Abstract
A major obstacle to the success of islet cell transplantation as a standard treatment for labile type 1 diabetes mellitus is the immediate loss of up to 70% of the transplanted islet mass. Activation of the complement cascade and coagulation factors has been implicated in initiating the destruction of the islet graft. In this study, we analyzed the gene expression changes in islet cells following exposure to type 1 diabetes mellitus serum (T1DM). Isolated human pancreatic islet cells were cultured for 2 d to stabilize islet cell gene expression. Cultured islets were divided into three groups for treatment as follows: group 1 was treated with autologous donor serum, while groups two and three were treated with sera from ABO-matched allogeneic donors or autoantibody positive type 1 diabetic patient, respectively. Complement was detected using anti-C3 FITC and CH50 assay. Islet gene expression was analyzed using Illumina micro-array technology. Results were confirmed using real-time PCR. Immunofluorescent imaging demonstrated complement deposition only in the T1DM condition. Gene array and class prediction analysis generated a list of 50 genes that were able to predict the effect of T1DM serum on islets. Quantitative PCR corroborated microarray results. Both techniques demonstrated upregulation of MMP9 (243%), IL-1β (255%), IL-11 (220%), IL-12A (132%), RAD (343%) and a concomitant downregulation of IL-1RN (64%) in islets treated with T1DM serum. Islets treated with T1DM serum overexpressed genes associated with angiogenesis while decreasing transcription of genes that protect islets from inflammatory cytokines and reactive oxygen species.
Keywords: islet cell transplantation, microarray, complement, inflammation, autoimmunity
Introduction
Islet cell transplantation has become an acceptable alternative treatment for patients with type 1 diabetes mellitus and hypoglycemic unawareness, and is also indicated in patients with glycemic lability.1 Islet cell transplant graft survival rates increased following the introduction of the steroid free immunosuppressive regimen at multiple centers2 however, despite measurable graft function, long-term maintenance of post-transplant insulin independent status is poor.3 A major drawback of current protocols in islet transplantation is the requirement of multiple donor pancreata to achieve insulin independence. One of the critical components for attaining insulin independence is achieving a large enough engrafted islet mass after transplantation to ensure long-term graft survival.4
Isolated pancreatic islets have been shown to be incompatible with human blood, eliciting a host of complications that destroy the transplanted islet mass within the first few hours post transplant.5 This incompatibility known as Instant Blood Mediated Inflammatory Reaction (IBMIR) involves coagulation and complement as well as cellular infiltration. IBMIR research has focused primarily on the outcome of coagulation and lymphocyte infiltration5 and strategies to prevent coagulation,6-13 while complement activation has not been studied as extensively.14 IBMIR elicits platelet and complement activation and deposition on the islets, and attracts infiltrating leukocytes to the transplanted islet resulting in the entrapment of islets within clots.5 Nilsson et al. have demonstrated that islets contribute to the initiation of IBMIR by expressing tissue factor.9,10,12 However, little research has been done to analyze the molecular events within islets that take place following exposure type 1 diabetic blood, which in turn could shed light on key molecules and signaling pathways involved in the destruction of islets during IBMIR.
We hypothesized that upon exposure of islets to serum from type 1 diabetic patients, activation of complement initiated by auto-antibody binding to islet antigens will ensue and this will mediate gene expression changes in islets. Understanding of alterations in gene expression in islets may predict the key molecules and signaling pathways involved in the destruction of the islet graft. Our preliminary work15 showed that islets treated with diabetic patient serum exhibited gene expression changes typical for cellular damage. In this study, we further investigated the effect of diabetic serum on islets using micro-array and real-time PCR analysis. Our results indicate that complement contributes to the destruction of the islet graft simultaneously followed by induction in expression of Il-1 β (IL-1β) and other cytokines that attract the cells of both the adaptive immune system and the innate immune system. These events are also accompanied by concomitant repression of the transcription of genes that protect against inflammatory cytokines and reactive oxygen species, further contributing to the destruction of the islet graft.
Results
Complement activation and binding by treatment of islets with type 1 diabetic serum
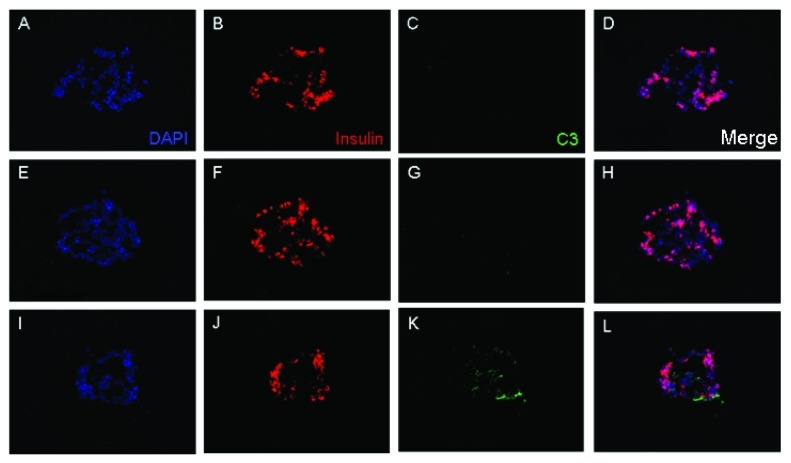
Following intraportal infusion, islets undergo IBMIR, which consists of activation of both a coagulation cascade and complement cascade. To investigate the role complement plays in the process of graft destruction that begins immediately post transplant, we analyzed the genetic changes that islet cells undergo when exposed to type 1 diabetic serum. To this end, type 1 diabetic serum known to be positive for at least one autoantibody was obtained from patients prior to islet transplant and was used to treat sectioned islets. The sectioned islets were then stained with an antibody for complement factor C3 to demonstrate the binding of complement. Islet sections that were treated with autoantibody positive type 1 diabetic serum alone, demonstrated the presence of complement (Fig. 1), while those sections that were exposed to autologous or non-diabetic allogeneic serum did not demonstrate any complement deposition.
