the ∆end3 mutation did not alter the localization of Kex2∆C-Chs3CT-GFP (Figure 9B), indicating that this construct does not reach the plasma membrane; it might still become a substrate for the ESCRT complex and be included into the intralumenal vesicles of the late endosome.
FIGURE 9:
(A) The C-terminus of Chs3p is not sufficient for exomer-dependent cargo export. Replacement of the cytosolic domain of Kex2p, a TGN-resident protein, with the Chs3p C-terminus did not direct Kex2ΔC-Chs3CT-GFP to the plasma membrane. Kex2ΔC-Chs3CT-GFP was localized to the vacuolar lumen, whereas Kex2ΔC-GFP localized to the vacuolar rim, indicating an influence of the Chs3p C-terminus on Kex2p sorting. Kex2-GFP, C-terminally truncated Kex2ΔC-GFP, and the Kex2ΔC-Chs3CT-GFP chimera were chromosomally expressed. Sec7-dsRed was used as a TGN marker. Scale bar, 5 μm. (B) Kex2ΔC-Chs3CT-GFP does not traffic to the plasma membrane, and its localization is exomer independent. To assess potential trafficking through the plasma membrane, the localization of chromosomally expressed Kex2ΔC-GFP and the Kex2ΔC-Chs3CT-GFP was assessed in a Δend3, endocytosis-deficient strain. Kex2ΔC-GFP and Kex2ΔC-Chs3CT-GFP exomer-dependent localization was assessed in a Δchs5 strain. Scale bars, 5 μm.
Taken together, these results suggest that although the C-terminus of Chs3p is necessary and sufficient to interact, albeit weakly, with Chs6p, it is not sufficient to drive the plasma membrane localization of another protein. Therefore, it is likely that other motifs in Chs3p exist that contact Chs6p and that these combined interactions temporally control the export of Chs3p from the TGN.
DISCUSSION
The late secretory pathway controls the trafficking of proteins to the cell surface and the endosomal system, but how the multitude of cargoes is correctly sorted to control their spatial and temporal localization is not well understood. In recent years, the exomer complex, comprising Chs5p and the ChAPs family, has emerged as a crucial sorting determinant for a subset of cargoes (Santos and Snyder, 1997, 2003; Trautwein et al., 2006). However, little is known about how exomer assembles at the TGN and recognizes specific cargo proteins. To gain insight into the exomer function and cargo interaction, we performed a structure–function analysis of the ChAP Chs6p. We chose Chs6p because it has one well-established cargo, the chitin synthase Chs3p, and is required for proper Chs3p localization at the bud neck early and late in the cell cycle (Zanolari et al., 2011).
The search for conserved structural motifs yielded a cluster of four TPRs in the center and one TPR toward the C-terminus of all ChAPs. TPR1–4 were required for interaction with Chs5p and other ChAPs, as well as for localization to the Golgi, probably through the interaction with Chs5p. In contrast, the fifth TPR, which is the most conserved one by sequence among the ChAPs, probably does not interact with Chs5p or other ChAPs directly and is not actively involved in cargo recognition. However, this TPR is still necessary for efficient Golgi recruitment. Because at least three TPRs appear to be necessary for biological relevant functions, that is, to serve as protein–protein interaction scaffolds (D'Andrea and Regan, 2003; Zeytuni and Zarivach, 2012), it is conceivable that the single TPR5 would contact TPR1–4. In this scenario, either TPR5 itself or a then-exposed sequence would interact with a thus-far-unknown factor at the Golgi to stabilize the TGN localization of ChAPs.
