Abstract
Ductal carcinoma in situ (DCIS) is a non-obligate precursor of invasive breast cancer. The current recognition that DCIS lesions exhibit inter- and intra-lesion diversity suggests that the process of evolution to invasive breast cancer is more complex than previously recognized. Here we demonstrate the reproducible growth of primary DCIS cells derived from patient’s surgical and biopsy samples by the mouse intraductal (MIND) model. MIND involves injection of cells into the NOD-SCID IL2Rgammanull (NSG) mouse mammary ducts. Twelve (8 unique and 4 repeats) DCIS and 2 atypical hyperplasia specimens, heterogeneous with respect to biomarker expression and histology, were injected into 48 mouse mammary glands and analyzed for successful xenotransplantation. Overall, 14/34 and 11/14 of MIND xenotransplanted glands contained human DCIS and atypical hyperplastic cells, respectively, after 8 weeks, which formed single and multi-layered epithelium inside the ducts, and were heterogeneous with respect to expression of human cytokeratins, estrogen receptor α (ER), and HER2. ER protein expression was recapitulated in MIND xenografts at ratios similar to the corresponding patient biopsies. In both patient biopsies and corresponding MIND xenografts HER2 protein expression and nuclear HER2 gene over-expression was restricted to the DCIS lesions and were not found in the surrounding stroma or normal ducts. The xenografted DCIS lesions recapitulate the pathology and heterogeneity of human disease thus providing a powerful tool for the characterization of the distinct cellular and molecular basis of inter- and intra-tumoral heterogeneity and the processes of DCIS to early invasive breast cancer progression.
Keywords: Ductal carcinoma in situ, hyperplasia, xenograft, model
Introduction
Ductal carcinoma in situ (DCIS) is characterized by the presence of hyperplastic epithelial cells within the ducts of the breast, contained by an unperturbed myoepithelial cell layer and intact basement membrane [1–3]. DCIS, while nonlethal, is an immediate precursor of invasive breast cancers (IBC), making the characterization of DCIS of critical importance [4–6]. Currently, the ability to predict the clinical outcome, with respect to progression or recurrence, of women with DCIS based on tumor characteristics, such as size, grade, estrogen receptor α (ER) or HER2 status, is hampered by the ability of these biomarkers to predict DCIS or IBC recurrence [7]. Clinically, similar prognostic approaches are employed for DCIS and IBC, and DCIS is, in fact, frequently over-treated. Supporting this, autopsy studies have shown that up to 16% of asymptomatic women have DCIS [8]. Additionally, in patients who were misdiagnosed with benign breast disease, untreated, and subsequently revealed to have had DCIS, 43% on average developed invasive cancer [9].
In recent years, several studies have improved our current understanding of the molecular mechanism underlying DCIS and its relationship to IBC [10, 11]. In 2000, Perou et al. published a transcriptional profiling study that classified most breast cancers into one of four distinct subtypes, including two ER-positive (luminal A and B) and two ER-negative (HER2 and basal) [12]. More recently gene expression analyses have compared profiles of normal breast tissue, DCIS and IBC [13–17]. Independent proteomic, SAGE, and microarray analyses, while failing to identify a universal gene signature of DCIS, do support the idea that the transition to DCIS is a critical switch in the tumorigenesis pathway [14, 15, 18]. Ma et al. demonstrated that these alterations in gene expression are not isolated to the epithelial compartment, but are also taking place in the surrounding stroma, even before invasion occurs [19]. However, while dramatic alterations occur in gene expression during the transition from normal tissue to DCIS, the molecular profiles of DCIS and IBC of the same histological subtype are remarkably similar, but not identical. This suggests that there may be distinct pathways of progression for each of the four subtypes. Unlike IBC, however, DCIS lesions often display significant intratumor heterogeneity, and evidence suggests that higher intratumor diversity may be associated with increased risk of invasive progression [13, 20]. Recently, Kerlikowske et al. published perhaps the most promising study identifying a biomarker expression pattern of DCIS invasion potential using a model emphasizing expression of p16, COX-2, and Ki67, as opposed to histopathology [21]. This, and studies looking closely at the role of the microenvironment in malignant progression, are areas of research that warrant considerable study [22, 23].
