Abstract
A variety of assays have been proposed to detect small quantities of nucleic acids at the point-of-care. One approach relies on target-induced aggregation of gold nanoparticles functionalized with oligonucleotide sequences complementary to adjacent regions on the targeted sequence. In the presence of the target sequence, the gold nanoparticles aggregate, producing an easily detectable shift in the optical scattering properties of the solution. The major limitations of this assay are that it requires heating, and that long incubation times are required to produce a result. This study aims to optimize the assay conditions and optical readout, with the goals of eliminating the need for heating and reducing the time to result without sacrificing sensitivity or dynamic range. By optimizing assay conditions and measuring the spectrum of scattered light at the endpoint of incubation, we find that the assay is capable of producing quantifiable results at room temperature in 30 minutes with a linear dynamic range spanning from 150 amoles to 15 fmoles of target. If changes in light scattering are measured dynamically during the incubation process, the linear range can be expanded 2-fold, spanning 50 amoles to 500 fmoles, while decreasing the time to result down to 10 minutes.
Keywords: Gold nanoparticles, oligonucleotides, optical scattering, quantification
Introduction
There is a growing interest in developing new nucleic acid based tests to detect and quantify infectious diseases at the point-of-care (POC) in low-resource settings.[1; 2] To be effective in these settings, tests should have a fast run-time, deliver unambiguous results, require minimal infrastructure, be extremely sensitive and specific to small quantities of the targeted nucleic acid sequence, and ideally quantify the amount of target present.[3] A number of approaches are being pursued to achieve these goals.[4; 5] These techniques often face two major challenges: the amplification of small quantities of target; and the detection and quantification of the amplified target.
Metallic nanoparticles conjugated to nucleic acid probe sequences have been identified as a promising method to address the challenge of target detection and quantification of very small quantities of nucleic acids due to their unique optical properties.[6; 7; 8; 9; 10] In this approach, two different DNA probe sequences (1 and 2) are conjugated to gold nanoparticles (AuNPs), which are complementary to adjacent regions of a target sequence (1'2'). When the target is added to a solution containing a mixture of the oligo-AuNP conjugates, the gold nanoparticles aggregate, causing a strong shift to the red in the scattering spectrum of the solution due to the plasmon resonance of the closely linked AuNPs (FIGURE 1).[11; 12; 13; 14; 15; 16; 17] This scattering shift can be easily visualized using a simple glass waveguide system which illuminates scattering by directing light through the plane of a standard microscope slide.[11; 15] The aggregation assay is specific to the targeted nucleic acid sequence, with an ability to detect single nucleotide polymorphisms, and produces results that are detectible by the naked eye.[18; 19] While the assay has been used with a variety of hybridization buffers, heating to 37°C during incubation, and a wide variation in incubation times (from 1 hour to 24 hours), it is not clear which conditions lead to an optimal limit of detection (LOD) or whether results can be read quantitatively across a meaningful dynamic range.[11; 12; 20; 21; 22] Previous studies investigating the dynamic range of other DNA-based AuNP aggregation assays have found them to be linear over a range of around 1–2 orders of magnitude of target concentration.[22; 23]
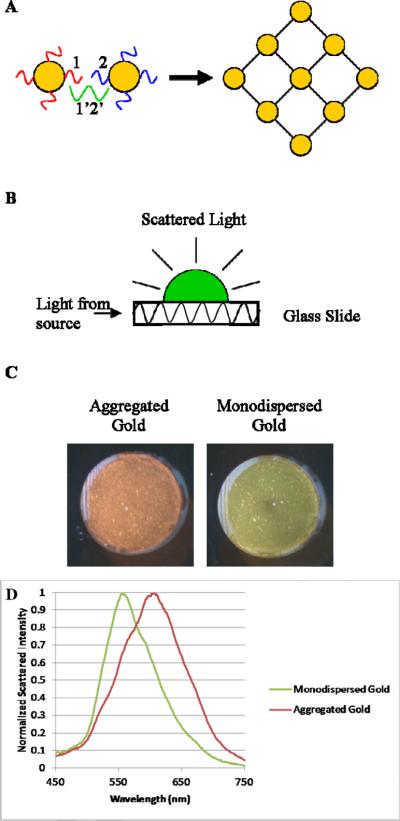
Figure 1.
