Abstract
Microtubule-associated proteins of the MAP1 family (MAP1A, MAP1B, and MAP1S) share, among other features, a highly conserved COOH-terminal domain approximately 125 amino acids in length. We conducted a yeast 2-hybrid screen to search for proteins interacting with this domain and identified α1-syntrophin, a member of a multigene family of adapter proteins involved in signal transduction. We further demonstrate that the interaction between the conserved COOH-terminal 125-amino acid domain (which is located in the light chains of MAP1A, MAP1B, and MAP1S) and α1-syntrophin is direct and occurs through the pleckstrin homology domain 2 (PH2) and the postsynaptic density protein 95/disk large/zonula occludens-1 protein homology domain (PDZ) of α1-syntrophin. We confirmed the interaction of MAP1B and α1-syntrophin by co-localization of the two proteins in transfected cells and by co-immunoprecipitation experiments from mouse brain. In addition, we show that MAP1B and α1-syntrophin partially co-localize in Schwann cells of the murine sciatic nerve during postnatal development and in the adult. However, intracellular localization of α1-syntrophin and other Schwann cell proteins such as ezrin and dystrophin-related protein 2 (DRP2) and the localization of the axonal node of Ranvier-associated protein Caspr1/paranodin were not affected in MAP1B null mice. Our findings add to a growing body of evidence that classical MAPs are likely to be involved in signal transduction not only by directly modulating microtubule function, but also through their interaction with signal transduction proteins.
Introduction
The vertebrate MAP1 family of microtubule-associated proteins consists of three members, MAP1A, MAP1B, and MAP1S. MAP1A and MAP1B are >300 kDA proteins and are expressed at high levels in the central and peripheral nervous system in the adult and during development, respectively [1]. MAP1S is smaller (120 kDa) and is ubiquitously expressed [2]. All three proteins share several defining features. They are synthesized as polyprotein precursors and are subsequently cleaved into a heavy and a light chain which bind to each other to form the respective MAP1 complex [1], [2]. Heavy and light chains of all MAP1 proteins contain structurally and functionally conserved domains that mediate heavy chain-light chain interaction, microtubule binding, and the potential to interact with F-actin [1]–[5].
The best characterized member of the MAP1 family is MAP1B, a 320-kDa protein which is expressed in the central nervous predominantly during development and in the peripheral nervous system throughout life [1], [6]. While originally thought to be expressed mainly in neurons, MAP1B was found to be expressed in Schwann cells [7] and oligodendrocytes [8]–[10] as well. Consistent with its expression in the nervous system, MAP1B deficient mice display defects in brain development [11]–[14]. In the peripheral nervous system, MAP1B deficiency results in a reduced number of large myelinated axons, the reduced thickness of myelin sheaths, and a decrease in nerve conduction velocity in the sciatic nerve [13].
In order to elucidate molecular mechanisms that might be involved in the function of MAP1B during development we performed a search for protein interaction partners using one of the domains conserved between MAP1A, MAP1B, and MAP1S as bait. Here we show that the COOH terminus of the light chain of MAP1B interacts with α1-syntrophin, a modular adapter protein associated with the dystrophin-glycoprotein complex (DGC) [15]–[18]. α1-syntrophin, a 58-kD protein highly expressed in the brain, belongs to a multigene family which consists of five isoforms α1, ß1 and ß2, γ1 and γ2. The syntrophins function by recruiting signaling molecules through their multiple protein interaction motifs. These consist of pleckstrin homology domains 1a, 1b, and 2 (PH1a, PH1b, PH2), a PDZ (postsynaptic density protein 95/ disk large/zonula occludens-1 protein homology) domain, and the syntrophin unique domain (SU). α1-syntrophin associates with the DGC in the plasma membrane of several cell types via direct binding of its PH2 and SU region to dystrophin, dystrobrevin or utrophin [19], [20]. The PDZ domain of α1-syntrophin binds to a variety of signaling molecules including sodium channels [21], [22], neuronal nitric oxide synthase [23]–[25], aquaporin-4 [26], [27] and serine/threonine kinases [28], [29]. Mice lacking α1-syntrophin display aberrations in neuromuscular synapses with undetectable levels of postsynaptic utrophin and reduced levels of acetylcholine receptor and acetylcholinesterase [30].
Materials and Methods
Ethics Statement
Tissues from mice were obtained in compliance with the Austrian law regulating the use of animals in biomedical research, Tierversuchsgesetz, BGBl. Nr. 501/1989 and BGBl. I Nr. 162/2005. The manuscript does not include experiments on live animals. The production and culling of mice in order to obtain tissues (as performed in this manuscript) does not require approval of the Austrian Ministry of Science and Research, the governmental body regulating the use of animals in biomedical research. Wild-type and MAP1B−/− mice were anesthetized and sacrificed by decapitation.
Yeast 2-hybrid Screen and Recombinant Clones
The Matchmaker 2-hybrid system (Clontech, Mountain View, California) was employed following the manufacturer’s recommendations. Using the small scale transformation procedure, strain EGY48 (p8oplacZ) carrying a plasmid encoding the COOH terminus of LC1 (CT-LC1 in Fig. 1a, corresponding to MH3B [4]) fused to the DNA binding domain of the LexA protein was transformed with 500 µg of a cDNA library prepared from 19-day old mouse embryo and cloned into a vector resulting in fusions of the cDNA clones with the transcription activation domain of the Matchmaker system.
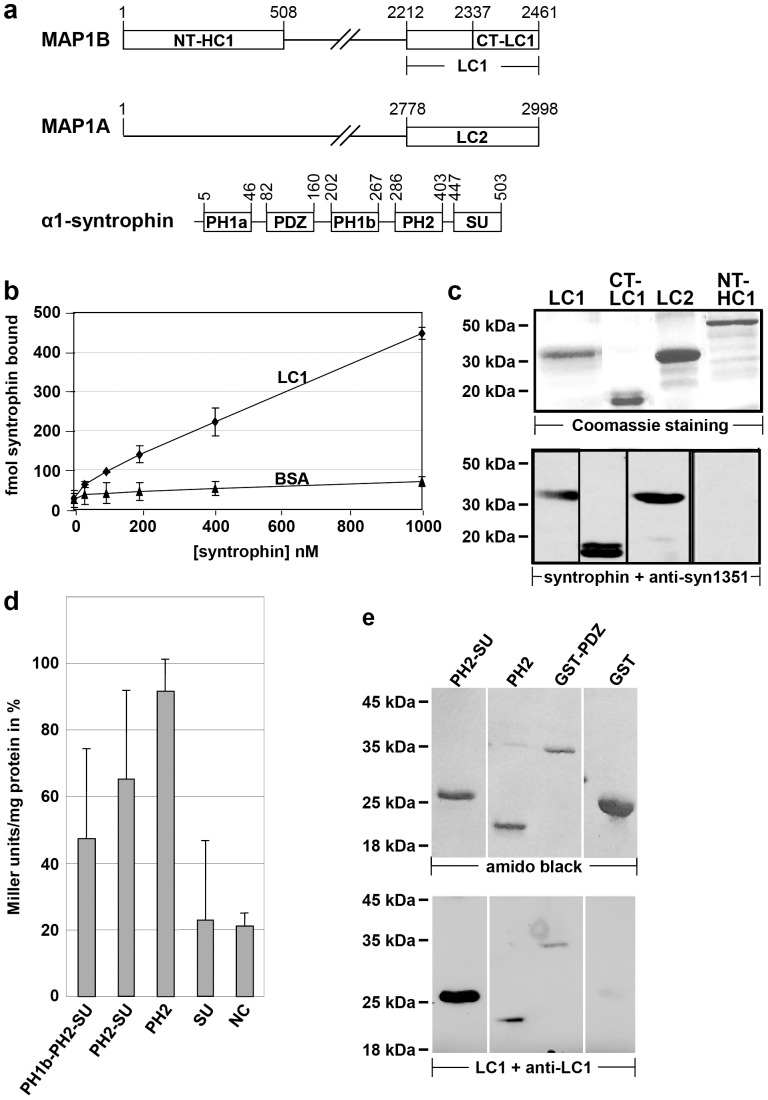
Figure 1. The light chains of MAP1B and MAP1A interact with α1-syntrophin.