Compared to IBC, the study of DCIS has been greatly inhibited by the lack of suitable experimental models. Existing models employ xenografts of DCIS cells placed outside of their normal microenvironment, which is not ideal for examining the molecular alternations that occur within the DCIS lesion and the surrounding microenvironment during invasion progression [24–28]. Further progress in understanding the molecular mechanisms underlying DCIS invasion potential requires better models of early breast cancer that represent the heterogeneity of human disease within the innate microenvironment, i.e. surrounded by the myoepithelial cells and basement membrane of the mammary duct.
Recently, we presented a novel xenograft model of DCIS: the mouse intraductal, or MIND, model which utilizes a unique intraductal transplantation approach to developing DCIS in immunocompromised mice [29]. This model offers a novel microenvironment (i.e. within the mammary ducts) for growth of human malignant lesions and is a suitable tool with which to study the molecular and biologic mechanisms of breast noninvasive-to-invasive progression. The aim of the current study was to demonstrate the feasibility of growing primary human DCIS utilizing MIND xenografts. The DCIS xenograft lines form single and multi-layered epithelial cells inside the mouse ducts thus recapitulating the human disease, histologically. Furthermore, the expression of biomarkers, ER and Her-2, and nuclear HER2 gene amplification are similar to the original patient’s biopsy samples. While we have not shown progression to invasion in any of the xenografts eight weeks following transplantation, there is the potential for this model to mimic human DCIS heterogeneity and that a fraction of DCIS lesions may progress to invasion with longer time follow up. If progression to IBC is validated with a longer time study, the model would provide a realistic tool for studying the sequence of early events in the epithelium and the stromal microenvironment that orchestrate the process of DCIS to malignant breast cancer progression. Furthermore, the hypothesis that DCIS lesions are unique and each follow a distinct evolutionary path to malignancy may now be tested.
Materials and Methods
Animals
Recipients were 8- to 10-week-old virgin female SCID-beige or NOD-SCID IL2Rgammanull (NSG) mice which were either bred or purchased from Jackson Laboratories. Animal and human experiments were conducted following protocols approved by the University of Kansas School of Medicine Animal Care and Use and Human Subjects Committee.
Specimen collection and transplantation
All patients gave written informed consent for participation in this research. Subject recruits included patients undergoing image-guided core needle biopsy or surgical excision due to suspicion of DCIS. In all cases, research specimens were obtained only after acquisition of diagnostic specimens. Following collection, biopsy tissue was placed in preservation media (LiforCell, Lifeblood Medical, Inc., Freehold, NJ) and stored at 4°C on ice until processing. Biopsy tissue was transferred to a Teflon block, finely minced with scalpels, and transferred to a 50 ml conical tube containing digestion media (per 100 mg of tissue: 5 mg collagenase (Roche Applied Science, Indianapolis, IN), 0.24 mg hyaluronidase (2140 units/mg; Sigma-Aldrich St. Louis, MO), 200 mg BSA, 100 μl Antibiotic-Antimycotic, 10 ml DMEM/F12) incubated 16 h at 50 rpm at 37°C. The specimens were removed, shaken briefly by hand and centrifuged 80 × g for 30 s. Pre-warmed Trpsin-EDTA (1ml; Stem Cell Technologies, Vancouver, BC) was added to the resulting pellet and gently pipetted up and down for 1 min. Cold HF (10 ml; HBSS, 2% FBS) was added and specimens were centrifuged at 350 × g for 5 min. The supernatant was removed and 2 ml pre-warmed 5 mg/ml Dispase (Stem Cell Technologies) and 200 μl of 1 mg/ml DNase I (Stem Cell Technologies) were added to the pellet. To resuspend the pellet the specimen was pipetted up and down for 1 min. An additional 10 ml cold HF was added to the cell suspension, which was filtered through a 40 μm cell strainer. The cell suspension was centrifuged at 350 × g for 5 min and the resulting cell pellet was resuspended in 200 μl PBS and counted. A 30-gauge Hamilton syringe, 50-μl capacity, with a blunt-ended 1/2-inch needle was used to deliver the cells as previously described [29]. Two μl of PBS (with 0.04% trypan blue) containing 25,000–35,000 cells were injected. A slow-releasing pellet containing 50μg estradiol and 20 mg progesterone was placed subcutaneously. After eight weeks, mice were sacrificed and mammary tissue was processed for embedding as previously described [29].