Interestingly, deletions in either TPR1–4 or TPR5 were still able to interact with Chs3p in an in vitro cross-linking approach, indicating that in both cases the ability of cargo recognition was maintained and the lack of steady-state localization of these truncations to the TGN was the reason for the defect in exporting Chs3p to the plasma membrane. It is intriguing that Chs6∆TPR1-4 was reproducibly more efficiently cross-linked to Chs3p than wild-type Chs6p in vitro, even in the absence of Chs5p. Because TPR1–4 are essential for the interaction with Chs5p, it is tempting to speculate that Chs5p, and potentially other ChAPs, may regulate the binding affinity of the Chs6p to the cargo. The affinity of the cargo and its receptor needs to be relatively low to allow readily dissociation of the cargo–receptor complex after either inclusion into the transport carrier or upon release at the target compartment. Although we cannot exclude a regulatory role of the TPRs in cargo binding, they are dispensable for cargo specificity: transplanting TPRs from Bch2p, which has no role in Chs3p trafficking, did not cause mislocalization of Chs3p, hence excluding a function in specific cargo recognition. This was a bit surprising at first because TPRs interact with their ligands through a combination of factors, such as hydrophobic pockets, residue type, charge, and electrostatics (Zeytuni and Zarivach, 2012), and we assumed that these repeats were uniquely suited to recognize a variety of cargoes that do not share sequence homology and are structurally very different, such as Chs3p and Fus1p. Instead, we find that cargo specificity and recognition are located outside the TPRs and are most likely rather complex (Figure 10). Our data indicate that Chs6p-specific sequences from the N-terminus, the central domain, and the C-terminus are involved in the spatial and temporal control of Chs3p localization. In an attempt to narrow down these allegedly large areas, we found that among the N-terminal 246 amino acids there must be many residues that are required for Chs3p export from the TGN and do not form a short linear motif. This analysis made it essentially impossible to go on to define a specific motif that would comprise the Chs3p-binding pocket because our chimera analysis would instead suggest that the folding of the N-terminus would provide a platform or binding pocket for at least part of Chs3p.
FIGURE 10:
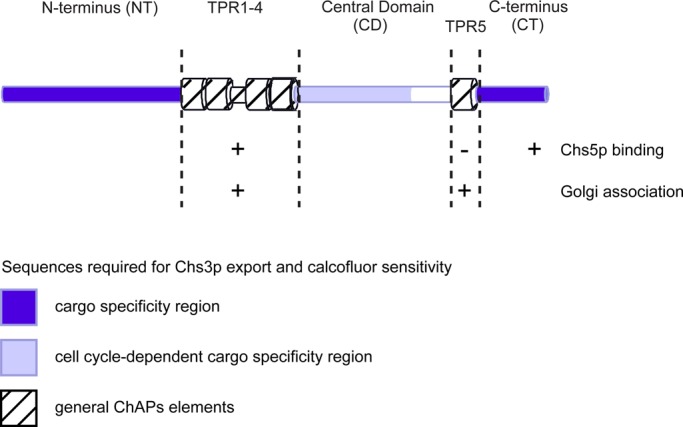
Summary scheme of Chs6p domains and their function in regard to cargo-specific interaction, interaction with Chs5p, the core exomer subunit, and TGN recruitment.
Although replacing the entire central domain of Chs6p by Bch2p sequences caused Chs6p to be nonfunctional in terms of Chs3p transport, systematic replacement of parts of the central domain revealed that amino acids 409–563, which are located just downstream of TPR1–4, have a cell cycle–specific role in Chs3p export; they are only required early in the cell cycle. This finding is consistent with the notion that traffic of Chs3p is differentially regulated in the cell cycle (Zanolari et al., 2011). Thus, Chs3p may also contact the central domain of Chs6p for transport. The cell cycle–specific requirements may be due to the posttranslational modifications known to occur in Chs3p (Peng et al., 2003; Valdivia and Schekman, 2003; Lam et al., 2006), some of which could be cell cycle–dependent. Alternatively, accessory proteins might specifically control the formation of the exomer–Chs3p complex at the TGN in a cell cycle–dependent manner. The reason we favor a second interaction site for Chs3p in the central domain is based on the findings that, first, replacing the entire central domain by Bch2p inhibits Chs3p export from the TGN throughout the cell cycle and, second, the N-terminus of Chs6p is not sufficient to drive export of Chs3p from the TGN. Our data indicate that there is even a third interaction site in the C-terminal region of Chs6p, as we need to transplant sequences from all three regions outside of the TPRs for efficient transport of Chs3p to the bud neck. In principle, our data would be consistent with two models: the first would suggest the presence of three individual binding sites/surfaces for Chs3p, each of which would be necessary but not sufficient. Alternatively, at least two if not all three regions would come together in the folded three-dimensional molecule and present one or two large interaction surfaces. At this point, we cannot distinguish between these two possibilities. However, we can exclude that a simple binding pocket provided by Chs6p that would bind one particular sequence of Chs3p would be sufficient for productive complex formation causing Chs3p plasma membrane localization. We identified a sequence in the C-terminus of Chs3p required for its TGN export, which bound weakly but specifically to Chs6p. Still, this sequence was not sufficient to cause an unrelated protein to become an exomer substrate or to be plasma membrane localized. The idea that the ChAPs do not just require a simple, linear sequence was suggested by Barfield et al. (2009), who found that a necessary exomer–interaction sequence was not sufficient to transform a nonexomer cargo into an exomer-dependent cargo. Yet in this case the interpretation was complicated by the simultaneous requirement of two different ChAPs for the transport of Fus1p. Moreover, the Fus1p motif is not contained in Chs3p, and the Chs3p tail is not matched by a homologous sequence in Fus1p. In this study, we were able to extend this notion to a more complex interaction mode between the ChAPs and their cargo. We showed that large interaction surface(s) and multiple sequences on both the cargo receptor and the cargo are required for cargo export through the exomer-dependent pathway.