Figure 1. Complement deposition on islets exposed to type 1 diabetic serum. Isolated islets were fixed and cryosectioned at 6 μm thickness and consecutive sections were used per condition. Islet sections were incubated in a 1:2 dilution of autologous serum (A–D), allogeneic serum (E–H) or type 1 diabetic serum (I–L) at 37°C for 2 h. Sections were washed and labeled with rabbit polyclonal anti-C3-FITC (green) or mouse monoclonal anti-insulin conjugated to AF647 (red). Only type 1 diabetic serum displayed signs of complement deposition (K), which corresponded to an islet cell as evidenced by positive insulin staining (J). DAPI was used for counterstaining (blue).
To analyze total complement activation following treatment of human islets with autoantibody positive type 1 diabetic serum, we performed CH50 assays using Microvue EIA kit. The type 1 diabetic sera used in this study showed CH50 values of 155–173 CH50 U Eq/mL when compared with values of 120–133 CH50U Eq/mL for non-diabetogenic sera. Approximately, 4000 islets were treated with 200 μl of serum and incubated for 1 h at 37°C. The supernatant after the incubation was tested for complement activity. Islet incubation reduced the CH50 activity of diabetic sera to 96–117 U/mL resulting in a 32–38% inhibition. Importantly, the post-incubation values were below the values observed for non-diabetic sera. These results clearly demonstrated the activation of classical pathway of complement cascade by the type 1 diabetic sera used in this study. We also tested the viability of islets following treatment with type 1 diabetic sera using PI-Hoe33342 staining. After 1 h incubation at 37°C, when compared to control islets in culture medium, the islet viability significantly reduced when treated with type 1 diabetic serum (80.5 ± 9.2 Vs 28.6 ± 12.3; p < 0.0001) and allogenic serum (80.5 ± 9.2 Vs 65,6 ± 9.3; p < 0.05) but did not change significantly when treated with autologous serum (80.5 ± 9.2 Vs 71.9 ± 18.0; p = 0.11). These data clearly demonstrated that complement activation significantly affected islet viability.
Since excess glucose levels in blood could influence gene expression in islets, we tested the glucose levels in the type 1 diabetic and non-diabetic sera used in the present study. Glucose levels in these two groups used were 128 ± 21 and 131 ± 29 mg/dL respectively. These results showed that glucose concentration did not contribute to change in the gene expression reported here.
Complement induces gene expression changes in isolated islets
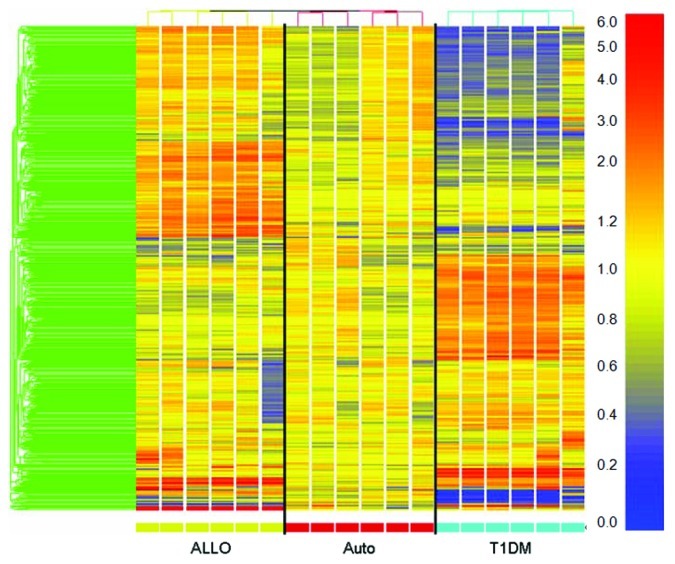
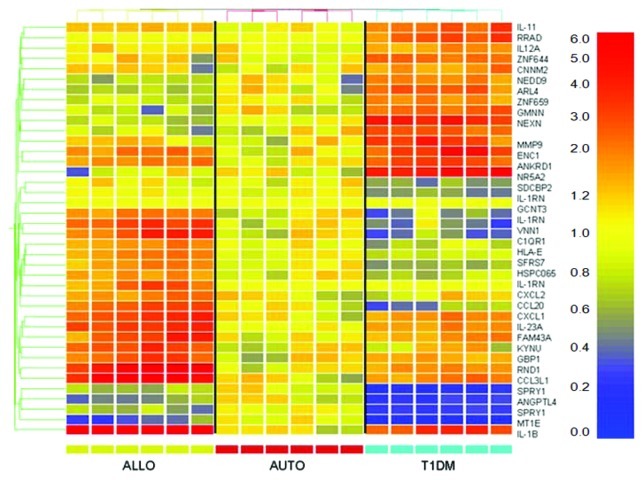
Isolated islets were treated with diabetic serum known to be autoantibody positive to further investigate gene expression changes induced by autoantibody and complement activation. Gene profiles that lacked any detectable signal intensity across the board were removed from analysis in an attempt to reduce false discovery. Flat expression profiles were removed from data analysis by filtering the data on the criteria of meeting the minimum fluorescence cutoff and having a 2-fold difference in expression in at least one sample when compared with the mean of the autologous condition for each gene. This dramatically reduced the number of genes from ~48,000 genes to an average of 3,415 ± 1,552 genes. Supervised clustering performed on the filtered microarray data demonstrated that the samples clustered by treatment condition (Fig. 2). Class prediction analysis was then performed in an attempt to compile a list of genes that could be attributed to complement deposition (Fig. 3). A representative experiment was used as the training set to establish whether the predictor genes could predict the correct treatment classification in itself. Genespring™ software generated a list of genes, both significant and insignificant, that differentiated between the three treatment conditions based upon gene expression. Once the predictor list was established as having 100% correct prediction in the training set, this list was imported to the other experiments for further analysis resulting in a mean prediction (autologous, allogeneic, T1DM, unable to predict) accuracy of 56% ± 17%. The predictor genes were analyzed for the expression profile for each experiment and the resulting data compiled for an overall expression pattern of islet cells treated with T1DM serum (Table 1). This table demonstrates that type 1 diabetic serum induces several significant gene expression changes in islets during the initial period of contact. When compared with normal autologous expression the following genes were significantly overexpressed: RRAD (343.17 ± 62.37, p = 0.0008), GMNN (284.59 ± 40.54, p < 0.0001), ANKRD1 (279.99 ± 45.87, p = 0.0003), IL-1β (255.66 ± 31.31, p < 0.0001), IL-23A (251.37 ± 35.85, p = 0.0002), MMP9 (243.37 ± 35.13, p = 0.0001), IL-11 (220.12 ± 45.46, p = 0.0135), ENC1 (219.3 ± 15.27, p < 0.0001), SDS (210.56 ± 26.46, p = 0.0004), GBP1 (181.43 ± 30.03, p < 0.0001), NEXN (179.03 ± 15.65, p < 0.0001), EPC1 (175.61 ± 16.43, p = 0.0001), CXCL1 (146.62 ± 19.66, p = 0.0256); while the following genes were significantly under-expressed: SDCBP2 (66.73 ± 8.78, p = 0.004), HSPC065 (64.27 ± 10.42, p = 0.0243), IL-1RN (64 ± 6.02, p = 0.0049), CCL20 (61.86 ± 8.36, p = 0.0008), ANGPTL4 (48.1 ± 7.76, p < 0.0001), SPRY1 (37.13 ± 6.24, p < 0.0001), MT1E (34.48 ± 10.71, p < 0.0001).