Immunofluorescent (IF) staining
IF was performed after tissue deparaffinization by clearance in xylene and hydration through graded ethanol series. Microwave antigen retrieval in Tris-HCl buffer (100 mM Tris-HCl, 0.1% Tween-20) was performed for 20 min. Washes were performed in IF buffer (130 mM NaCl, 7 mM Na2HPO4, 3.5 mM NaH2PO4, 7.7 mM NaN3, 0.1% bovine albumin, 0.2% Triton X-100, 0.5% Tween-20). Antibodies and dilutions are listed in supplementary table 1. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA. Confocal microscopy was performed by using a laser-scanning confocal microscope (model 510; Carl Zeiss MicroImaging, Inc., Thornwood, NY). The acquisition software used was LSM image browser (Carl Zeiss MicroImaging, Inc.).
Fluorescent in Situ Hybridization (FISH)
Mouse and human DNA probes were prepared using Cot-1 DNA (Invitrogen, Carlsbad, CA) and the FISH Tag DNA Kit per protocol (Molecular Probes). The labeled DNA probes were resuspended in Tris-EDTA buffer at a concentration of 4 ng/μl. Hybridization and washes were carried out as previously described [29]. Slides were coverslipped with mounting media containing DAPI, and imaged as described above.
HER2 ImmunoFISH
HER2 FISH pharmDx™ Kit (Dako) staining was performed according to manufacturer’s recommendation with the following modifications. Following final wash in Wash Buffer (Dako), tissue sections were covered with Blocking Reagent (MOM Kit, Vector Laboratories) diluted in IF buffer for 60 min. Tissue sections were incubated for 5 min in MOM Diluent Working Solution (MOM Kit, Vector Laboratories) before being covered in primary antibodies (anti-HER2 and anti-human CK 19) diluted in MOM Diluent. Sections were incubated overnight with primary antibodies at room temperature. Slides were washed 3 times for 10 min each in IF buffer. Dilute fluorescently conjugated secondary antibodies (supplementary table 1) in MOM Diluent working solution were added to each section and incubated 1 h at room temperature. Slides were washed 3 times for 10 min in IF buffer, counterstained with DAPI (Vector laboratories) and coverslipped.
Results
Patient and biopsy characteristics
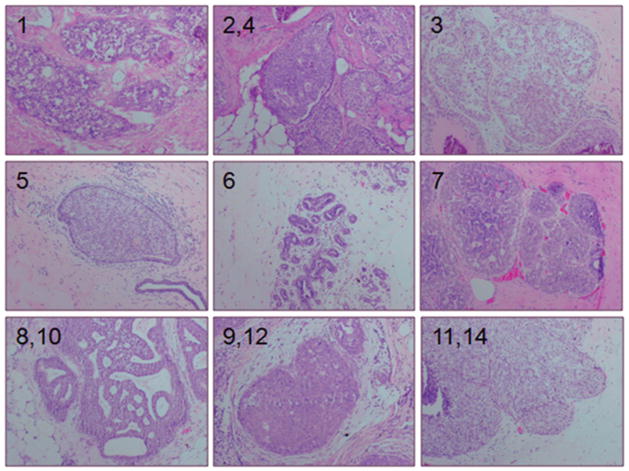
The 12 DCIS biopsies came from 8 unique patients, with 4 having repeated procedures at later dates. Patients with biopsies containing hyperplasia and DCIS were younger compared to patients with normal biopsies (44 and 48 vs. 58, respectively; p≤ 0.05). Of the 36 biopsies received, 22 were considered normal, 2 had atypical hyperplasia and 12 had DCIS, based on the pathology report of adjacent biopsies (table 1). The DCIS biopsies weighed more than the normal biopsies (464 vs. 184 mg; p< 0.05) and yielded significantly more cells compared to normal biopsies (1,385,015 vs. 14,218; p≤ 0.05) (table 1). The biopsy samples containing DCIS were heterogeneous with respect to ER, PR, Ki67, HER2 and histological grade (table 2 and figure 1). Seven of these were ER+ and PR+, including one specimen from a patient with a BRCA2 mutation (Case 5). Two biopsies (Cases 2 and 4), taken from the same patient at different dates, were ER+, PR+, and HER2+. One biopsy was only ER+, and two were only HER2+. No biopsies collected were negative for all three of these biomarkers, ER, PR, and HER2. All but one DCIS biopsy had nuclear grades of 2 or 3. While 10/12 of the DCIS biopsies contained more than one histological pattern, a testament to the heterogeneous nature of DCIS, 7/12 exhibited at least some area of cells with a comedo pattern of growth.