Why would the ChAPs have evolved to recognize complex trafficking motifs? One could speculate that similar to the Sec23/24 complex of the COPII coat at the endoplasmic reticulum, a multitude of cargoes have to be transported (Kuehn and Schekman, 1997; Kurihara et al., 2000; Miller et al., 2002, 2003) and therefore large interaction surface or multiple binding sites might be useful. However, the exomer-dependent transport route is not the major export pathway from the TGN. In fact, only two clients for the exomer route have been identified (Sanchatjate and Schekman, 2006; Trautwein et al., 2006; Barfield et al., 2009). Thus the purpose might be different: Chs3p and Fus1p are both proteins that require temporal and spatial regulation of their transport. Therefore, exomer-dependent cargoes may have a very specific role at the plasma membrane that would require a relatively tight control of discharge at the plasma membrane and subsequent endocytosis. At least Chs3p requires endocytic recycling for proper bud-neck localization (Reyes et al., 2007; Sacristan et al., 2012). However, N = 1 is obviously too tiny a data set to allow general conclusions. Therefore, the identification and characterization of other exomer-dependent cargoes will shed more light on the function and selectivity of the exomer complex.
MATERIALS AND METHODS
Strains and growth conditions
Yeast strains used in this study are listed in Supplemental Table S1. Standard yeast media were prepared as described (Sherman, 1991). Calcofluor plates were based on minimal medium containing additionally 0.1% yeast extract, 1% 2-(N-morpholino)ethanesulfonic acid buffer, pH 6.0, and 0.1 mg/ml Calcofluor White (Sigma-Aldrich, St. Louis, MO).
Yeast genetic methods
Standard genetic techniques were used throughout (Sherman, 1991). Chromosomal tagging and deletions were performed as described (Knop et al., 1999; Gueldener et al., 2002). For C-terminal tagging with 3×GFP, the plasmid pYM-3GFP was used (Zanolari et al., 2011). All PCR-based chromosomal manipulations were confirmed by analytical colony PCR. The Sec7p-dsRed plasmid (pTPQ128) was described previously (Proszynski et al., 2005). Marker-free chromosomal deletions were performed using the delitto perfetto method (Storici and Resnick, 2006) and confirmed by sequencing. Genetic chimeras were constructed using a modified version of the same technique: After insertion of the CORE cassette, the desired foreign genetic element was amplified from genomic DNA using chimeric primers, which were homologous to the 45 base pairs upstream and downstream of the delitto perfetto site. This PCR product was then directly used for transformation, thus recombining with the locus and replacing the CORE cassette.
Western blot detection
Epitope tags and proteins were detected using the following antibodies: anti-myc (9E10, 1:1000; Sigma-Aldrich); anti-hemagglutinin (HA; HA11, 1:1000; Eurogentec, Seraing, Belgium); anti-FLAG (M2, 1:1000; Sigma-Aldrich); anti-Chs5p (affinity purified, 1:500); anti-Chs3p (affinity purified, 1:1000); anti-GFP (1:5000, Torrey Pines Biolabs, Secaucus, NJ; or anti-GFP 7.1 and 13.1, 1:500, Roche, Indianapolis, IN); anti‑Pgk1 (#A-6457, 1:1000; Invitrogen), anti-Anp1p (1:1000 working solution supplemented with extract from Δanp1 yeast cells; a gift from S. Munro, MRC Laboratory of Molecular Biology, Cambridge, United Kingdom), and anti-Sec61p serum (1:10,000; a gift from M. Spiess, Biozentrum Basel, Basel, Switzerland). ECL (GE Healthcare, Piscataway, NJ) was used for detection.