Figure 2. Complement activation initiated by auto-antibodies in type 1 diabetic serum induce a specific profile of gene expression. Islets exposed to complement in diabetic serum or control sera (allogenic or autologous) were compared using Illumina micro-array analysis. The resulting data was analyzed using the GeneSpring Software. Supervised clustering revealed 679 differentially expressed genes in diabetic serum when compared with autologous serum. Data from one representative experiment out of five independent experiments is shown.
Figure 3. class prediction analysis of islets exposed to type 1 diabetic serum. One representative experiment was chosen for class prediction analysis to establish a list of genes that could differentiate the serum conditions based on gene expression changes. Samples were normalized to the mean of the autologous condition. Yellow reflects unchanged expression, blue reflects downregulation and red reflects upregulation.
Table 1. Average signal intensity for class predictor genes in T1DM serum-treated islets.
|
Gene ID |
Average |
p |
Gene ID |
Average |
p |
| RRAD* |
343.17±62.37 |
0.0008 |
ARL4 |
131.02±11.24 |
0.0083 |
| GMNN |
284.59±40.54 |
<0.0001 |
ZNF644 |
130.73±10.43 |
0.0016 |
| ANKRD1 |
279.99±45.87 |
0.0003 |
ZNF658 |
129.53±20.99 |
0.2211 |
| CCL4L1 |
266.89±57.03 |
0.1038 |
CCL3 |
122.87±18.69 |
0.2104 |
| 1L-1B* |
255.66±31.31 |
<0.0001 |
CCL3L3 |
120.05±18.04 |
0.2372 |
| IL-23A |
251.37±35.85 |
0.0002 |
CNNM2 |
119.42±15.84 |
0.347 |
| MMP9* |
243.37±35.13 |
0.0001 |
RND1 |
117.11±14.88 |
0.3477 |
| 1L11* |
220.12±45.46 |
0.0135 |
KYNU |
116.29±16.86 |
0.3803 |
| ENC1 |
219.3±15.27 |
<0.0001 |
CCL3L1 |
112.77±16.41 |
0.462 |
| SDS |
210.56±26.46 |
0.0004 |
SERPINB2 |
109.23±17.98 |
0.5174 |
| 1780523 |
192.66±32.69 |
0.01 |
SLC25A36 |
101.29±11.33 |
0.9154 |
| HLA-E |
187.16±32.35 |
0.2664 |
NR1D2 |
97.71±10.52 |
0.3283 |
| STAT5A |
187.03±35.81 |
0.2182 |
SFRS7 |
96.49±10.16 |
0.7461 |
| FAM43A |
184.9±37.17 |
0.0263 |
TLCD1 |
85.71±7.8 |
0.0828 |
| ZNF659 |
183.94±30.03 |
0.0049 |
NR5A2 |
83.99±14.84 |
0.2859 |
| GBP1 |
181.43±16.84 |
<0.0001 |
GCNT3 |
79.71±13.39 |
0.115 |
| NEXN |
179.03±15.65 |
<0.0001 |
ARRDC3 |
72.9±11.45 |
0.0576 |
| EPC1 |
175.61±16.43 |
0.0001 |
CXCL2 |
68.26±11.37 |
0.6288 |
| C1QR1 |
158.63±36.78 |
0.7955 |
SDCBP2 |
66.73±8.78 |
0.004 |
| VNN1 |
148.68±108.07 |
0.2311 |
HSPC065 |
64.27±10.42 |
0.0243 |
| CXCL1 |
146.62±19.66 |
0.0256 |
IL-1RN* |
64±6.02 |
0.0049 |
| NCOA7 |
145.57±19.01 |
0.0216 |
CCL20 |
61.86±8.36 |
0.0008 |
| NEDD9 |
137.78±18.92 |
0.0161 |
ANGPTL4 |
48.1±7.76 |
<0.0001 |
| IL-18R1 |
136.37±30.01 |
0.8777 |
SPRY1 |
37.13±6.24 |
<0.0001 |
| SLC25A3 |
133.19±15.01 |
0.0434 |
MT1E |
34.48±10.71 |
<0.0001 |
| IL-12A* | 132.94±13.7 | 0.0578 | LAMB3 | 33.93±18.56 | 0.3128 |
Indicates gene expression corroborated with Q-PCR
Real-time QPCR of select genes corroborates changes indicated by micro-array analysis
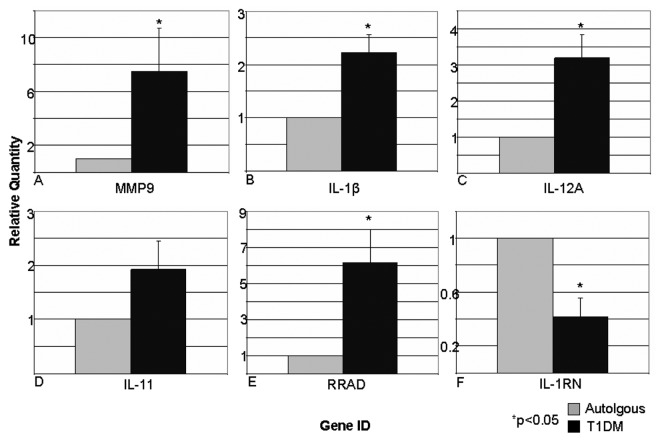
In an attempt to substantiate the data collected by micro-array analysis, representative genes were chosen for further analysis by real-time quantitative PCR. TaqMan probes specific for matrix metalloproteinase -9 (MMP-9), interleukin 1-β (IL-1β), interleukin 11 (IL-11), interleukin 12A (IL-12A), ras-related associated with diabetes (RRAD), and interleukin 1 receptor antagonist (IL-1RN) were used to amplify these genes. Due to practical reasons, a representative sample for each condition was chosen from each experiment, totaling 15 samples. Total RNA left over from the micro-array analysis was used to generate cDNA. Relative quantification of the T1DM condition with respect to the autologous condition confirmed the respective change in gene expression (Fig. 4). MMP-9 demonstrated a 7.50 fold upregulation in T1DM when compared with autologous control. IL-1β likewise showed a significant 2.23-fold upregulation (p < 0.01) in the T1DM condition. Although not significant, IL-11 was also upregulated by 1.92-fold in the diabetic condition. IL-12A underwent a significant 3.19-fold upregulation when treated with diabetic serum (p < 0.05). RRAD expression was significantly upregulated by 6.13 fold by T1DM serum (p < 0.05) when compared with autologous serum. The expression of IL-1RN in T1DM treated islets was significantly reduced to 0.42 of the autologous expression level (p < 0.01).