a) Schematic representation of MAP1B, MAP1A, and α1-syntrophin to indicate the relevant domains. The drawings are not to scale. Numbers indicate amino acid positions for rat MAP1A [71], [72], rat MAP1B (according to database entry XM_215469), and mouse α1-syntrophin [16], [32]. MAP1B and MAP1A are synthesized as polyprotein precursors which are proteolytically cleaved to yield heavy chains (HC1 and HC2, respectively) and light chains (LC1 and LC2, starting at residue 2212 and 2778 of the polyprotein precursor, respectively). NT-HC1, NH2 terminus of HC1; CT-LC1, COOH terminus of LC1; PH1a, PH1b, and PH2, pleckstrin homology domains; PDZ, postsynaptic density protein 95/disk large/zonula occludens-1 protein homology domain; SU, syntrophin unique domain; b) Microtiter plates coated with 100 nM recombinant LC1 (LC1) or BSA (BSA) were overlaid with increasing concentrations of Eu3+-labeled recombinant α1-syntrophin. α1-syntrophin was found to bind specifically to LC1, whereas binding to BSA was weak and considered to be unspecific background of the assay. The values represent the mean ± standard deviation of three independent experiments. c) The indicated recombinant proteins were subjected to SDS-PAGE, stained with Coomassie brilliant blue or blotted onto nitrocellulose and probed with recombinant α1-syntrophin protein. For immunological detection of syntrophin bound to blotted proteins the pan-syntrophin antibody (anti-syn1351) was used. Syntrophin was found to bind to LC1, LC2 and the COOH-terminal half of LC1, but not to the NH2-terminal fragment of HC1 which served as a negative control. d) Yeast 2-hybrid β-galactosidase activity in homogenates of yeast transformants co-expressing the LC1 COOH-terminal domain CT-LC1 (fused to the DNA-binding domain of the LexA protein) and various deletion mutants of α1-syntrophin as indicated, or a fragment of RACK-1 as negative control (NC; each fused to the transcription activation domain). β-galactosidase activity in Miller units is given in percent relative to activity obtained with the NH2 terminus of the MAP1B heavy chain which has been demonstrated previously to interact with the LC1 COOH-terminal domain [4] and was used here as positive control. The largest α1-syntrophin fragment containing the PH1b, PH2, and SU domains was identified in the original yeast 2-hybrid screen as interaction partner of LC1. Elevated levels of β-galactosidase activity were detected with all α1-syntrophin fragments containing the PH2 domain. Autoactivation of the α1-syntrophin deletion mutant proteins was excluded (data not shown). Two independent yeast colonies were assessed for each two-hybrid pair in 4 independent experiments. Values represent the mean ± standard deviation. e) The indicated recombinant α1-syntrophin fragments were subjected to SDS-PAGE, blotted onto nitrocellulose and either stained with amido black or probed with recombinant LC1 protein. For immunological detection of LC1 bound to blotted proteins anti-LC1 was used. LC1 was found to bind to syntrophin fragments containing the PH2 or PDZ domain, but not to GST which served as a negative control. Anti-LC1 did not directly react with the blotted proteins when prior incubation of the blot with LC1 was omitted (not shown).
Constructs
Constructs for use in yeast: Construction of the plasmid encoding the bait protein for the screen, the COOH terminus of rat MAP1B LC1 (CT-LC1 in Fig. 1a, corresponding to MH3B [4]) fused to the DNA binding domain of the 2-hybrid system, was described previously [4]. A mouse α1-syntrophin fragment comprising the PH1b, PH2, and SU domains (amino acids 172–503; Fig. 1a) fused to the transcription activator domain in vector pB42AD of the 2-hybrid system was obtained in the screen; this clone was used as template to generate α1-syntrophin deletion mutants by PCR such that the PCR fragments were amenable to restriction with EcoRI and XhoI to be inserted into the EcoRI and XhoI restriction sites of pB42AD, resulting in the respective α1-syntrophin domain fused to the transcription activator domain. The following deletion mutants were generated: PH2-SU, amino acids 284–503, using primers 5′-CCGGAATTCGGGAGCCAGGACATCAAGCAGATTGGC-3′ and 5′-GGTAGACAAGCCGACAACCTTGATTGGA-3′; PH2, amino acids 284–441, using primers 5′-CCGGAATTCGGGAGCCAGGACATCAAGCAGATTGGC-3′ and 5′-CCGCTCGAGCGGCTCGGGCTGCTCCAG-3′; SU, amino acids 433–503, using primers 5′-CCGGAATTCGCAGCTGAGCCTGGAGCAGCCCGAGCC-3′ and 5′-GGTAGACAAGCCGACAACCTTGATTGGA-3′. For a negative control (NC; Fig. 1d) we used a COOH-terminal fragment of murine RACK-1 (amino acids 173–317) fused to the transcription activator domain in vector pB42AD [31].