For myc epitope detection in cross-linker immunoprecipitation experiments and GST pull-downs, anti-myc 9E10 (1:4000; Sigma-Aldrich) and TrueBlot anti-mouse horseradish peroxidase secondary antibody (1:2500; eBioscience, San Diego, CA) were used, and ECL Advance (GE Healthcare) was used for detection according to the manufacturer's instructions.
Microscopy
Cells were grown to log phase in rich or selective medium supplemented with adenine, then harvested, washed, and mounted. Images were acquired with an AxioCam mounted on a Zeiss Axioplan 2 fluorescence microscope (Carl Zeiss, Jena, Germany), using filters for GFP, dsRed, or 4′,6-diamidino-2-phenylindole.
Chitin staining was carried out as described (Lord et al., 2002). Briefly, cells grown for at least 16 h to late log phase were stained after formaldehyde fixation in 1 mg/ml calcofluor, washed three times in water, and imaged directly.
Subcellular fractionation
Ten OD600 of mid–log cells were incubated in 1 ml of dithiothreitol (DTT) buffer (10 mM Tris, pH 9.4, 10 mM DTT) for 5 min at 30°C, spun down, and resuspended in 1 ml of SP buffer (75% yeast extract, peptone [YP], 0.7 M sorbitol, 0.5% glucose, 10 mM Tris, pH 7.5). Thirty microliters of zymolyase T20 (10 mg/ml) was added, and the cells were spheroplasted at 30°C for 40 min. Cells were washed once in zymolyase-free SP buffer, resuspended in the same buffer, and incubated at 30°C for 30 min. Regenerated cells were gently spun down and lysed in 1 ml of 50 mM Tris, pH 7.5, 1 mM EDTA, 50 mM NaCl, and protease inhibitors by pipetting up and down. The lysate was cleared at 500 × g for 2 min, and the supernatant (“total cell lysate” [TCL]) subjected to centrifugation at 13,000 × g (10 min). The supernatant (S13) was carefully taken off with a pipette and subjected to centrifugation at 100,000 × g (1 h). Both pellets (P13 and P100) were rinsed with lysis buffer and then resuspended in 1 ml of lysis buffer. All steps were carried out at 4°C. Samples were taken from all final fractions and subjected to immunoblot analysis.
Coimmunoprecipitation
Yeast lysates from 10 OD600 of cells were prepared by spheroplasting as described. Spheroplasts were sedimented (2 min, 1000 × g), lysed in B150Tw20 buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 6.8, 150 mM K acetate (Ac), 5 mM Mg(Ac)2, and 1% Tween-20) with protease inhibitors, and cleared by centrifugation (10 min, 16,000 × g). Immunoprecipitations were performed with 5 μg of affinity-purified rabbit immunoglobulin G (Dianova, Hamburg, Germany), 5 μg of affinity-purified anti-Chs5p antibody, 5 μg of anti-HA (HA.11; Eurogentec), 5 μg of anti-myc (9E10; Sigma-Aldrich), or 5 μg of anti-AU5 (Abcam, Cambridge, MA) and 100 μl of 20% protein A–Sepharose per 1 ml of cleared lysate for 1 h at 4°C. The beads were washed and resuspended in sample buffer, and bound proteins were analyzed by immunoblot.
Cross-linker immunoprecipitation
For each sample, 10 OD600 of yeast cells were resuspended in 220 μl of B88 buffer (20 mM HEPES, pH 6.8, 150 mM KAc, 5 mM Mg(Ac)2, 250 mM sorbitol) with protease inhibitors and subjected to FastPrep lysis (MP Biomedicals, Illkirch, France). The lysate was cleared by centrifugation at 13,000 × g for 5 min at 4°C. DSP (2 mM final concentration; Pierce, Rockford, IL) dissolved in dimethly sulfoxide was added to 140 μl of lysate. The cross-linking reaction took place for 30 min at room temperature and was stopped with 7 μl of 1 M Tris (pH 7.5) for 15 min. Then 8 μl of 20% SDS was added, and the sample was incubated at 65°C for 15 min. A 1.35-ml amount of IP buffer (50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 1% TX-100, 1 μg/μl bovine serum albumin) was added, and the sample was centrifuged for 10 min at 16,000 × g. The supernatant was subjected to immunoprecipitation overnight at 4°C using 5 μg of monoclonal anti-GFP antibody (clones 7.1 and 13.1; Roche) cross-linked to protein A–Sepharose with DMP. Control immunoprecipitations with 5 μg of monoclonal anti-HA antibody (HA.11 clone 16B12; Covance, Berkeley, CA) were preformed in parallel. Precipitates were washed once in 50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 1% TX-100, and 0.1% SDS and twice in the same buffer containing 250 mM NaCl. Precipitates were resuspended in 50 mM Tris/HCl, pH 7.5 and 250 mM NaCl and transferred into new tubes. The washed precipitates were incubated at 65°C for 30 min in SDS sample buffer containing 100 mM DTT and analyzed by immunoblot. Alternatively, extracts were prepared by bead lysis, and immunoprecipitations were preformed with 5 μg of affinity-purified anti-Chs3p antibody.