A) A schematic showing the basic process of the aggregation reaction. Oligo-AuNP conjugates 1 and 2 are bound together into a large aggregated network by the target sequence 1'2'. B) A schematic of the principles of the side-illumination waveguide system used to illuminate the scattering of the samples. C) Photograph of representative samples on the side-illumination system, showing the visible red-shift of aggregated vs. monodispersed AuNP. D) This shift can also be observed in the spectroscopy of the samples.
This paper reports a series of studies designed to optimize the aggregation assay for decreased LOD, increased dynamic range, minimal run time, and to eliminate the need for external heating by varying reaction conditions known to affect metallic nanoparticle aggregation such as incubation temperature, incubation duration and salt content of the buffer.[21; 24; 25; 26] We compare the limit of detection and dynamic range of the assay using three different read out methods: photographic assessment of color, spectrophotometric measurement of light scattering at a single time point, and temporal variations in the spectrum of scattered light. We also present a new method for dynamic measurement of the aggregation process which allows for quantification of the targeted nucleic acids. We find that monitoring dynamic changes in lights scattering increases the quantifiable dynamic range of the assay by 2 orders of magnitude without compromising the limit of detection, as well as decreases the time before the assay returns a result by 3 fold.
Materials and Methods
Materials
DNA probe and target sequences were ordered from Integrated DNA Technologies (Coralville, IA), and resuspended in water at 100 μM. 50 nm diameter gold nanoparticles were obtained from Ted Pella (Redding, CA). Formamide, dextran sulfate, sodium chloride, DNase-free water, PBS, mineral oil, and magnesium chloride were all obtained from Sigma Aldrich (Saint Louis, MO). Quant-It Oligreen was obtained from Invitrogen (Grand Island, NY). Aldehyde coated glass slides were obtained from ArrayIT Corp. (Sunnyvale, CA).
Methods
Conjugating Gold Nanoparticles and Oligonucleotide Sequences
As an example of a clinically relevant target, we selected the eukaryotic parasite genus Plasmodium which causes malaria. There is a growing interest in developing new nucleic acid based diagnostics for malaria in order to target markers of resistance, and to increase the sensitivity and specificity over current diagnostic methods. Oligonucleotide probe sequences were designed with the NCBI's Basic Local Alignment Search Tools (BLAST) to be specific to the genus Plasmodium and to not cross react with human DNA. A poly-A tail and PEG spacer were used to increase the flexibility of the sequence. A thiol group was attached to the 5' end of each DNA probe to allow for conjugation with the AuNP. Sequences for the two probes and target were as follows:
Probe 1: 5' – ThioMC6 – A15 – PEG18 – CAT CAA AAG CTG ATA GGT CA – 3';
Probe 2: 5' – ThioMC6 – A15 – PEG18 – GAA ACT CGA TTG ATA CAC ACT A – 3';
Target (from Plasmodium 18S gene): 5' – TAG TGT GTA TCA ATC GAG TTT CTG ACC TAT CAG CTT TTG ATG – 3'.
Oligonucleotide sequences were attached to the gold particles using methods adapted from the literature.[12] In brief, 200 μL of 15 μM disulfide-oligonucleotide sequences were mixed with 5 μL of 1M tris(2-carboxyethyl)phosphine (TCEP) for 30 minutes at room temperature to reduce the disulfide bond. 800 μL of 75 pM 50 nm gold colloid was added and the mixture was allowed to sit overnight. Over the next 48 hours the salt concentration of the solution was raised, in steps of 0.1×, to 0.5× PBS to increase the loading of oligonucleotides onto the surface of the AuNP. To remove excess oligo sequences, the oligo-AuNP conjugates were then washed 3 times by centrifuging at 4000g for 10min and resuspending in 1× PBS. After each batch of probe conjugation, the transmission spectrum of the oligo-AuNP conjugates was measured using a Cary 50 spectrophotometer; a single extinction peak at 535 nm was taken as evidence that no aggregation occurred during the conjugation step. 50nm AuNP were chosen for this study because they have been shown to offer a compromise between a strong optical signature and stability of the colloids in solution.[11; 19] When characterizing probe creation, a series of experiments were carried out to determine the average number of oligonucleotide sequences on each gold particle. This was accomplished by mixing 1.3M β-mercaptoethanol with the AuNP-oligo probes overnight to release the DNA from the gold. The amount of DNA in the supernatant was then quantified using a commercial fluorescent oligonucleotide quantification system (Quant-It Oligreen). We found that on average 460 copies of Probe 1 and 530 copies of Probe 2 attached to each of their respective AuNPs.