Constructs for the expression of recombinant proteins in E. coli: The construction of plasmids encoding 6xHis-tagged LC1, LC2, CT-LC1, and NT-HC1 (MH1B [4]) has been described [3], [4]. 6xHis-tagged full length α1-syntrophin was generated by PCR using primers 5′-CCGCAATTGATGGCGTCAGGCAGGCGC-3′ and 5′-GTCCCAGCCAACGGAGGTCCC-3′ and a mouse full length α1-syntrophin cDNA [32] as template. The PCR fragment was restricted with MunI and EcoRI yielding a fragment comprising amino acids 1–172 of α1-syntrophin. This fragment was joined in a triple ligation to the EcoRI/XhoI fragment of the α1-syntrophin cDNA obtained in the 2-hybrid screen containing the rest of the coding sequence (amino acids 172–503) and a MunI/XhoI restricted derivative of pQE-60 (Qiagen, Valencia, California) to yield construct pQEsyn encoding full length mouse α1-syntrophin with appropriate flanking restriction sites. The full length α1-syntrophin cDNA was cut out with Mun1 and XhoI and inserted into the EcoRI and XhoI sites of a derivative of pET15b (Novagen, Madison, Wisconsin) resulting in a construct encoding NH2-terminally 6xHis-tagged α1-syntrophin. Likewise, fragments containing α1-syntrophin domains PH2 plus SU (PH2-SU) or PH2 only were obtained by digestion with EcoRI and XhoI of the respective pB42AD vectors (see above) and inserted into the EcoRI and XhoI sites of the derivative of pET15b resulting in constructs encoding NH2-terminally 6xHis-tagged PH2-SU and PH2 domains, respectively. To express the PDZ domain of α1-syntrophin (amino acids 80–164), a PCR fragment containing the PDZ domain was generated using the full length α1-syntrophin cDNA (see above) as template and primers 5′-ACTGGAATTCCGCCGCGTGACGGTGCGCAAGGC-3′ and 5′-ATCGCTCGAGCTTCATGTACTTAACCTCCAACACAACCTCCTTGCCTGTCTTC-3′. The resulting PCR fragment was inserted by blunt end ligation into the EcoRV site of pBluescript (Fermentas, Glen Burnie, Maryland), cut out with EcoRI and XhoI and inserted into plasmid pGEX-4T-1 (GE Healthcare Biosciences AB, Uppsala, Sweden) to yield a construct for the expression of the NH2-terminally GST-tagged PDZ domain of α1-syntrophin.
For expression in vertebrate cells we used a construct encoding COOH-terminally myc-tagged LC1 described previously [5]. For the expression of α1-syntrophin we fused the MunI/XhoI fragment of pQEsyn encoding full length α1-syntrophin (see above) in frame to NH2-terminal GFP in an EcoRI/XhoI restricted derivative of pEGFP-C1 (Clontech) which contains a human cytomegalovirus immediate early promoter.
The correct sequence of all constructs was confirmed.
Antibodies
Affinity-purified rabbit polyclonal anti-LC1 antibody [5] and anti-HC1 antibody (anti-HC750) [33]; affinity-purified rabbit polyclonal anti-myc [5]; monoclonal pan anti-syntrophin antibody anti-syn1351 [34]; polyclonal rabbit anti-DRP2 [35] kindly provided by P. Brophy; polyclonal rabbit anti-paranodin antibody SL51 [36] kindly provided by J. A. Girault; polyclonal rabbit anti-ezrin (Upstate, Lake Placid, NY); monoclonal mouse anti-α-tubulin B-5-1-2 (Sigma, St. Louis, MO) and monoclonal rat anti-α-tubulin (YL1-2; Abcam, Cambridge, UK); Secondary antibodies: Alexa Fluor 488- or Fluor 568-labelled goat anti-rabbit, anti-rat, or anti-mouse (Molecular Probes, Leiden, The Netherlands), rhodamine-red-labeled goat anti-mouse or anti-rabbit (Jackson, West Grove, PA) and Cy5-labelled donkey anti-rat (Jackson). Secondary antibodies for immunoblots: HRP-conjugated goat anti-rabbit or anti-mouse (Jackson) or alkaline phosphatase-conjugated anti-rabbit or anti-mouse (Promega, Mannheim, Germany).
Immunohistochemistry, Cell Culture, Transfection, and Microscopy
Wild-type and MAP1B−/− mice [13] were anesthetized and sacrificed by decapitation. Sciatic nerves were dissected and fixed with 4% PFA in PBS for 10 min at room temperature. After washing with PBS, nerves were teased on SuperFrost Plus glass slides (Thermo Fisher Scientific, Waltham, MA), air-dried and stained as described [37]. PtK2 cells were grown, transiently transfected, and stained for fluorescence microscopy with a confocal Zeiss Axiovert microscope with LSM 510 software (Zeiss, Oberkochen, Germany) as described [3].
Protein Analysis
His-tagged recombinant proteins were expressed in E. Coli and purified as described [3].
Protein Interaction Assay with Europium Labeled Proteins
Europium labeling of recombinant α-syntrophin and binding assays were performed as described previously [38]. Briefly, 96-well microtiter plates were coated with 100 nM LC1 or BSA type H1 (Gerbu, Gaiberg, Germany) as a control. Following blocking with 4% BSA, plates were overlaid with increasing amounts of Eu3+-labeled α1-syntrophin. Plates were washed and protein bound was determined by releasing the complexed Eu3+ with enhancement solution and measuring fluorescence with a Delfia time-resolved fluorometer (Wallac, Turku, Finland). Binding of α-syntrophin to BSA was considered to be non-specific.
For blot overlay assays, recombinant proteins were fractionated by SDS–PAGE. Blots (nitrocellulose membrane, 0.2 µm; Schleicher & Schuell, Dassel, Germany) were blocked in buffer A (0.25% Tween 20 in phosphate-buffered saline) containing 2% bovine serum albumin (BSA) for 1 h, washed 3 times for 5 min in buffer A, incubated with 10–100 µg/ml recombinant protein in buffer A containing 2% BSA for 2 h, washed again, and probed with an appropriate primary antibody against the recombinant protein in buffer A containing 1% BSA. After additional washing, the recombinant protein-antibody complexes were detected using alkaline phosphatase-conjugated secondary antibodies (Promega, Mannheim, Germany) and a detection system described previously [39] or horse radish peroxidase-conjugated secondary antibodies (Jackson, West Grove, Pennsylvania) and the chemiluminescence detection system (Pierce, Rockford, Illinois) according to the manufacturer’s recommendations.
Immunoprecipitation
For immunoprecipitations, whole brains of 3-week-old or sciatic nerves of 6-day-old wild-type or transgenic mice expressing myc-tagged LC1 in the nervous system [40] were homogenized on ice in TEN buffer (100 mM Tris-HCl (pH 7.5), 100 mM NaCl, 10 mM EDTA, 0.1 mM DTT) containing a mixture of protease inhibitors (Roche, Vienna, Austria). Immunoprecipitates [5] were analyzed by immunoblotting as described [39].
β-galactosidase Assay
Ortho-nitrophenyl-β-d-galactopyranoside was used as the substrate for β-galactosidase for the liquid culture assay (Clontech). For each two-hybrid pair, two independent yeast colonies were selected, grown to an OD600 of 0.5–0.8, harvested, resuspended in Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, pH 7.0) with 0.27% (v/v) 2-mercaptoethanol, and lysed by vigorous shaking in a “Merkenschlager” cell mill using glass beads. Protein concentration was determined by the method of Bradford [41] and β-galactosidase activity was measured and calculated in modified Miller units: 1 U = 1000*OD420/t*V*mg protein. Values were expressed as percent relative to the activity obtained with the positive control reaction indicated.
Results
The Light Chains of MAP1B and MAP1A Interact with α1-syntrophin
The COOH-terminal domain of MAP1 proteins is conserved in all members of this protein family from drosophila to man. To identify proteins interacting with this conserved domain which is located in the light chains of mammalian MAP1A, MAP1B and MAP1S, we performed a yeast 2-hybrid screen using this domain of LC1 as bait and a mouse 19-day embryo cDNA library as target. One of the candidate proteins identified in this screen was α1-syntrophin, a modular adapter protein with multiple protein interaction motifs associated with the dystrophin protein family [15]–[18].