BLAST analysis and TPR prediction
The Chs6p primary protein sequence was subjected to fungal BLAST search (available at Saccharomyces Genome Database, www.yeastgenome.org) using the default settings of the BLASTP algorithm on all available fungal nuclear genomes, excluding S. cerevisiae. TPRs were predicted with the TPRPRED algorithm (Karpenahalli et al., 2007), using the standard settings.
GST pull-downs
The C-terminal full-length tail of Chs3p or C-terminally truncated versions (Δ21 and Δ37) were cloned into pGEX-6P-1 using EcoRI and XhoI restriction sites. The full-length tail comprised the last 55 aa following the last predicted TM domain, whereas truncations of this tail lacked the C-terminal 21 and 37 aa, respectively. Expression in Rosetta Escherichia coli cells was induced by the addition of 1 mM isopropyl-β-d-thiogalactoside and growth in Luria Broth (LB) medium at 37°C for 4 h. Cells were lysed in phosphate-buffered saline (PBS)/5% glycerol, and GST fusions were purified with glutathione (GSH) agarose (Sigma-Aldrich), eluted with 40 mM GSH, and dialyzed against PBS/5% glycerol.
GST and GST-tagged Chs3p C-terminus were bound to GSH agarose. For each sample, 10 OD600 of yeast cells were resuspended in 250 μl of B88 buffer with protease inhibitors and subjected to bead lysis or resuspended in 220 μl of B88 buffer and subjected to fast prep lysis. Yeast lysates were diluted six times in B150Tw20 to a final protein concentration of approximately 0.5 μg/μl. The lysates were incubated with the coupled resin for 1 h at 4°C. The beads were washed twice with B150Tw20 buffer and once with B150Tw20 buffer supplemented with 150 mM NaCl and then resuspended in 40 μl of SDS sample buffer, followed by incubation at 95°C for 10 min. Bound proteins were analyzed by immunoblot.
For quantification of Chs6-9myc binding to full-length or truncated C-terminal Chs3p tails. Images of scanned blots were inverted, and intensity values were determined for each band using ImageJ (National Institutes of Health, Bethesda, MD) by drawing a box of fixed size around each band and using the “integrated density” function. Each band was background corrected against the intensity value of the gel lane (below the band). Absolute values were then normalized relative to GST.
Supplementary Material
Acknowledgments
We thank B. Zanolari and M. Trautwein for their initial observation of the lithium-sensitivity phenotype and M. Spiess and S. Munro for the anti-Sec61p and anti-Anp1p sera, respectively. We also thank all members of the Spang lab for technical advice and helpful discussion. This work was supported by a graduate student fellowship of the Werner Siemens Foundation to U.R., the University of Basel (A.S.), and the Swiss National Science Foundation (A.S.).
Abbreviations used:
- TGN
trans-Golgi network
- TPR
tetratricopeptide repeat
- TM
transmembrane
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-12-1015) on September 26, 2012.
A.S. and U.R. conceived the study, U.R. and A.D. performed the experiments (U.R.: Figures 1, 2; 3, B–D; 4; 6; 7, A–C; and 8, A–D; and Supplemental Figures S1–S3; A.D.: Figures 3A; 5; 7, D and E; 8E; 9; and 10; and Supplemental Figures S3D and S4), C.S. and R.C. contributed the Chs3 truncations and the information that this was an interaction site, A.S., U.R., and A.D. wrote the manuscript, and all authors commented on the manuscript.