Aggregation Assay
To test the performance of the assay, a set of target concentrations were prepared by serial dilution ranging from 50 pmoles to 15 amoles of target in steps of half a log. Target amounts are expressed as the total number of target DNA strands added to the final reaction mixture. In 5μL of target added to the final mixture, this works out to a concentration range of 10μM to 3pM. A hybridization buffer was prepared consisting of 20% formamide, 16% dextran sulfate, and 3.75 mM MgCl2. For each experiment, a working solution was created by mixing 60 μL of each probe and 80 μL of the hybridization buffer. 10 μL of this working solution was mixed with 5 μL of a target solution to give a final reaction mixture. The standard assay conditions reported most commonly in the literature were defined to be a 2 hour incubation in a 37°C waterbath using the hybridization buffer described above.[11; 18; 20; 21]
To optimize reaction conditions for decreased time, eliminating heating and maintaining or perhaps improving the LOD and dynamic range, we varied the assay runtime, temperature and salt concentration in the buffer. A master solution was prepared for each target concentration, and then split into aliquots for each condition. Temperature of the incubation step was varied using a programmable water bath, and the solutions were incubated for 2 hours with the hybridization buffer described above. The time of the incubation was varied by creating a master solution for each target concentration and then splitting it into separate aliquots for each time point. These aliquots were then all placed simultaneously in a 37°C waterbath and then removed after the chosen incubation time, from 10 minutes to 3 hours. To vary the salt concentration of the assay, different hybridization buffers were created with varying added volumes of a 5M stock of NaCl to achieve NaCl concentrations in the final solution of 0.3M, 0.6M and 1M. These samples were incubated for 2 hours at 37°C. The need for heating was investigated by running the aggregation assay with a 2 hour incubation at 37°C and a 2 hour room temperature incubation. Under each set of conditions we compared the LOD and contrast between positive (aggregated) and negative (monodispersed) samples by recording the color of the spots with photography and spectrophotometry.
Measurement
Assay results were quantified by spotting 1μL of the final reaction mixture onto an aldehyde-coated glass slide and capturing a digital photograph of scattered white light and by measuring the spectrum of scattered light across the visible spectrum. Illumination of the scattering spectrum for both measurements was accomplished by placing the slide with the samples on it into a side-illuminated illumination system consisting of a Edmund Optics MI-150 light source, fiber optic light spreader, and a custom built mount to couple the light into the edge of the slide, using it as a waveguide.[15] To record image data, a dissection microscope (Olympus SZ61) and Zeiss Axiovision MRc5 camera were used with the Zeiss Axiovision software with an exposure time of 1.25 sec for each image. To record spectra, an Ocean Optics USB 4000 portable spectrophotometer was used with a fiber optic probe (400μm core diameter) and data were recorded with an exposure time of 100 msec and a boxcar averaging width of 5. The fiber was clamped into a mount to keep it a uniform distance from the slide and normal to the surface of the slide during and between experiments. The fiber was positioned under the center of each spot for data collection. Preliminary experiments testing the repeatability of the system found <5% difference between repeated measurements of the same spots. Images and spectra were recorded immediately after spotting each sample on the slide. Spectral data was processed by subtracting the spectra of a solvent blank, and normalizing the subtracted spectra to a peak height of one. To measure light scattering images and spectra dynamically, a 1.5 μL drop of mineral oil was layered over the 1 μL spot of sample to retard drying and images and spectra were taken immediately after spotting and then every 2.5 minutes up to 10 minutes.