We first confirmed that the light chains of MAP1B and MAP1A directly interact with α1-syntrophin. Purified recombinant α1-syntrophin bound specifically to LC1 in a microtiter plate overlay assay (Fig. 1b). Likewise, in a blot overlay assay, recombinant α1-syntrophin bound to LC1, LC2, and the conserved COOH-terminal domain which was used as bait in the original screen (Fig. 1c). In contrast, α1-syntrophin did not interact with the NH2-terminal domain of MAP1B (Fig. 1c). This 508-amino acid domain is also conserved in all proteins of the MAP1 family and was used here as negative control.
To identify which domain(s) of α1-syntrophin interact with LC1 we first performed a yeast 2-hybrid β-galactosidase assay. Starting with the α1-syntrophin cDNA fragment that interacted with LC1 in the original screen and contained the PH1b, PH2, and SU domains we analyzed the interaction with LC1 of several α1-syntrophin deletion mutants. We found that the COOH terminus of LC1 (the bait protein of the screen) interacted with all α1-syntrophin deletion mutants that contained the PH2 domain (Fig. 1d), revealing that this domain contains an LC1 binding site. This interaction of LC1 with the PH2 domain was confirmed in blot overlay assays (Fig. 1e). Since α1-syntrophin also contains a PDZ domain and LC1 and LC2 have been reported to interact with PDZ domains of other proteins [40], [42] we tested interaction of LC1 with a recombinant protein containing the PDZ domain of α1-syntrophin fused to glutathione S-transferase (GST). In a blot overlay assay, LC1 interacted specifically with the PDZ domain but not with GST (negative control, Fig. 1e).
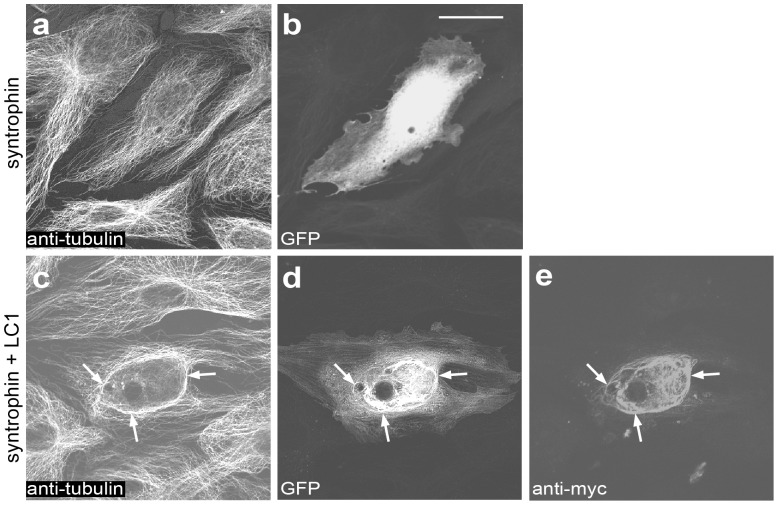
To confirm cellular interaction of α1-syntrophin with LC1 we ectopically expressed tagged versions of the two proteins in PtK2 cells. In the absence of LC1, GFP-tagged α1-syntrophin displayed diffuse distribution (Fig. 2). We did not observe binding of α1-syntrophin to actin in epitheloid PtK2 cells as has been described for endothelial, smooth muscle, and Chinese hamster ovary cells [43]. Upon co-expression of LC1, α1-syntrophin was found to co-localize with LC1 on microtubules. The truncated version of α1-syntrophin comprising the PH1b, PH2, and syntrophin unique domains which bound to LC1 in vitro (Fig. 1) also interacted with LC1 in PtK2 cells (not shown). Cells expressing GFP only in the presence of LC1 displayed diffuse distribution of GFP, ruling out the possibility that the co-localization of α1-syntrophin with LC1 is due to the GFP-tag (not shown).
Figure 2. α1-syntrophin binds to microtubules in cells expressing LC1.
PtK2 cells were transiently transfected to express either EGF-tagged α1-syntrophin alone (a and b) or α1-syntrophin and myc-tagged LC1 (c–e). Cells were fixed, co-stained for tubulin (anti-tubulin) and LC1 (anti-myc) and analyzed by fluorescence microscopy. In the absence of ectopically expressed LC1, α1-syntrophin was diffusely distributed throughout the cytoplasm (b). When co-expressed with LC1, α1-syntrophin was found to co-localize with LC1 on microtubules (c-e, arrows). Expression of LC1 causes microtubules to bundle, as has been described previously [5]. Scale bar, 20 µm.
Further confirmation for the association of α1-syntrophin with LC1 in vivo was obtained by co-immunoprecipitation experiments. Using wild-type mouse brain extracts and anti-LC1 antibodies, a small amount of α1-syntrophin was co-precipitated (not shown). Since the epitope of the anti-LC1 antibody is located in the COOH-terminal part of LC1 that has been identified as interaction domain for α1-syntrophin (Fig. 1), binding of α1-syntrophin in this region might reduce interaction of anti-LC1 with the LC1-syntrophin complex. Therefore, we next used protein extracts of brains from three-week-old transgenic mice which express myc-tagged LC1 in the brain [40]. Using anti-myc antibodies, syntrophin was co-precipitated with myc-tagged LC1, demonstrating that the proteins associate with each other in vivo (Fig. 3a).
Figure 3. α1-syntrophin is found in a complex with LC1 in the central and peripheral nervous system.
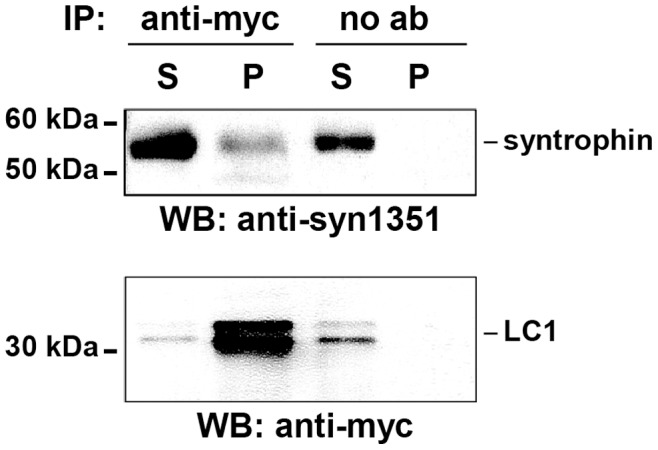
Brain protein extracts obtained from transgenic mice expressing myc-tagged LC1 were immunoprecipitated (IP) either with anti-myc antibodies (anti-myc) or without antibody (no ab; negative control). Pellets (P) and the corresponding supernatants (S) were fractionated by SDS-PAGE and analyzed by immunoblotting (WB) using anti-syntrophin (anti-syn1351) or anti-myc antibodies (anti-myc). The positions of protein size markers, syntrophin, and LC1 are indicated. The double band corresponding to LC1 resulted from insufficient denaturation prior to gel electrophoresis.