Table 1.
Characteristics of patient biopsies. Data is presented as mean ± standard error. Numbers without similar letters differ (p≤ 0.05)
| Pathology | Normal | Hyperplasia | DCIS |
|---|---|---|---|
| No. of specimens | 22 | 2 | 12 |
| Age of patient | 57 ± 2a | 44 ± 1b | 47 ± 3b |
| Weight of specimens (mg) | 184± 27a | 279 ± 180ab | 464 ± 135b |
| No. of cells recovered | 14,218 ± 4788a | 1,768,000 ± 1,432,000ab | 1,385,015 ± 1,432,000b |
Table 2.
Summary of MIND xenotransplantation cases. Cases with the same letter were collected from the same subject at different dates
| Case no. | Primary Biopsy Pathology | Wt. of specimen (mg) | Recovered cells/mg tissue | Strain of mouse | No. of glands with growth | ||
|---|---|---|---|---|---|---|---|
| Characteristics | Classification | Histology | |||||
| 1 | DCIS: 100% ER+ 80% PR+ 68% Ki-67 |
Nuclear grade 2 &3 | Solid and cribriform | 205 | 1159 | SCID-beige | 0/5 |
| 2a | DCIS: 10% ER+ 2% PR+ 3+ HER2+ 18% Ki-67 |
High grade; nuclear grade 3 | Comedo, solid, and cribriform | 188 | 1914 | NSG | 3/3 |
| 3 | DCIS: 88% ER+ 5% PR+ 10% Ki-67 |
Intermediate grade; nuclear grade 2 | Solid and cribriform | 225 | 495 | NSG | 1/2 |
| 4a | DCIS: 10% ER+ 2% PR+ 3+ HER2+ 18% Ki-67 |
High grade; nuclear grade 3 | Comedo | 152 | 503 | NSG | 2/2 |
| 5 | DCIS: 98% ER+ 95% PR+ BRCA2 mutation 2% Ki-67 |
High grade; nuclear grade 3 | Cribriform, comedo and solid | 1740 | 57 | NSG | 0/2 |
| 6 | Hyperplasia | NA | NA | 100 | 3360 | NSG | 7/10 |
| 7 | DCIS: 98% ER+ | Nuclear grade 1 | Solid | 210 | 809 | NSG | 2/6 |
| 8b | DCIS: 99% ER+ 100% PR+ 5% Ki-67 |
Low grade; nuclear grade 2 | Micropapillary and cribriform | 259 | 4517 | NSG | 1/4 |
| 9c | DCIS: 3+ HER 2 25% Ki-67 |
High grade; nuclear grade 3 | Comedo, Micropapillary and cribriform | 245 | 345 | NSG | 0/2 |
| 10b | DCIS: 99% ER+ 100% PR+ 5% Ki-67 |
Low grade; nuclear grade 2 | Micropapillary and cribriform | 790 | 14528 | NSG | 1/2 |
| 11d | DCIS: 100% ER+ 40% PR+ 5% Ki-67 |
High grade; nuclear grade 3 | Comedo, cribriform and solid | 770 | 2078 | NSG | 0/2 |
| 12c | DCIS: 3+ HER2 25% Ki-67 |
High grade; nuclear grade 3 | Comedo, micropapillary and cribriform | 170 | 1941 | NSG | 2/2 |
| 13 | Hyperplasia | NA | NA | 459 | 6972 | NSG | 4/4 |
| 14d | DCIS: 100% ER+ 40% PR+ 5% Ki-67 |
High grade; nuclear grade 3 | Comedo, cribriform and solid | 617 | 3204 | NSG | 2/2 |
Figure 1.