Results and Discussion
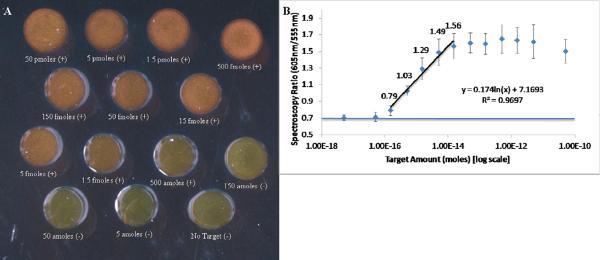
Using the standard conditions, the aggregation assay was run 5 times for a wide range of target concentrations. Using visual inspection of scattered light photographs to determine when the color of the spot changed from green (negative) to orange (positive), the limit of detection of the assay was between 150 amole and 500 amole of target in the final reaction mixture (FIGURE 2A). To quantify spectrophotometric data, the ratio of red to green scattered light at 605 nm to that at 555 nm was calculated; these wavelengths were selected because they correspond to the peak scattering wavelengths of monodispersed and aggregated oligo-AuNPs respectively. Spectroscopy ratios below 0.8 were considered to be negative (containing mostly monodispersed AuNP), while samples with spectroscopy ratios equal to or greater than this were considered positive (contains aggregated gold). The assay red to green ratio was linearly (R2>0.95) related to the log10 of the amount of target with a dynamic range spanning 150 amoles of target and 15 fmoles of target (FIGURE 2B). Together, these results suggest that the oligo-AuNP conjugation method and the aggregation process are repeatable, and that using the spectroscopy ratio of the two peak wavelengths is a reliable way of quantifying the amount of target.
Figure 2.
A) Photograph of one run of the aggregation assay using `standard' reaction conditions. Each sample is marked with whether it would be considered positive (aggregated) or negative (monodispersed) by visual inspection. B) Averaged peak spectroscopy ratios (n=5) as a function of target concentration. The spectroscopy ratio is the ration between the amount of scattered light recorded at 605nm and 555nm after each of these spectra have been normalized to their respective peak value. The spectroscopic ratios have been labeled for samples in the linear range of this assay. Note that the 150 amole sample can be determined to be positive by spectroscopy, but not by visual inspection. A horizontal line has been plotted on the graph to show the average red/green ratio of the no target control sample. Error bars represent 1 standard deviation.
We then aimed to optimize the reaction conditions of the assay (temperature, time and buffer composition) to improve its sensitivity and yield an assay more amenable for POC use. These conditions were chosen because of their well known effects on AuNP aggregation. During these optimization experiments we used three different target amounts: 50 fmoles target, which was found to consistently cause complete aggregation; 500 amoles, an intermediate amount which was found to consistently lead to partial aggregation of the oligo-AuNPs; and a negative control containing no target to show the effects of the varied conditions on un-aggregated samples.
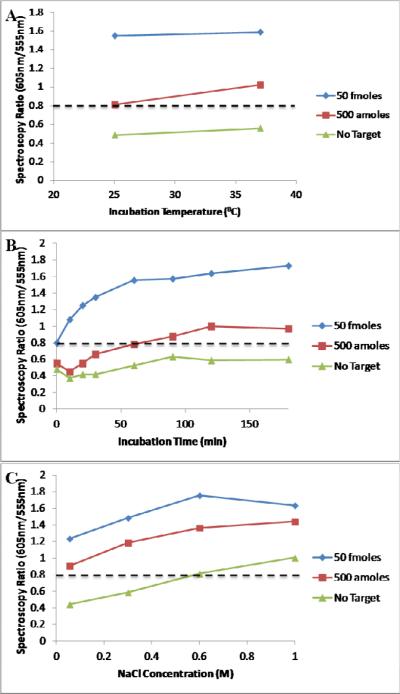
We first tested the effect of temperature to determine the effect of carrying out the aggregation assay at room temperature. Other studies have used room temperature incubation in this assay format but in this study we sought to specifically isolate its effect on the limit of detection of the assay.[18; 21] Increased temperature (a 45°C incubation) was also investigated and was found to cause aggregation in all samples including the no target control (data not shown). We hypothesize that this is caused by non-DNA mediated aggregation of the AuNP. At 37°C incubation the assay had a LOD of 500 amoles of target as determined by both spectroscopy and visual inspection (FIGURE 3A). Under room temperature incubation conditions, the color difference between aggregated and monodispersed AuNPs is too subtle for visual detection, but spectroscopy maintains a limit of detection of 500 amoles. By using spectroscopy it is possible to maintain a low limit of detection even when no heating apparatus is used.
Figure 3.
A) The effect of incubation temperature on the spectroscopy results of the aggregation assay. B) The effect of different incubation durations. C) The effect of the salt concentration of the reaction buffer. A dashed line has been drawn for the red/green ratio = 0.8 above which spectral data was considered positive.