The above experiments clearly demonstrate that the light chains of MAP1B and MAP1A can directly interact with α1-syntrophin and associate with syntrophin in the nervous system in vivo. Furthermore, we localized the light chain interaction sites to the PH2 and PDZ domains of α1-syntrophin.
MAP1B and Syntrophin Co-localize in Schwann Cells in Adult Peripheral Nerve
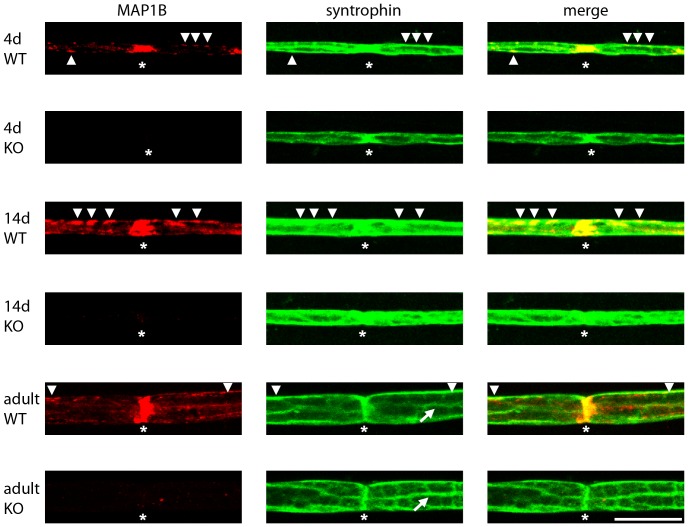
We next analyzed the subcellular localization of MAP1B and syntrophin in peripheral nerve by staining teased fibers of sciatic nerves of wild-type and MAP1B deficient mice at different stages during postnatal development (Fig. 4). At all ages tested (4 days, 14 days, and adult) MAP1B appeared to be highly concentrated at the nodes of Ranvier (Fig. 4). Specific MAP1B staining was also observed in the abaxonal Schwann cell membrane. At postnatal day 14 this staining was particularly prominent. Syntrophin was found at the nodes of Ranvier and in the abaxonal membrane. At both locations it partially co-localized with MAP1B. Confirming previous results, we observed that, in the adult, syntrophin was localized at Cajal bands [44]. Teased fibers of wild-type and MAP1B knockout mice did not display consistent differences in syntrophin localization (Fig. 4).
Figure 4. MAP1B and syntrophin co-localize at the nodes of Ranvier and the abaxonal Schwann cell membrane.
Sciatic nerves were prepared from 4-day (4d) or 14-day (14d) old or adult (adult) wild-type (WT) and MAP1B−/− (KO) mice. Individual myelinated axons were isolated and stained for MAP1B (antibody anti-HC750) or syntrophin (pan syntrophin antibody anti-syn1351) as indicated. The pictures represent projections of confocal Z-stacks. The staining for MAP1B in postnatal and adult Schwann cells is specific as it is absent in Schwann cells of MAP1B−/− mice. At all ages MAP1B was found to be concentrated at the nodes of Ranvier (asterisks). It also localized at the abaxonal membrane (arrow heads), particularly strong at postnatal day 14. Syntrophin was also found at nodes of Ranvier and the abaxonal membrane. In the adult, it was found to be localized to Cajal bands (arrows) in agreement with previous results [44]. Co-localization of MAP1B and syntrophin was most prominent at the nodes of Ranvier and partial co-localization was found at the abaxonal membrane (arrow heads). Scale bar, 20 µm.
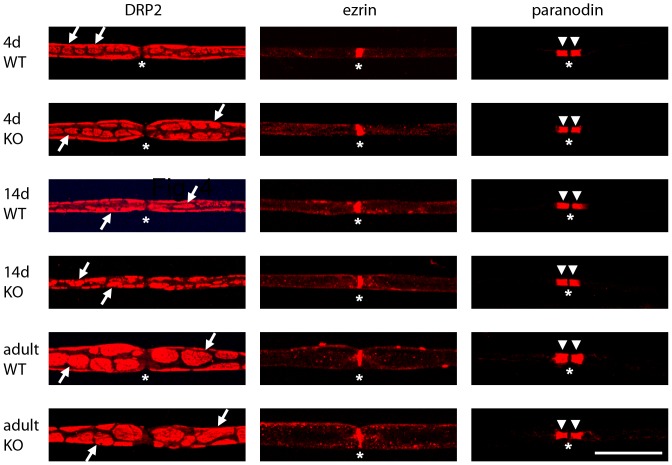
We also analyzed whether other Schwann cell proteins such as DRP2 and ezrin and the axonal protein Caspr1/paranodin were affected by deficiency in MAP1B (Fig. 5). During postnatal development and in the adult, DRP2 was expressed in characteristic clusters at the abaxonal Schwann cell membrane [35]. Ezrin was found to be expressed in defined regions which sharpened during postnatal development. These regions represent Schwann cell microvilli at the nodes of Ranvier [45]. Caspr1/paranodin marked the paranodal axonal compartments [36], [46] at early and later stages (Fig. 5). No differences in DRP2, ezrin, or Caspr1/paranodin staining were observed between wild-type and MAP1B deficient fibers during postnatal development and in the adult. Moreover, no significant differences could be detected between wild-type and MAP1B deficient fibers in the distance between consecutive nodes of Ranvier (normalized to fiber diameter), in the number and spacing of Schmidt-Lantermann incisures, or in the distance from the nodes of Ranvier to the first Schmidt-Lantermann incisure in the internodal region (not shown).
Figure 5. DRP2, ezrin and Caspr1/paranodin localization in peripheral nerve is not affected by MAP1B deficiency.
Sciatic nerves were prepared from 4-day (4d) or 14-day (14d) old or adult (adult) wild-type (WT) and MAP1B−/− (KO) mice. Individual myelinated axons were isolated and stained for DRP2, ezrin, or Caspr1/paranodin as indicated. The pictures represent projections of confocal Z-stacks. The nodes of Ranvier (asterisks) and the DRP2 clusters (arrows) are indicated where it was possible to define them. The gap in Caspr1/paranodin staining at the node is due to the absence of the protein in the actual node region which is flanked by a proximal and a distal paranodal compartment (arrow heads). Scale bar, 20 µm.
Discussion
Our results demonstrate that the light chains of MAP1A and MAP1B interact with the modular adapter protein α1-syntrophin in the central and peripheral nervous system. We identified the conserved COOH termini of the light chains and the PH2 and PDZ domains of syntrophin as interacting domains. The light chains of MAP1A and MAP1B have previously been reported to bind to PDZ domains of glutamate receptor interacting protein 1 [47], PDZrhoGEF [42], and neuronal nitric oxide synthase [40]. Thus, interaction with PDZ domains of target proteins involved in signal transduction emerges as a characteristic function of the COOH-terminal domain of the light chains of MAP1 proteins.