REFERENCES
- Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S, Endo T, Kohda D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. doi: 10.1016/s0092-8674(00)80691-1. [DOI] [PubMed] [Google Scholar]
- Appenzeller C, Andersson H, Kappeler F, Hauri HP. The lectin ERGIC-53 is a cargo transport receptor for glycoproteins. Nat Cell Biol. 1999;1:330–334. doi: 10.1038/14020. [DOI] [PubMed] [Google Scholar]
- Bard F, Malhotra V. The formation of TGN-to-plasma-membrane transport carriers. Annu Rev Cell Dev Biol. 2006;22:439–455. doi: 10.1146/annurev.cellbio.21.012704.133126. [DOI] [PubMed] [Google Scholar]
- Barfield RM, Fromme JC, Schekman R. The exomer coat complex transports Fus1p to the plasma membrane via a novel plasma membrane sorting signal in yeast. Mol Biol Cell. 2009;20:4985–4996. doi: 10.1091/mbc.E09-04-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biegert A, Mayer C, Remmert M, Soding J, Lupas AN. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 2006;34:W335–W339. doi: 10.1093/nar/gkl217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Cos T, Ford RA, Trilla JA, Duran A, Cabib E, Roncero C. Molecular analysis of Chs3p participation in chitin synthase III activity. Eur J Biochem. 1998;256:419–426. doi: 10.1046/j.1432-1327.1998.2560419.x. [DOI] [PubMed] [Google Scholar]
- D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Gatto GJ Jr, Geisbrecht BV, Gould SJ, Berg JM. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat Struct Biol. 2000;7:1091–1095. doi: 10.1038/81930. [DOI] [PubMed] [Google Scholar]
- Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JW, Griffin K, Jih GT, Stuckey J, Verhey KJ. Co-operative versus independent transport of different cargoes by kinesin-1. Traffic. 2008;9:725–741. doi: 10.1111/j.1600-0854.2008.00722.x. [DOI] [PubMed] [Google Scholar]
- Harsay E, Bretscher A. Parallel secretory pathways to the cell surface in yeast. J Cell Biol. 1995;131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsay E, Schekman R. A subset of yeast vacuolar protein sorting mutants is blocked in one branch of the exocytic pathway. J Cell Biol. 2002;156:271–285. doi: 10.1083/jcb.200109077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia KC, Hoelz A. Crystal structure of alpha-COP in complex with epsilon-COP provides insight into the architecture of the COPI vesicular coat. Proc Natl Acad Sci USA. 2010;107:11271–11276. doi: 10.1073/pnas.1006297107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal A, Stokin GB, Yang Z, Xia CH, Goldstein LS. Axonal transport of amyloid precursor protein is mediated by direct binding to the kinesin light chain subunit of kinesin-I. Neuron. 2000;28:449–459. doi: 10.1016/s0896-6273(00)00124-0. [DOI] [PubMed] [Google Scholar]
- Karpenahalli MR, Lupas AN, Soding J. TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics. 2007;8:2. doi: 10.1186/1471-2105-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kuehn MJ, Schekman R. COPII and secretory cargo capture into transport vesicles. Curr Opin Cell Biol. 1997;9:477–483. doi: 10.1016/s0955-0674(97)80022-1. [DOI] [PubMed] [Google Scholar]
- Kurihara T, Hamamoto S, Gimeno RE, Kaiser CA, Schekman R, Yoshihisa T. Sec24p and Iss1p function interchangeably in transport vesicle formation from the endoplasmic reticulum in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:983–998. doi: 10.1091/mbc.11.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KK, Davey M, Sun B, Roth AF, Davis NG, Conibear E. Palmitoylation by the DHHC protein Pfa4 regulates the ER exit of Chs3. J Cell Biol. 2006;174:19–25. doi: 10.1083/jcb.200602049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M, Chen T, Fujita A, Chant J. Analysis of budding patterns. Methods Enzymol. 2002;350:131–141. doi: 10.1016/s0076-6879(02)50960-5. [DOI] [PubMed] [Google Scholar]