We then tested the effect of incubation time on oligo-AuNP aggregation in order to determine the minimum aggregation time, since a short time to result is critical in POC settings. The aggregation assay begins producing results after a relatively short time frame (FIGURE 3B). The 50 fmole sample became positive by spectroscopy and visual inspection after 10 minutes. The 500 amole sample became positive between 20–30 minutes. Up to 180 minutes of incubation, the no target sample remained mostly monodispersed. These data suggest that aggregation can happen on a short timescale and that the assay can produce results at the full limit of detection in under an hour, compared to the 2 hours or longer often reported for this assay.
Finally, we changed the buffer composition, increasing the concentration of salt ions to favor the electrostatic interactions that drive oligonucleotide annealing and to decrease the surface charge repulsion experienced by the AuNPs.[21] Using 1M NaCl in the final mixture was found to cause aggregation in all target concentrations including the no target control (FIGURE 3C). The use of 0.3M and 0.6M NaCl was found to improve the contrast between positive (aggregated) and negative (monodispersed) samples as well as to improve the LOD over the standard salt concentration by making it easier to classify the 500 amole sample as positive by visual inspection.
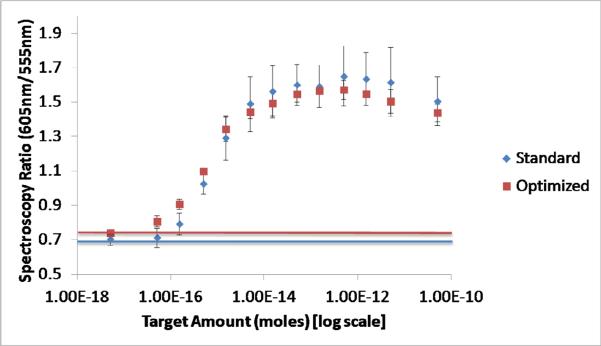
Combining the findings of the previous experiments, the optimal conditions for carrying out the aggregation assay were found to be a 30 minute incubation at room temperature, in a reaction solution containing 0.6M NaCl. A shortened time was used since the higher salt concentration was found increase the speed at which samples aggregated. These optimized conditions were tested 5 times on the same range of sample dilutions as the standard condition experiments (FIGURE 4). The spectroscopy ratio had a strong linear relationship with the log10 of the target concentration over a dynamic range from 50 amoles to 15 fmoles (R2 = 0.95). A non-targeted DNA sequence was also tested with the optimized conditions over a range of 500 fmoles to 5 amoles. The assay was found to be negative at all concentrations of the non-targeted sequence, demonstrating the selectivity of the process (data not shown).
Figure 4.
Comparison of the spectroscopy ratios of the `standard' and `optimized' reaction conditions as a function of the amount of target. Horizontal lines have been plotted on the graph to show the average red/green ratio of the no target control samples.
Compared to the standard conditions, the optimized conditions decreased the LOD, maintained a similar dynamic range and decreased the contrast between positive and negative samples. The increased spectroscopy ratio of the baseline is thought to be due to the increased salt in the reaction causing some non-DNA mediated aggregation. The decreased spectroscopy ratio of the high target samples is thought to be due to the decreased reaction time not allowing for as much aggregation to occur as in the standard assay. Both sets of conditions have similar linear ranges. Given 5μL of target input to the reaction, the optimized reaction has a limit of detection of 10pM. The optimized conditions maintain the performance and repeatability of the standard aggregation assay, while being much more useful as a POC diagnostic technique, since results can be obtained in a quarter of the time, and with no heating apparatus required.