The direct comparison of MAP1B expression in sciatic nerve fibers of wild-type and MAP1B−/− mice by immunohistochemistry allowed us to specifically detect MAP1B expression in myelinating Schwann cells of the adult, uninjured nerve and at higher levels in Schwann cells during postnatal development (Fig. 4). MAP1B was found at the nodes of Ranvier and at the internodal abaxonal membrane. At both locations MAP1B partially co-localized with syntrophin. Syntrophins including the α-, β1-, β2- and γ2-isoforms have previously been shown to be present in the internodal abaxonal Schwann cell membrane, in particular localizing to the Cajal bands where they confer specific scaffolding properties to the dystrophin glycoprotein complex [44]. In the present study we used a pan syntrophin antibody reacting with α-, β and γ-isoforms for the analyses in peripheral nerve (Fig. 3 and 4). Thus, while our biochemical analysis (Fig. 1) clearly shows that the light chains of MAP1A and MAP1B interact with α1-syntrophin, we cannot exclude the possibility that the light chains also interact with other isoforms of syntrophins in the peripheral nerve.
MAP1B deficiency did not appear to alter syntrophin expression or localization in Schwann cells, nor did it alter the characteristic organization of DRP2 in clusters at the abaxonal Schwann cell membrane, nor the internodal distance (not shown) or the organization of the nodes of Ranvier as analyzed by staining for ezrin and Caspr1/paranodin (Fig. 5). Thus, the previously observed reduction in nerve conductance velocity and the reduced myelination in MAP1B deficient sciatic nerves [13] do not appear to be reflected by changes in syntrophin expression or intracellular organization.
Originally, MAPs were thought to regulate neuronal microtubule dynamics, stability, and spacing between individual microtubules in a microtubule bundle [1] as well as modulating access and activity of microtubule-dependent motor proteins and thus axonal and dendritic transport [48]–[55] through their direct interaction with microtubules. In a more recent development, classical MAPs have been found to bind to a wide variety of proteins with diverse functions. For example, proteins of the MAP1 family bind to receptors and ion channels [56]–[58], postsynaptic density (PSD) proteins PSD-93 and PSD-95 [59], [60], signaling molecules (EPAC, PDZrhoGEF, Tiam1, casein kinase 1d, RASSF1a [42], [61]–[66]) and proteins involved in intracellular traffic [67]–[70]. Thus, our findings presented here add to a growing body of evidence that classical MAPs can play a role in signal transduction not only by directly modulating microtubule function, but also through their interaction with a variety of signal transduction proteins.
Acknowledgments
We are grateful to P. Brophy (University of Edinburgh) for kindly providing the anti-DRP2 antibody and I. Fischer (Drexel University, Philadelphia) for the anti-HC750 antibody.
Funding Statement
This research was supported by grant F607 from the Austrian Science Fund (FWF) and grant J-15/05 of the City of Vienna and the Austrian Academy of Sciences (to F.P.) and National Institutes of Health grant NS33145 (to S.C.F.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Schoenfeld TA, Obar RA (1994) Diverse distribution and function of fibrous microtubule-associated proteins in the nervous system. Int Rev Cytol 151: 67–137. [DOI] [PubMed] [Google Scholar]
- 2. Orban-Nemeth Z, Simader H, Badurek S, Trancikova A, Propst F (2005) Microtubule-associated protein 1S, a short and ubiquitously expressed member of the microtubule-associated protein 1 family. J Biol Chem 280: 2257–2265. [DOI] [PubMed] [Google Scholar]
- 3. Noiges R, Eichinger R, Kutschera W, Fischer I, Németh Z, et al. (2002) MAP1A and MAP1B: light chains determine distinct functional properties. J Neurosci 22: 2106–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Noiges R, Stroissnigg H, Trancikova A, Kalny I, Eichinger R, et al. (2006) Heterotypic complex formation between subunits of microtubule-associated proteins 1A and 1B is due to interaction of conserved domains. Biochim Biophys Acta 1763: 1011–1016. [DOI] [PubMed] [Google Scholar]
- 5. Tögel M, Wiche G, Propst F (1998) Novel features of the light chain of microtubule-associated protein MAP1B: microtubule stabilization, self interaction, actin filament binding, and regulation by the heavy chain. J Cell Biol 143: 695–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gordon-Weeks PR, Fischer I (2000) MAP1B expression and microtubule stability in growing and regenerating axons. Microsc Res Tech 48: 63–74. [DOI] [PubMed] [Google Scholar]
- 7. Ma D, Chow S, Obrocka M, Connors T, Fischer I (1999) Induction of microtubule-associated protein 1B expression in Schwann cells during nerve regeneration. Brain Res 823: 141–153. [DOI] [PubMed] [Google Scholar]
- 8. Fischer I, Konola J, Cochary E (1990) Microtubule associated protein (MAP1B) is present in cultured oligodendrocytes and co-localizes with tubulin. J Neurosci Res 27: 112–124. [DOI] [PubMed] [Google Scholar]
- 9. Ulloa L, Ibarrola N, Avila J, Díez-Guerra FJ (1994) Microtubule-associated protein 1B (MAP1B) is present in glial cells phosphorylated different than in neurones. Glia 10: 266–275. [DOI] [PubMed] [Google Scholar]
- 10. Vouyiouklis DA, Brophy PJ (1993) Microtubule-associated protein MAP1B expression precedes the morphological differentiation of oligodendrocytes. J Neurosci Res 35: 257–267. [DOI] [PubMed] [Google Scholar]
- 11. Edelmann W, Zervas M, Costello P, Roback L, Fischer I, et al. (1996) Neuronal abnormalities in microtubule-associated protein 1B mutant mice. Proc Natl Acad Sci USA 93: 1270–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gonzalez-Billault C, Demandt E, Wandosell F, Torres M, Bonaldo P, et al. (2000) Perinatal lethality of microtubule-associated protein 1B-deficient mice expressing alternative isoforms of the protein at low levels. Mol Cell Neurosci 16: 408–421. [DOI] [PubMed] [Google Scholar]
- 13. Meixner A, Haverkamp S, Wässle H, Führer S, Thalhammer J, et al. (2000) MAP1B is required for axon guidance and is involved in the development of the central and peripheral nervous system. J Cell Biol 151: 1169–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takei Y, Kondo S, Harada A, Inomata S, Noda T, et al. (1997) Delayed development of nervous system in mice homozygous for disrupted microtubule-associated protein 1B (MAP1B) gene. J Cell Biol 137: 1615–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Adams ME, Butler MH, Dwyer TM, Peters MF, Murnane AA, et al. (1993) Two forms of mouse syntrophin, a 58 kd dystrophin-associated protein, differ in primary structure and tissue distribution. Neuron 11: 531–540. [DOI] [PubMed] [Google Scholar]