Figure 4. Real time quantitative PCR analysis confirms micro-array data in representative genes. Representative genes were chosen for corroboration by using relative quantification real-time PCR. TaqMan probes specific for MMP-9 (A), IL-1β (B), IL-12A (C), IL-11 (D), RAD (E), and IL-1RN (F) were used to produce cDNA from representative mRNA samples from each experiment. Real-time PCR detected significant gene expression changes of 7.50 ± 3.17 (p = 0.07), 2.23 ± 0.33 (p = 0.005), 3.19 ± 0.66 (p = 0.01), 1.92 ± 0.53 (p = 0.12), 6.13 ± 1.87 (p = 0.03), 0.42 ± 0.14 (p = 0.003) respectively. Data expressed as mean ± SEM.
Discussion
Instant Blood-Mediated Inflammatory Reaction (IBMIR) is known to involve both coagulation and complement cascades.5,14,16 In this study, we investigated the molecular mechanism(s) by which soluble factors from type 1 diabetic serum contributes to the destruction of the islet cell graft. Pancreatic islets are isolated mostly from brain dead donors experiencing cytokine storm.17 Hence donor serum was used as a control to establish the background effects of cytokines on isolated islet cells. Similarly ABO matched allogeneic organ donor serum was also used to control for mismatch in HLA compatibility and both served as negative controls for the presence of autoantibodies. Sera from a living type 1 diabetic patient known to be positive for islet autoantibodies was used to simulate islet transplantation.18 Furthermore, the glucose concentrations in the sera used from diabetic patients and non-diabetic controls did not differ significantly and hence is unlikely to have contributed to the gene expression data shown. The resulting gene expression profile data reflects the conditions of clinically transplantable islet cells undergoing gene alterations due to the soluble complement factors, as demonstrated by the deposition of complement factor C3 presented herein. We can eliminate the contribution of coagulation in these changes because serum was used for the treatment of islets rather than plasma. However, there are other soluble factors known to be present in diabetic serum,19,20 which cannot be excluded from contributing to the result. Similarly changes influenced by autoantibody alone could not be identified from the present data, since the sera samples were not heat inactivated to eliminate complement activation. CH50 assays also demonstrated major role for complement activation by interaction between serum and purified islets. During IBMIR, both cellular and soluble components are involved in causing damage to the transplanted islets. Due to limitations in separation of islets from blood cells after mixing, it is technically not possible to design experiments to analyze gene expression in islets alone during IBMIR.
Micro array analysis generated an average list of genes (3,415 ± 1552) that were differentially expressed between the three conditions in the five experiments analyzed (data not shown). In an attempt to reduce the amount of data to a workable level, class prediction analysis was implemented using a representative experiment. The resulting data set of 50 genes was now easily manageable. The list of predictor genes generated from experiment five was imported to the analysis of each other experiment for expression analysis. The expression values for each gene for each sample from each experiment were averaged. The resulting data demonstrated a trend of increasing transcription of genes that have activity toward inflammation and angiogenesis while turning off transcription of genes that protect the islets from inflammatory cytokines and reactive oxygen species. These changes could also represent the necrosis mediated by complement-mediated cytotoxicity. The microarray data seems to suggest that complement activation brought on by autoantibody recognition of islet β cells induces a paracrine effect resulting in the transcription of inflammatory genes and the inhibition of survival genes. This is of interest, because this pattern of increased inflammatory gene and decreased survival gene expression resembles that of activated macrophages.21
Gene array and real-time QPCR analyses showed major changes in six genes (Fig. 4). Despite the short period of treatment of islets with serum, expression of these genes showed significant alterations. Matrix Metalloproteinase-9 (MMP-9) is an enzyme that plays a pivotal role in the migration of immune cells.22 MMPs have been demonstrated to be present on rodent islets after isolation.23 A previous study using a mouse model of islet transplantation has demonstrated that pre-treatment of islets with MMP-9 improved islet graft revascularization, with improved vascular density, blood flow and oxygen tension.24 It is plausible that islet cells upregulate transcription of MMP-9 in an attempt to initiate angiogenesis to improve their oxygen supply.