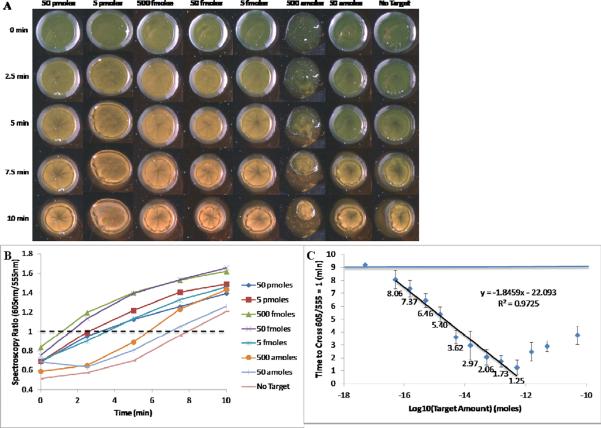
We hypothesized that monitoring dynamic changes in light scattering would offer faster results and a greater dynamic range. Samples spotted onto a microscope slide dry out after around 7 minutes, causing irreversible aggregation. Using a drop of mineral oil to cover the sample droplet on the surface of slide was found to delay this drying by up to 15 minutes. An aliquot of the reaction mixture was spotted on a slide immediately after preparation and covered with oil; spectra were acquired immediately after spotting and at 2.5, 5, 7.5 and 10 minutes after spotting (FIGURE 5A). The red to green ratio was plotted versus time and the time at which the ratio reached 1 was estimated using linear interpolation (FIGURE 5B). A variety of red/green ratio thresholds were investigated from 0.8 to 1.3 and a threshold of 1 was found to give the most reliable relationship between target concentration and the time to cross the threshold (data not shown). The experiment was carried out 5 times and the ratios at each concentration were averaged. There was found to be a strong linear region (R2 > 0.95) for target concentrations with a dynamic range of 50 amoles to 500 fmoles (FIGURE 5C). As samples dried on the slide, they were found to aggregate, including the no target controls which achieved a red/green ratio of 1 after 9 minutes. Although the no target controls aggregated, sample containing target were found to aggregate significantly faster, allowing for measurement of the amount of target in the solution. This offers an alternative measurement method for the aggregation assay, which not only decreases the time to a result from 30 minutes to 10 minutes, but also increases by two orders of magnitude the range over which the assay is linear. The assay conditions and results explored in this study are summarized in Table 1.
Figure 5.
A) Visual results of monitoring aggregation kinetics on the side-illumination system, using oil coverings to prevent sample drying. B) Spectroscopy ratios calculated as a function of time. A dashed line has been drawn at 605nm/555nm = 1 and the time each sample took to cross this threshold was calculated. C) The average of 5 samples of the kinetics experiment on the side-illumination system. Error bars represent 1 standard deviation. A horizontal line has been plotted to show the average red/green ratio of the no target control samples. A linear fit line showing the relationship between the log10 of the target amount and the time to cross is shown. The spectroscopic ratios have been labeled for samples in the linear range of this assay.
Table 1.
Summary of Key Experimental Conditions and Results
| PARAMETERS | Standard | Optimized | Oil-protected Dynamic Measurement |
|---|---|---|---|
| Time to Result | 2 hours | 30 min | 10 min |
| Temperature | 37°C | Room Temp. | Room Temp. |
| NaCl Concentration | 0.055M | 0.6M | 0.6M |
| RESULTS | |||
| Limit of Detection | 150 amole – 500 amole | 50 amole | 50 amoles |
| Linear Dynamic Range | 150 amoles to 15 fmoles | 50 amoles to 15 fmoles | 50 amoles to 500 fmoles |
In the dynamically measured spectroscopy data, we observed a marked decrease in the red to green ratios of samples containing greater than 500 fmoles of target. We believe that above this concentration the abundance of target sequences are saturating the recognition sequences on the AuNP-oligos, lowering the odds that a given target strand will bind to both recognition sequences in order to bring two AuNPs together. We believe that this effect is more apparent in the dynamically measured data because it operates over a shorter time frame and there is less of an opportunity for the diffusion limited aggregation reaction to run to completion at higher target concentrations. In order to resolve the target concentration of an unknown sample, the target could be diluted by powers of 10 to drop the concentration below the 500 fmole cutoff, which causes decreased time to aggregation of the assay. Using the example of malaria, which our DNA sequences are specific to, as a clinically relevant target we would expect this assay, with a limit of detection of 50 amoles in 5μL, to be able to detect around 6×106 DNA copies/μL. By using an isothermal amplification method suited for the POC, such as LAMP, which has been shown to yield 108-fold amplification, this assay could potentially detect below 0.5 parasites/μL the current limit of PCR diagnostics.[4; 27]
Conclusions
We demonstrated a method to use the AuNP aggregation assay developed by Mirkin and others to quantify the amount of an oligonucleotide target using the red/green ratio of the samples measured by spectroscopy.[12] We find that using conditions commonly found in the literature, the assay has a LOD of 150 amoles of target and a dynamic linear range of 150 amoles to 15 fmoles. We optimized the parameters of the assay and were able to decrease the runtime of the assay from 2 hours to 30 minutes, and eliminate the need for heating. This optimized reaction lowered the LOD to 50 amoles while maintaining a similar dynamic range to the standard assay. We also present a method for dynamic measurement of the aggregation which was found to greatly decrease the assay runtime to 10 minutes, maintain the 50 amole LOD of the optimized conditions, and double the dynamic range of the assay to cover 50 amoles to 500 fmoles of target. These findings suggest that by dynamic measurement of the aggregation assay it is possible to use it as a relatively simple tool to detect small quantities of nucleic acids and quantify them over a range of 4 logs.