- 16. Ahn AH, Freener CA, Gussoni E, Yoshida M, Ozawa E, et al. (1996) The three human syntrophin genes are expressed in diverse tissues, have distinct chromosomal locations, and each bind to dystrophin and its relatives. J Biol Chem 271: 2724–2730. [DOI] [PubMed] [Google Scholar]
- 17. Peters MF, Adams ME, Froehner SC (1997) Differential association of syntrophin pairs with the dystrophin complex. J Cell Biol 138: 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang B, Ibraghimov-Beskrovnaya O, Moomaw CR, Slaughter CA, Campbell KP (1994) Heterogeneity of the 59-kDa dystrophin-associated protein revealed by cDNA cloning and expression. J Biol Chem 269: 6040–6044. [PubMed] [Google Scholar]
- 19. Ahn AH, Kunkel LM (1995) Syntrophin binds to an alternatively spliced exon of dystrophin. J Cell Biol 128: 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang B, Jung D, Rafael JA, Chamberlain JS, Campbell KP (1995) Identification of alpha-syntrophin binding to syntrophin triplet, dystrophin, and utrophin. J Biol Chem 270: 4975–4978. [DOI] [PubMed] [Google Scholar]
- 21. Gee SH, Madhavan R, Levinson SR, Caldwell JH, Sealock R, et al. (1998) Interaction of muscle and brain sodium channels with multiple members of the syntrophin family of dystrophin-associated proteins. J Neurosci 18: 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schultz J, Hoffmuller U, Krause G, Ashurst J, Macias MJ, et al. (1998) Specific interactions between the syntrophin PDZ domain and voltage-gated sodium channels. Nat Struct Biol 5: 19–24. [DOI] [PubMed] [Google Scholar]
- 23. Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, et al. (1996) Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domains. Cell 84: 757–767. [DOI] [PubMed] [Google Scholar]
- 24. Hashida-Okumura A, Okumura N, Iwamatsu A, Buijs RM, Romijn HJ, et al. (1999) Interaction of neuronal nitric-oxide synthase with α1-syntrophin in rat brain. J Biol Chem 274: 11736–11741. [DOI] [PubMed] [Google Scholar]
- 25. Hillier BJ, Christopherson KS, Prehoda KE, Bredt DS, Lim WA (1999) Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex. Science 284: 812–815. [PubMed] [Google Scholar]
- 26. Amiry-Moghaddam M, Frydenlund DS, Ottersen OP (2004) Anchoring of aquaporin-4 in brain: molecular mechanisms and implications for the physiology and pathophysiology of water transport. Neuroscience 129: 999–1010. [DOI] [PubMed] [Google Scholar]
- 27. Amiry-Moghaddam M, Otsuka T, Hurn PD, Traystman RJ, Haug FM, et al. (2003) An alpha-syntrophin-dependent pool of AQP4 in astroglial end-feet confers bidirectional water flow between blood and brain. Proc Natl Acad Sci USA 100: 2106–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hasegawa M, Cuenda A, Spillantini MG, Thomas GM, Buee-Scherrer V, et al. (1999) Stress-activated protein kinase-3 interacts with the PDZ domain of alpha1-syntrophin. A mechanism for specific substrate recognition. J Biol Chem 274: 12626–12631. [DOI] [PubMed] [Google Scholar]
- 29. Lumeng C, Phelps S, Crawford GE, Walden PD, Barald K, et al. (1999) Interactions between beta 2-syntrophin and a family of microtubule- associated serine/threonine kinases. Nat Neurosci 2: 611–617. [DOI] [PubMed] [Google Scholar]
- 30. Adams ME, Kramarcy N, Krall SP, Rossi SG, Rotundo RL, et al. (2000) Absence of alpha-syntrophin leads to structurally aberrant neuromuscular synapses deficient in utrophin. J Cell Biol 150: 1385–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Osmanagic-Myers S, Wiche G (2004) Plectin-RACK1 (receptor for activated C kinase 1) scaffolding: a novel mechanism to regulate protein kinase C activity. J Biol Chem 279: 18701–18710. [DOI] [PubMed] [Google Scholar]
- 32. Adams ME, Dwyer TM, Dowler LL, White RA, Froehner SC (1995) Mouse alpha 1- and beta 2-syntrophin gene structure, chromosome localization, and homology with a discs large domain. J Biol Chem 270: 25859–25865. [DOI] [PubMed] [Google Scholar]
- 33. Fischer I, Romano-Clarke G (1990) Changes in microtubule-associated protein MAP1B phosphorylation during rat brain development. J Neurochem 55: 328–333. [DOI] [PubMed] [Google Scholar]
- 34. Froehner SC, Murnane AA, Tobler M, Peng HB, Sealock R (1987) A postsynaptic Mr 58,000 (58K) protein concentrated at acetylcholine receptor-rich sites in Torpedo electroplaques and skeletal muscle. J Cell Biol 104: 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sherman DL, Fabrizi C, Gillespie CS, Brophy PJ (2001) Specific disruption of a Schwann cell dystrophin-related protein complex in a demyelinating neuropathy. Neuron 30: 677–687. [DOI] [PubMed] [Google Scholar]
- 36. Menegoz M, Gaspar P, Le Bert M, Galvez T, Burgaya F, et al. (1997) Paranodin, a glycoprotein of neuronal paranodal membranes. Neuron 19: 319–331. [DOI] [PubMed] [Google Scholar]
- 37. Ackerl R, Walko G, Fuchs P, Fischer I, Schmuth M, et al. (2007) Conditional targeting of plectin in prenatal and adult mouse stratified epithelia causes keratinocyte fragility and lesional epidermal barrier defects. J Cell Sci 120: 2435–2443. [DOI] [PubMed] [Google Scholar]
- 38. Steinböck FA, Nikolic B, Coulombe PA, Fuchs E, Traub P, et al. (2000) Dose-dependent linkage, assembly inhibition and disassembly of vimentin and cytokeratin 5/14 filaments through plectin’s intermediate filament- binding domain. J Cell Sci 113: 483–491. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- 40. Stroissnigg H, Trančíková A, Descovich L, Fuhrmann J, Kutschera W, et al. (2007) S-nitrosylation of microtubule-associated protein 1B mediates nitric oxide induced axon retraction. Nat Cell Biol 9: 1035–1045. [DOI] [PubMed] [Google Scholar]
- 41. Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- 42. Longhurst DM, Watanabe M, Rothstein JD, Jackson M (2006) Interaction of PDZRhoGEF with microtubule-associated protein 1 light chains: link between microtubules, actin cytoskeleton, and neuronal polarity. J Biol Chem 281: 12030–12040. [DOI] [PubMed] [Google Scholar]
- 43. Iwata Y, Sampaolesi M, Shigekawa M, Wakabayashi S (2004) Syntrophin is an actin-binding protein the cellular localization of which is regulated through cytoskeletal reorganization in skeletal muscle cells. Eur J Cell Biol 83: 555–565. [DOI] [PubMed] [Google Scholar]
- 44. Albrecht DE, Sherman DL, Brophy PJ, Froehner SC (2008) The ABCA1 cholesterol transporter associates with one of two distinct dystrophin-based scaffolds in Schwann cells. Glia 56: 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Scherer SS, Xu T, Crino P, Arroyo EJ, Gutmann DH (2001) Ezrin, radixin, and moesin are components of Schwann cell microvilli. J Neurosci Res 65: 150–164. [DOI] [PubMed] [Google Scholar]