Interleukin 1 β (IL-1β) is an inflammatory cytokine primarily involved in inflammation and immune responses.25 IL-1β has also been demonstrated to cause the nuclear exportation of N-CoR corepressor complex, resulting in the depression of NFκB regulated genes.26 It is possible that islets transplanted into the hepatic portal vein upregulate IL-1β along with other pro-angiogenesis factors while inhibiting transcription of blockers of angiogenesis (IL-1RN) factors to promote revascularization of the islet graft.
Interleukin 11 is known to stimulate T cell dependent B cell development as well as plasmacytoma proliferation.27 In addition to IL-11’s hematopoietic regulatory function, IL-11 has also been demonstrated to induce synthesis of the tissue inhibitor of metalloptroteinase-1 (TIMP-1) preventing the breakdown of extracellular matrix.28 Interestingly IL-12A (IL-12p35) and IL-23A (IL-23p19), which share a common IL-12p40 subunit, were both upregulated by diabetic serum in islets. When the p35 and p19 subunits couple together with p40, they form IL-12 and IL-23 respectively. These two cytokines have opposite effects, with IL-12 promoting cell mediated immunity29 and IL-23 promoting memory T cells, regulatory T cells, angiogenesis and MMP-9 activity.29,30 Ras associated with diabetes (RAD), a member of the Ras family of small GTPases, has been reported to be overexpressed in muscle tissue of patients diagnosed with type 2 diabetes mellitus.31 Ras is normally involved in signal transduction through many pathways including the MAP Kinase pathway. Ras is known to control such cellular mechanisms as proliferation, differentiation and apoptosis.32,33
IL-1 receptor antagonist (IL-1RN) has been demonstrated to be downregulated in patients with T2DM.34 In vitro studies have shown that reduced expression of IL-1RN coupled with increased expression of IL-1β lead to impaired β cell function and increased activation of apoptosis.34 Our data indicates that serum from T1DM patients has the ability to downregulate IL-1RN in vitro, indicating that a similar phenomenon may be occurring during islet cell transplantation. This would be detrimental to the survival of the transplanted islet mass, as IL-1β is produced during post-transplant inflammation. A recombinant form IL-1RN, recently used as a treatment for T2DM, has shown some promise by improving glycemic control and β cell secretory function.35
Taken together, our data indicate that treatment of islets with type 1 diabetic serum induces a paracrine effect on the transplanted islet cells leading to the transcription of genes that promote angiogenesis and inhibit survival. The gene expression changes identified in this study could be prominently due to complement activation, while the role of other soluble factors present in diabetic serum cannot be ruled out in contributing to this effect. It should also be noted that IL-1β exposure to pancreatic β cells has resulted in the upregulation of iNOS and its effect has been well documented.36-38 The results of this study demonstrate several genes and pathways of possible intervention to improve the current islet transplantation outcome.
Materials and Methods
Pancreas procurement and islet isolation
Donor pancreata were obtained through the local organ procurement organization after consent for research was obtained and the experimental protocol was approved by the Institutional Review Board (IRB).
Pancreas removal was performed by Baylor Regional Transplant Institute (BRTI) surgeons using a standardized whole organ procurement technique to minimize warm ischemia, and preserved in the two layer method.39 Pancreatic islet cells were isolated following the semi automated method originally described by Ricordi et al.40 Upon completion of the isolation procedure, the islets were assessed for purity and quantity by staining with dithizone. Islets isolated from five different donors were used for this study.
Islet viability was measured by staining with Hoechst 33342 (10 µg/mL) and propidium iodide (20 µg/mL) for 10 min at 37°C before imaging via fluorescent microscopy. Fluorescent micrographs were merged in ImageJ and the PI-positive area was divided by the Hoechst-positive area to calculate islet viability.
Serum treatment of islets
Freshly isolated islet cells were cultured in islet culture medium, CMRL (Cellgro) supplemented with 10% heat inactivated fetal bovine serum (FBS, Atlanta Biologicals) in 5% CO2 air at 22°C41 for 24–48 h to remove damaged cells and allow recovery from the stresses of isolation. After culture, islets were counted and viability assessed as described above. Islets were divided into three groups. Serum samples from previous pancreas donors, and diabetic patients were stored at -80°C and thawed prior to treatment of groups. Each serum sample was thawed and used only once to ensure the presence of intact IgM. To group 1, 1.0 ml of autologous donor serum was added in triplicate to 1,000 islet equivalents (IEQ). To group 2, 1.0 ml of ABO-matched allogeneic serum from a previous organ donor was added in triplicate to 1,000 IEQ. To group 3, 1.0 ml of newly diagnosed type 1 diabetic serum, kindly provided by Dr. Ellen Kaizer-Grishman, known to be positive for at least one autoantibody against insulin, glutamic acid decarboxylase 65 (GAD65) or tyrosine phosphatase-like protein (IA-2) was added in triplicate to 1,000 IEQ. Five replicate experiments were performed using different batches of islets and different serum samples. Groups 1, 2 and 3 from experiments one through four were placed in triplicate in a 24 well plate and incubated at 37°C for 3 h. Experiment five was plated similarly but with six samples per condition. Upon completion of incubation, the islets were transferred to separate microfuge tubes and washed with sterile PBS. Islet cells were then placed in RLT lysis buffer (Qiagen) containing 0.1% β-ME and stored at -80°C for RNA extraction or fixed for immunofluorescence analysis.
Complement classical pathway activity was assessed by measuring hemolysis of sheep erythrocytes using a MicroVue CH50 Eq EIA kit (Quidel Corporation) following the manufacturer’s instructions.