Acknowledgements
The authors wish to thank Sarah Glazer for her assistance quantifying the amount of oligonucleotide bound to each gold nanoparticle. This work was supported in part by a grant from the Bill and Melinda Gates Foundation through the Grand Challenges Explorations Initiative. The work was also supported by Award Numbers U54AI057156 and R21AI087104 from the National Institute Of Allergy And Infectious Diseases. The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute Of Allergy And Infectious Diseases or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- [1].Girosi F, Olmsted SS, Keeler E, Hay Burgess DC, Lim YW, Aledort JE, Rafael ME, Ricci KA, Boer R, Hilborne L, Derose KP, Shea MV, Beighley CM, Dahl CA, Wasserman J. Developing and interpreting models to improve diagnostics in developing countries. Nature. 2006;444(Suppl 1):3–8. doi: 10.1038/nature05441. [DOI] [PubMed] [Google Scholar]
- [2].Urdea M, Penny LA, Olmsted SS, Giovanni MY, Kaspar P, Shepherd A, Wilson P, Dahl CA, Buchsbaum S, Moeller G, Hay Burgess DC. Requirements for high impact diagnostics in the developing world. Nature. 2006;444(Suppl 1):73–9. doi: 10.1038/nature05448. [DOI] [PubMed] [Google Scholar]
- [3].Peeling RW, Holmes KK, Mabey D, Ronald A. Rapid tests for sexually transmitted infections (STIs): the way forward. Sexually Transmitted Infections. 2006;82:v1–v6. doi: 10.1136/sti.2006.024265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mens PF, van Amerongen A, Sawa P, Kager PA, Schallig HDFH. Molecular diagnosis of malaria in the field: development of a novel 1-step nucleic acid lateral flow immunoassay for the detection of all 4 human Plasmodium spp. and its evaluation in Mbita, Kenya. Diagnostic Microbiology and Infectious Disease. 2008;61:421–427. doi: 10.1016/j.diagmicrobio.2008.03.009. [DOI] [PubMed] [Google Scholar]
- [6].Jain KK. Nanodiagnostics: application of nanotechnology in molecular diagnostics. Expert Review of Molecular Diagnostics. 2003;3:153–161. doi: 10.1586/14737159.3.2.153. [DOI] [PubMed] [Google Scholar]
- [7].Penn SG, He L, Natan MJ. Nanoparticles for bioanalysis. Current Opinion in Chemical Biology. 2003;7:609–615. doi: 10.1016/j.cbpa.2003.08.013. [DOI] [PubMed] [Google Scholar]
- [8].Jain KK. Nanotechnology in clinical laboratory diagnostics. Clinica Chimica Acta. 2005;358:37–54. doi: 10.1016/j.cccn.2005.03.014. [DOI] [PubMed] [Google Scholar]
- [9].Paciotti GF, Kingston DGI, Tamarkin L. Colloidal gold nanoparticles: a novel nanoparticle platform for developing multifunctional tumor-targeted drug delivery vectors. Drug Development Research. 2006;67:47–54. [Google Scholar]
- [10].Thaxton CS, Georganopoulou DG, Mirkin CA. Gold nanoparticle probes for the detection of nucleic acid targets. Clinica Chimica Acta. 2006;363:120–126. doi: 10.1016/j.cccn.2005.05.042. [DOI] [PubMed] [Google Scholar]
- [11].Storhoff JJ, Lucas AD, Garimella V, Bao YP, Muller UR. Homogeneous detection of unamplified genomic DNA sequences based on colorimetric scatter of gold nanoparticle probes. Nat Biotechnol. 2004;22:883–7. doi: 10.1038/nbt977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607–9. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- [13].Ghosh SK, Pal T. Interparticle Coupling Effect on the Surface Plasmon Resonance of Gold Nanoparticles: From Theory to Applications. Chemical Reviews. 2007;107:4797–4862. doi: 10.1021/cr0680282. [DOI] [PubMed] [Google Scholar]