- Magliery TJ, Regan L. Beyond consensus: statistical free energies reveal hidden interactions in the design of a TPR motif. J Mol Biol. 2004;343:731–745. doi: 10.1016/j.jmb.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Meissner D, Odman-Naresh J, Vogelpohl I, Merzendorfer H. A novel role of the yeast CaaX protease Ste24 in chitin synthesis. Mol Biol Cell. 2010;21:2425–2433. doi: 10.1091/mbc.E10-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E, Antonny B, Hamamoto S, Schekman R. Cargo selection into COPII vesicles is driven by the Sec24p subunit. EMBO J. 2002;21:6105–6113. doi: 10.1093/emboj/cdf605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EA, Beilharz TH, Malkus PN, Lee MC, Hamamoto S, Orci L, Schekman R. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell. 2003;114:497–509. doi: 10.1016/s0092-8674(03)00609-3. [DOI] [PubMed] [Google Scholar]
- Muniz M, Nuoffer C, Hauri HP, Riezman H. The Emp24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles. J Cell Biol. 2000;148:925–930. doi: 10.1083/jcb.148.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Proszynski TJ, et al. A genome-wide visual screen reveals a role for sphingolipids and ergosterol in cell surface delivery in yeast. Proc Natl Acad Sci USA. 2005;102:17981–17986. doi: 10.1073/pnas.0509107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Sanz M, Duran A, Roncero C. Chitin synthase III requires Chs4p-dependent translocation of Chs3p into the plasma membrane. J Cell Sci. 2007;120:1998–2009. doi: 10.1242/jcs.005124. [DOI] [PubMed] [Google Scholar]
- Sacristan C, Reyes A, Roncero C. Neck compartmentalization as the molecular basis for the different endocytic behaviour of Chs3 during budding or hyperpolarized growth in yeast cells. Mol Microbiol. 2012;83:1124–1135. doi: 10.1111/j.1365-2958.2012.07995.x. [DOI] [PubMed] [Google Scholar]
- Sanchatjate S, Schekman R. Chs5/6 complex: a multiprotein complex that interacts with and conveys chitin synthase III from the trans-Golgi network to the cell surface. Mol Biol Cell. 2006;17:4157–4166. doi: 10.1091/mbc.E06-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B, Snyder M. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J Cell Biol. 1997;136:95–110. doi: 10.1083/jcb.136.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B, Snyder M. Specific protein targeting during cell differentiation: polarized localization of Fus1p during mating depends on Chs5p in Saccharomyces cerevisiae. Eukaryot Cell. 2003;2:821–825. doi: 10.1128/EC.2.4.821-825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Storici F, Resnick MA. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 2006;409:329–345. doi: 10.1016/S0076-6879(05)09019-1. [DOI] [PubMed] [Google Scholar]
- Trautwein M, Schindler C, Gauss R, Dengjel J, Hartmann E, Spang A. Arf1p, Chs5p and the ChAPs are required for export of specialized cargo from the Golgi. EMBO J. 2006;25:943–954. doi: 10.1038/sj.emboj.7601007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia RH, Schekman R. The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc Natl Acad Sci USA. 2003;100:10287–10292. doi: 10.1073/pnas.1834246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CW, Hamamoto S, Orci L, Schekman R. Exomer: A coat complex for transport of select membrane proteins from the trans-Golgi network to the plasma membrane in yeast. J Cell Biol. 2006;174:973–983. doi: 10.1083/jcb.200605106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanolari B, Rockenbauch U, Trautwein M, Clay L, Barral Y, Spang A. Transport to the plasma membrane is regulated differently early and late in the cell cycle in Saccharomyces cerevisiae. J Cell Sci. 2011;124:1055–1066. doi: 10.1242/jcs.072371. [DOI] [PubMed] [Google Scholar]
- Zeytuni N, Zarivach R. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure. 2012;20:397–405. doi: 10.1016/j.str.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Zhang B, Kaufman RJ, Ginsburg D. LMAN1 and MCFD2 form a cargo receptor complex and interact with coagulation factor VIII in the early secretory pathway. J Biol Chem. 2005;280:25881–25886. doi: 10.1074/jbc.M502160200. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Kulkarni K, Hanrahan SJ, Thompson AJ, Barford D. The APC/C subunit Cdc16/Cut9 is a contiguous tetratricopeptide repeat superhelix with a homo-dimer interface similar to Cdc27. EMBO J. 2010;29:3733–3744. doi: 10.1038/emboj.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M, Chuang JS, Tsung M, Hamamoto S, Schekman R. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1565–1576. doi: 10.1091/mbc.9.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.