Immunofluorescent imaging
An aliquot of islet cells was taken and fixed in 4% paraformaldehyde (Sigma) in PBS (Gibco). Fixed islets were then centrifuged and placed in Tissue Tek OCT compound (Sakura) for cyrosectioning. Cryosections were cut at 6 μm thickness, and placed on charged slides beginning with section 1. Sixteen sections were used in total. Sections were then treated with either autologous, allogeneic, or type 1 diabetic serum for two hours at 37°C. Sections were then washed in PBS. Sheep polyclonal anti-human-C3-FITC (Abcam) was added in 1:100 PBS to each section and incubated at room temperature for 30 min. Sections were then washed in PBS, followed by a rinse in permeabilization buffer [3% BSA (Sigma), 0.1% Triton X-100 (Sigma), 0.1% Azide (Sigma) in PBS]. Sections were then placed in a permeabilization buffer for 15 min at room temperature. Mouse monoclonal anti-insulin (Abcam) conjugated to AF647 (Invitrogen) was diluted 1:100 in permeabilization buffer and added to each section for 30 min at room temperature for the purpose of islet identification. Sections were washed twice in permeabilization buffer, followed by a third rinse in PBS. DAPI (Invitrogen) was added as a counterstain to each section in a 1:5000 dilution in PBS and incubated at room temperature for five minutes. Sections were washed twice in PBS, dried briefly and covered with Fluoromount-G (Southern Biotech). All incubations with fluorescently labeled antibodies were done in the dark. Sheep IgG (Abcam) conjugated to FITC (Sigma) and mouse IgG1 (R&D Systems) conjugated to AF647 (Invitrogen) served as isotype controls. Images were taken on an upright fluorescent microscope using MetaMorph 6.2r6 (Molecular Devices) for image acquisition and analysis.
Microarray analysis
Total RNA was isolated using TRI Reagent (Ambion) or RNeasy mini kit (Qiagen) following the manufacturers’ instructions. Coding mRNA from total RNA was used to generate double stranded cDNA by reverse transcription. Double stranded cDNA served as a template for the production of biotinylated cRNA using the Illumina Total Prep mRNA Isolation kit (Ambion). Biotinylated cRNA was hybridized to Illumina human whole geneome Hu6 V2 or V3 (experiment 4) gene chips (Illumina) following the manufacturers’ directions. Hybridized gene chips were then scanned on an Illumina BeadStation 500 following the manufacturer’s protocols (Illumina). Illumina’s BeadStudio software was used to generate signal intensity values from the scans, subtract background, and scale each microarray to the median average intensity for all samples (per-chip normalization). Data analysis was performed using GeneSpring GX 7.3 software (Agilent). Samples were clustered independently to determine if gene expression reflected treatment condition. Upon completion of unsupervised clustering analysis, a representative experiment was chosen for further class prediction analysis. Class prediction was performed using a supervised learning algorithm the K-nearest neighbors method as previously described.42 Genespring generated a list of 50 genes that predicted gene condition 100% correctly in the representative experiment. This list of genes was then tested against the other experiments. From this list, significant gene expression changes were determined by ANOVA analysis with multiple error correction using Benjamini-Hochberg false discovery rate as previously described.18 Significant genes were chosen for further analysis among all experiments.
Real time PCR
One representative sample from each condition in each experiment was randomly chosen for confirmation of the data obtained in micro array analysis. Total RNA was reverse transcribed to generate cDNA using RetroScript cDNA synthesis kit (Ambion). 100ng of cDNA was plated in triplicate for each condition and each gene of interest. TaqMan (Applied Biosystems) probes specific for the genes of interest were used to specifically amplify the genes of interest. GAPDH served as a normalizing housekeeping gene. Relative quantity of the gene of interest was compared with the autologous (calibrator) condition for both allogeneic and diabetic conditions by the 2∆∆Ct method for relative quantification. Replicates were averaged from each experimental condition for each gene. Each experiment was then averaged for each gene of interest. Students two way t-test was used to determine statistical significance for the average of each gene compared with autologous control.
Statistical methods
All data is expressed as mean ± SEM. Grubbs method of detecting statistical outliers was used to remove outliers from further statistical analysis. One-way ANOVA T testing with multiple error correction, as previously described,42 was used for class comparison to determine statistical significance of the micro-array data, with statistical significance being p < 0.01. For PCR analysis Student’s two-way unpaired t-test was used to determine significance. Results were considered significant when p < 0.05.
Acknowledgments
This work was supported by a grant (#5-2010-668) from the Juvenile Diabetes Research Foundation (to B.N.) and by the Baylor Health Care System. Technical support by Ana Rahman and Morihito Takita, MD PhD is gratefully acknowledged.