- [14].Park SY, Stroud D. Theory of the optical properties of a DNA-modified gold nanoparticle system. Physica B: Condensed Matter. 2003;338:353–356. [Google Scholar]
- [15].Sonnichsen C, Geier S, Hecker NE, Plessen G.v., Feldmann J, Ditlbacher H, Lamprecht B, Krenn JR, Aussenegg FR, Chan VZH, Spatz JP, Moller M. Spectroscopy of single metallic nanoparticles using total internal reflection microscopy. Applied Physics Letters. 2000;77:2949–2951. [Google Scholar]
- [16].Storhoff JJ, Lazarides AA, Mucic RC, Mirkin CA, Letsinger RL, Schatz GC. What Controls the Optical Properties of DNA-Linked Gold Nanoparticle Assemblies? Journal of the American Chemical Society. 2000;122:4640–4650. [Google Scholar]
- [17].Lazarides AA, Kelly KL, Schatz GC, C. Northwestern Univ Evanston Il Dept Of, Effective Medium Theory of DNA-linked Gold Nanoparticle Aggregates: Effect of Aggregate Shape. Defense Technical Information Center; Ft. Belvoir: 2001. [Google Scholar]
- [18].Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. One-Pot Colorimetric Differentiation of Polynucleotides with Single Base Imperfections Using Gold Nanoparticle Probes. Journal of the American Chemical Society. 1998;120:1959–1964. [Google Scholar]
- [19].Reynolds RA, Mirkin CA, Letsinger RL. Homogeneous, Nanoparticle-Based Quantitative Colorimetric Detection of Oligonucleotides. Journal of the American Chemical Society. 2000;122:3795–3796. [Google Scholar]
- [20].Aslan K, Geddes CD. Microwave-accelerated ultrafast nanoparticle aggregation assays using gold colloids. Anal Chem. 2007;79:2131–6. doi: 10.1021/ac0620967. [DOI] [PubMed] [Google Scholar]
- [21].Jin R, Wu G, Li Z, Mirkin CA, Schatz GC. What controls the melting properties of DNA-linked gold nanoparticle assemblies? J Am Chem Soc. 2003;125:1643–54. doi: 10.1021/ja021096v. [DOI] [PubMed] [Google Scholar]
- [22].Sato K, Hosokawa K, Maeda M. Non-cross-linking gold nanoparticle aggregation as a detection method for single-base substitutions. Nucl. Acids Res. 2005;33:e4. doi: 10.1093/nar/gni007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhao W, Chiuman W, Brook MA, Li Y. Simple and Rapid Colorimetric Biosensors Based on DNA Aptamer and Noncrosslinking Gold Nanoparticle Aggregation. ChemBioChem. 2007;8:727–731. doi: 10.1002/cbic.200700014. [DOI] [PubMed] [Google Scholar]
- [24].Meakin P. Aggregation kinetics. Physica Scripta. 1992:295. [Google Scholar]
- [25].Lytton-Jean AK, Mirkin CA. A thermodynamic investigation into the binding properties of DNA functionalized gold nanoparticle probes and molecular fluorophore probes. J Am Chem Soc. 2005;127:12754–5. doi: 10.1021/ja052255o. [DOI] [PubMed] [Google Scholar]
- [26].Kim T, Lee CH, Joo SW, Lee K. Kinetics of gold nanoparticle aggregation: experiments and modeling. J Colloid Interface Sci. 2008;318:238–43. doi: 10.1016/j.jcis.2007.10.029. [DOI] [PubMed] [Google Scholar]
- [27].Paris DH, Imwong M, Faiz AM, Hasan M, Yunus EB, Silamut K, Lee SJ, Day NP, Dondorp AM. Loop-mediated isothermal PCR (LAMP) for the diagnosis of falciparum malaria. Am J Trop Med Hyg. 2007;77:972–6. [PubMed] [Google Scholar]