- 46. Einheber S, Zanazzi G, Ching W, Scherer S, Milner TA, et al. (1997) The axonal membrane protein Caspr, a homologue of neurexin IV, is a component of the septate-like paranodal junctions that assemble during myelination. J Cell Biol 139: 1495–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Seog DH (2004) Glutamate receptor-interacting protein 1 protein binds to the microtubule-associated protein. Biosci Biotechnol Biochem 68: 1808–1810. [DOI] [PubMed] [Google Scholar]
- 48. Hagiwara H, Yorifuji H, Sato-Yoshitake R, Hirokawa N (1994) Competition between motor molecules (kinesin and cytoplasmic dynein) and fibrous microtubule-associated proteins in binding to microtubules. J Biol Chem 269: 3581–3589. [PubMed] [Google Scholar]
- 49. Paschal BM, Obar RA, Vallee RB (1989) Interaction of brain cytoplasmic dynein and MAP2 with a common sequence at the C terminus of tubulin. Nature 342: 569–572. [DOI] [PubMed] [Google Scholar]
- 50. Trinczek B, Ebneth A, Mandelkow EM, Mandelkow E (1999) Tau regulates the attachment/detachment but not the speed of motors in microtubule-dependent transport of single vesicles and organelles. J Cell Sci 112 (Pt 14): 2355–2367. [DOI] [PubMed] [Google Scholar]
- 51. Dixit R, Ross JL, Goldman YE, Holzbaur EL (2008) Differential regulation of dynein and kinesin motor proteins by tau. Science 319: 1086–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Magnani E, Fan J, Gasparini L, Golding M, Williams M, et al. (2007) Interaction of tau protein with the dynactin complex. EMBO J 26: 4546–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vershinin M, Xu J, Razafsky DS, King SJ, Gross SP (2008) Tuning microtubule-based transport through filamentous MAPs: the problem of dynein. Traffic 9: 882–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ding J, Liu JJ, Kowal AS, Nardine T, Bhattacharya P, et al. (2002) Microtubule-associated protein 1B: a neuronal binding partner for gigaxonin. J Cell Biol 158: 427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ding J, Allen E, Wang W, Valle A, Wu C, et al. (2006) Gene targeting of GAN in mouse causes a toxic accumulation of microtubule-associated protein 8 and impaired retrograde axonal transport. Hum Mol Genet 15: 1451–1463. [DOI] [PubMed] [Google Scholar]
- 56. Hanley JG, Koulen P, Bedford F, Gordon-Weeks PR, Moss SJ (1999) The protein MAP-1B links GABAC receptors to the cytoskeleton at retinal synapses. Nature 397: 66–69. [DOI] [PubMed] [Google Scholar]
- 57. Park SM, Liu G, Kubal A, Fury M, Cao L, et al. (2004) Direct interaction between BKCa potassium channel and microtubule-associated protein 1A. FEBS Lett 570: 143–148. [DOI] [PubMed] [Google Scholar]
- 58. Maurer MH, Grunewald S, Gassler N, Rossner M, Propst F, et al. (2004) Cloning of a novel neuronally expressed orphan G-protein-coupled receptor which is up-regulated by erythropoietin, interacts with microtubule-associated protein 1b and colocalizes with the 5-hydroxytryptamine 2a receptor. J Neurochem 91: 1007–1017. [DOI] [PubMed] [Google Scholar]
- 59. Brenman JE, Topinka JR, Cooper EC, McGee AW, Rosen J, et al. (1998) Localization of postsynaptic density-93 to dendritic microtubules and interaction with microtubule-associated protein 1A. J Neurosci 18: 8805–8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ikeda A, Zheng QY, Zuberi AR, Johnson KR, Naggert JK, et al. (2002) Microtubule-associated protein 1A is a modifier of tubby hearing (moth1). Nat Genet 30: 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Magiera MM, Gupta M, Rundell CJ, Satish N, Ernens I, et al. (2004) Exchange protein directly activated by cAMP (EPAC) interacts with the light chain (LC) 2 of MAP1A. Biochem J 382: 803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wolff S, Xiao Z, Wittau M, Sussner N, Stoter M, et al. (2005) Interaction of casein kinase 1 delta (CK1delta) with the light chain LC2 of microtubule associated protein 1A (MAP1A). Biochim Biophys Acta 1745: 196–206. [DOI] [PubMed] [Google Scholar]
- 63. Dallol A, Agathanggelou A, Fenton SL, Ahmed-Choudhury J, Hesson L, et al. (2004) RASSF1A interacts with microtubule-associated proteins and modulates microtubule dynamics. Cancer Res 64: 4112–4116. [DOI] [PubMed] [Google Scholar]
- 64. Song MS, Chang JS, Song SJ, Yang TH, Lee H, et al. (2005) The centrosomal protein RABP1 regulates mitotic progression by recruiting RASSF1A to spindle poles. J Biol Chem 280: 3920–3927. [DOI] [PubMed] [Google Scholar]
- 65. Liu L, Amy V, Liu G, McKeehan WL (2002) Novel complex integrating mitochondria and the microtubular cytoskeleton with chromosome remodeling and tumor suppressor RASSF1 deduced by in silico homology analysis, interaction cloning in yeast, and colocalization in cultured cells. In Vitro Cell Dev Biol Anim 38: 582–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Montenegro-Venegas C, Tortosa E, Rosso S, Peretti D, Bollati F, et al. (2010) MAP1B regulates axonal development by modulating Rho-GTPase Rac1 activity. Mol Biol Cell 21: 3518–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Opal P, Garcia JJ, Propst F, Matilla A, Orr HT, et al. (2003) Mapmodulin/leucine-rich acidic nuclear protein binds the light chain of microtubule-associated protein 1B and modulates neuritogenesis. J Biol Chem 278: 34691–34699. [DOI] [PubMed] [Google Scholar]
- 68. Ives JH, Fung S, Tiwari P, Payne HL, Thompson CL (2004) Microtubule-associated protein light chain 2 is a stargazin-AMPA receptor complex-interacting protein in vivo. J Biol Chem 279: 31002–31009. [DOI] [PubMed] [Google Scholar]
- 69. Tretyakova I, Zolotukhin AS, Tan W, Bear J, Propst F, et al. (2005) NXF family protein participates in cytoplasmic mRNA trafficking. J Biol Chem 280: 31981–31990. [DOI] [PubMed] [Google Scholar]
- 70. Liu L, McKeehan WL (2002) Sequence analysis of LRPPRC and its SEC1 domain interaction partners suggests roles in cytoskeletal organization, vesicular trafficking, nucleocytosolic shuttling, and chromosome activity. Genomics 79: 124–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Langkopf A, Hammarback JA, Müller R, Vallee RB, Garner CC (1992) Microtubule-associated proteins 1A and LC2. J Biol Chem 267: 16561–16566. [PubMed] [Google Scholar]
- 72. Nakayama A, Odajima T, Murakami H, Mori N, Takahashi M (2001) Characterization of two promoters that regulate alternative transcripts in the microtubule-associated protein (MAP) 1A gene. Biochim Biophys Acta 1518: 260–266. [DOI] [PubMed] [Google Scholar]