Glossary
Abbreviations:
- T1DM
type 1 diabetes mellitus
- T2DM
type 2 diabetes mellitus
- IBMIR
instant blood-mediated inflammatory reaction
- IL
interleukin
- TGF
transforming growth factor
- NK
Natural Killer cell
- MMP
matrix metalloproteinase
- RAD
Ras associated with diabetes
- IL-1RN
interleukin 1 receptor antagonist
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/islets/article/21510
References
- 1.Shapiro AMJ, Lakey JRT, Ryan EA, Korbutt GS, Toth E, Warnock GL, et al. Islet transplantation in seven patients with type 1 diabetes mellitus using a glucocorticoid-free immunosuppressive regimen. N Engl J Med. 2000;343:230–8. doi: 10.1056/NEJM200007273430401. [DOI] [PubMed] [Google Scholar]
- 2.Shapiro AMJ, Ricordi C, Hering BJ, Auchincloss H, Lindblad R, Robertson RP, et al. International trial of the Edmonton protocol for islet transplantation. N Engl J Med. 2006;355:1318–30. doi: 10.1056/NEJMoa061267. [DOI] [PubMed] [Google Scholar]
- 3.Ryan EA, Paty BW, Senior PA, Bigam D, Alfadhli E, Kneteman NM, et al. Five-year follow-up after clinical islet transplantation. Diabetes. 2005;54:2060–9. doi: 10.2337/diabetes.54.7.2060. [DOI] [PubMed] [Google Scholar]
- 4.Hering BJ, Kandaswamy R, Ansite JD, Eckman PM, Nakano M, Sawada T, et al. Single-donor, marginal-dose islet transplantation in patients with type 1 diabetes. JAMA. 2005;293:830–5. doi: 10.1001/jama.293.7.830. [DOI] [PubMed] [Google Scholar]
- 5.Bennet W, Sundberg B, Groth CG, Brendel MD, Brandhorst D, Brandhorst H, et al. Incompatibility between human blood and isolated islets of Langerhans: a finding with implications for clinical intraportal islet transplantation? Diabetes. 1999;48:1907–14. doi: 10.2337/diabetes.48.10.1907. [DOI] [PubMed] [Google Scholar]
- 6.Goto M, Johansson H, Maeda A, Elgue G, Korsgren O, Nilsson B. Low molecular weight dextran sulfate prevents the instant blood-mediated inflammatory reaction induced by adult porcine islets. Transplantation. 2004;77:741–7. doi: 10.1097/01.TP.0000114872.26990.4F. [DOI] [PubMed] [Google Scholar]
- 7.Goto M, Johansson H, Maeda A, Elgue G, Korsgren O, Nilsson B. Low-molecular weight dextran sulfate abrogates the instant blood-mediated inflammatory reaction induced by adult porcine islets both in vitro and in vivo. Transplant Proc. 2004;36:1186–7. doi: 10.1016/j.transproceed.2004.04.028. [DOI] [PubMed] [Google Scholar]
- 8.Johansson H, Goto M, Dufrane D, Siegbahn A, Elgue G, Gianello P, et al. Low molecular weight dextran sulfate: a strong candidate drug to block IBMIR in clinical islet transplantation. Am J Transplant. 2006;6:305–12. doi: 10.1111/j.1600-6143.2005.01186.x. [DOI] [PubMed] [Google Scholar]
- 9.Johansson H, Lukinius A, Moberg L, Lundgren T, Berne C, Foss A, et al. Tissue factor produced by the endocrine cells of the islets of Langerhans is associated with a negative outcome of clinical islet transplantation. Diabetes. 2005;54:1755–62. doi: 10.2337/diabetes.54.6.1755. [DOI] [PubMed] [Google Scholar]
- 10.Moberg L, Johansson H, Lukinius A, Berne C, Foss A, Källen R, et al. Production of tissue factor by pancreatic islet cells as a trigger of detrimental thrombotic reactions in clinical islet transplantation. Lancet. 2002;360:2039–45. doi: 10.1016/S0140-6736(02)12020-4. [DOI] [PubMed] [Google Scholar]
- 11.Moberg L, Korsgren O, Nilsson B. Neutrophilic granulocytes are the predominant cell type infiltrating pancreatic islets in contact with ABO-compatible blood. Clin Exp Immunol. 2005;142:125–31. doi: 10.1111/j.1365-2249.2005.02883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moberg L, Olsson A, Berne C, Felldin M, Foss A, Källen R, et al. Nicotinamide inhibits tissue factor expression in isolated human pancreatic islets: implications for clinical islet transplantation. Transplantation. 2003;76:1285–8. doi: 10.1097/01.TP.0000098905.86445.0F. [DOI] [PubMed] [Google Scholar]
- 13.Özmen L, Ekdahl KN, Elgue G, Larsson R, Korsgren O, Nilsson B. Inhibition of thrombin abrogates the instant blood-mediated inflammatory reaction triggered by isolated human islets: possible application of the thrombin inhibitor melagatran in clinical islet transplantation. Diabetes. 2002;51:1779–84. doi: 10.2337/diabetes.51.6.1779. [DOI] [PubMed] [Google Scholar]
- 14.Tjernberg J, Ekdahl KN, Lambris JD, Korsgren O, Nilsson B. Acute antibody-mediated complement activation mediates lysis of pancreatic islets cells and may cause tissue loss in clinical islet transplantation. Transplantation. 2008;85:1193–9. doi: 10.1097/TP.0b013e31816b22f3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson A, McWilliams C, Kaizer E, Chaussabel D, Glaser C, Noguchi H, et al. Gene expression profiling of human pancreatic islets undergoing a simulated process of instant blood-mediated inflammatory reaction. Transplant Proc. 2008;40:430–2. doi: 10.1016/j.transproceed.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 16.Goto M, Tjernberg J, Dufrane D, Elgue G, Brandhorst D, Ekdahl KN, et al. Dissecting the instant blood-mediated inflammatory reaction in islet xenotransplantation. Xenotransplantation. 2008;15:225–34. doi: 10.1111/j.1399-3089.2008.00482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Contreras JL, Eckstein C, Smyth CA, Sellers MT, Vilatoba M, Bilbao G, et al. Brain death significantly reduces isolated pancreatic islet yields and functionality in vitro and in vivo after transplantation in rats. Diabetes. 2003;52:2935–42. doi: 10.2337/diabetes.52.12.2935. [DOI] [PubMed] [Google Scholar]
- 18.Kaizer EC, Glaser CL, Chaussabel D, Banchereau J, Pascual V, White PC. Gene expression in peripheral blood mononuclear cells from children with diabetes. J Clin Endocrinol Metab. 2007;92:3705–11. doi: 10.1210/jc.2007-0979. [DOI] [PubMed] [Google Scholar]
- 19.Kannel WB, D’Agostino RB, Wilson PWF, Belanger AJ, Gagnon DR. Diabetes, fibrinogen, and risk of cardiovascular disease: the Framingham experience. Am Heart J. 1990;120:672–6. doi: 10.1016/0002-8703(90)90026-T. [DOI] [PubMed] [Google Scholar]
- 20.Lechleitner M, Koch T, Herold M, Dzien A, Hoppichler F. Tumour necrosis factor-alpha plasma level in patients with type 1 diabetes mellitus and its association with glycaemic control and cardiovascular risk factors. J Intern Med. 2000;248:67–76. doi: 10.1046/j.1365-2796.2000.00705.x. [DOI] [PubMed] [Google Scholar]
- 21.Roach JC, Smith KD, Strobe KL, Nissen SM, Haudenschild CD, Zhou D, et al. Transcription factor expression in lipopolysaccharide-activated peripheral-blood-derived mononuclear cells. Proc Natl Acad Sci U S A. 2007;104:16245–50. doi: 10.1073/pnas.0707757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osman M, Tortorella M, Londei M, Quaratino S. Expression of matrix metalloproteinases and tissue inhibitors of metalloproteinases define the migratory characteristics of human monocyte-derived dendritic cells. Immunology. 2002;105:73–82. doi: 10.1046/j.0019-2805.2001.01349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barro C, Zaoui P, Morel F, Benhamou PY. Matrix metalloproteinase expression in rat pancreatic islets. Pancreas. 1998;17:378–82. doi: 10.1097/00006676-199811000-00008. [DOI] [PubMed] [Google Scholar]
- 24.Olsson R, Maxhuni A, Carlsson PO. Revascularization of transplanted pancreatic islets following culture with stimulators of angiogenesis. Transplantation. 2006;82:340–7. doi: 10.1097/01.tp.0000229418.60236.87. [DOI] [PubMed] [Google Scholar]
- 25.Voronov E, Shouval DS, Krelin Y, Cagnano E, Benharroch D, Iwakura Y, et al. IL-1 is required for tumor invasiveness and angiogenesis. Proc Natl Acad Sci U S A. 2003;100:2645–50. doi: 10.1073/pnas.0437939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and β-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/S0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- 27.Paul SR, Bennett F, Calvetti JA, Kelleher K, Wood CR, O’Hara RM, Jr., et al. Molecular cloning of a cDNA encoding interleukin 11, a stromal cell-derived lymphopoietic and hematopoietic cytokine. Proc Natl Acad Sci U S A. 1990;87:7512–6. doi: 10.1073/pnas.87.19.7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du XX, Williams DA. Interleukin-11: a multifunctional growth factor derived from the hematopoietic microenvironment. Blood. 1994;83:2023–30. [PubMed] [Google Scholar]
- 29.Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, Hunte B, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–25. doi: 10.1016/S1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 30.Langowski JL, Zhang X, Wu L, Mattson JD, Chen T, Smith K, et al. IL-23 promotes tumour incidence and growth. Nature. 2006;442:461–5. doi: 10.1038/nature04808. [DOI] [PubMed] [Google Scholar]
- 31.Reynet C, Kahn CR. Rad: a member of the Ras family overexpressed in muscle of type II diabetic humans. Science. 1993;262:1441–4. doi: 10.1126/science.8248782. [DOI] [PubMed] [Google Scholar]
- 32.Kauffmann-Zeh A, Rodriguez-Viciana P, Ulrich E, Gilbert C, Coffer P, Downward J, et al. Suppression of c-Myc-induced apoptosis by Ras signalling through PI(3)K and PKB. Nature. 1997;385:544–8. doi: 10.1038/385544a0. [DOI] [PubMed] [Google Scholar]
- 33.Vojtek AB, Der CJ. Increasing complexity of the Ras signaling pathway. J Biol Chem. 1998;273:19925–8. doi: 10.1074/jbc.273.32.19925. [DOI] [PubMed] [Google Scholar]
- 34.Maedler K, Sergeev P, Ehses JA, Mathe Z, Bosco D, Berney T, et al. Leptin modulates β cell expression of IL-1 receptor antagonist and release of IL-1β in human islets. Proc Natl Acad Sci U S A. 2004;101:8138–43. doi: 10.1073/pnas.0305683101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larsen CM, Faulenbach M, Vaag A, Vølund A, Ehses JA, Seifert B, et al. Interleukin-1-receptor antagonist in type 2 diabetes mellitus. N Engl J Med. 2007;356:1517–26. doi: 10.1056/NEJMoa065213. [DOI] [PubMed] [Google Scholar]
- 36.Papaccio G, Graziano A, D’Aquino R, Valiante S, Naro F. A biphasic role of nuclear transcription factor (NF)-kappaB in the islet β-cell apoptosis induced by interleukin (IL)-1β. J Cell Physiol. 2005;204:124–30. doi: 10.1002/jcp.20276. [DOI] [PubMed] [Google Scholar]
- 37.Tabatabaie T, Vasquez-Weldon A, Moore DR, Kotake Y. Free radicals and the pathogenesis of type 1 diabetes: beta-cell cytokine-mediated free radical generation via cyclooxygenase-2. Diabetes. 2003;52:1994–9. doi: 10.2337/diabetes.52.8.1994. [DOI] [PubMed] [Google Scholar]
- 38.Wilson GL, Patton NJ, LeDoux SP. Mitochondrial DNA in beta-cells is a sensitive target for damage by nitric oxide. Diabetes. 1997;46:1291–5. doi: 10.2337/diabetes.46.8.1291. [DOI] [PubMed] [Google Scholar]
- 39.Kuroda Y, Fujino Y, Morita A, Tanioka Y, Ku Y, Saitoh Y. Oxygenation of the human pancreas during preservation by a two-layer (University of Wisconsin solution/perfluorochemical) cold-storage method. Transplantation. 1992;54:561–2. doi: 10.1097/00007890-199209000-00036. [DOI] [PubMed] [Google Scholar]
- 40.Ricordi C, Lacy PE, Finke EH, Olack BJ, Scharp DW. Automated method for isolation of human pancreatic islets. Diabetes. 1988;37:413–20. doi: 10.2337/diabetes.37.4.413. [DOI] [PubMed] [Google Scholar]
- 41.Kin T, Senior P, O’Gorman D, Richer B, Salam A, Shapiro AMJ. Risk factors for islet loss during culture prior to transplantation. Transpl Int. 2008;21:1029–35. doi: 10.1111/j.1432-2277.2008.00719.x. [DOI] [PubMed] [Google Scholar]
- 42.Allantaz F, Chaussabel D, Stichweh D, Bennett L, Allman W, Mejias A, et al. Blood leukocyte microarrays to diagnose systemic onset juvenile idiopathic arthritis and follow the response to IL-1 blockade. J Exp Med. 2007;204:2131–44. doi: 10.1084/jem.20070070. [DOI] [PMC free article] [PubMed] [Google Scholar]