H&E staining of patient biopsies of DCIS. Numbers indicate the case designation. Cases 2 and 4, 8 and 10, 9 and 12, and 11 and 14 each were from the same patients collected on different procedure dates.
Intraductal transplantation of primary human specimens
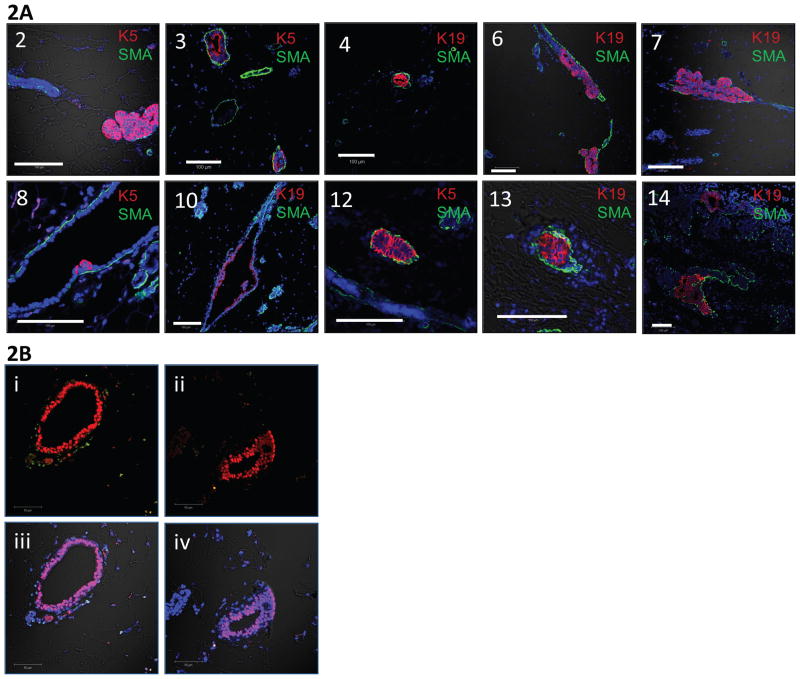
MIND xenotransplanted glands were collected after eight weeks and processed for assessment of successful transplantation. Overall, 14/34 (cases 1–5, 7–12, and 14) and 11/14 (cases 6 and 13) mouse mammary glands contained human DCIS and hyperplastic cells, respectively, within the mammary ducts at eight weeks post transplantation, as shown by IF staining for human cytokeratin 5 or 19 and smooth muscle actin (table 2 and figure 2). None of the five glands from case 1, which were transplanted into SCID-beige mice, showed evidence of growth (tables 2 and 3). When only MIND transplants that utilized NSG mice are considered the ratio of successful growth of human DCIS increases to 14/29 (tables 2 and 3). The other three cases of DCIS which did not grow tended to yield a lower number of cells following digestion of the corresponding biopsy compared to the DCIS cases which did grow (827 vs. 3212 cells/mg of tissue, respectively), including the case from the patient with a BRCA2 mutation (table 3). Human cells formed single and multi-layered epithelium inside the ducts and were heterogeneous with respect to the expression of human specific cytokeratins 5 and 19 (figure 2A). The flattened myoepithelial cell layer, which contains smooth muscle action, appears to be comprised of mouse cells, as evidenced by the lack of human cytokeratin staining and FISH staining (figures 2A and B). The specificity of the human specific cytokeratins is evident by the presence of unstained mouse mammary duct, as seen in each case.
Figure 2.
Staining techniques were used to confirm the presence of primary human cells within the transplanted mouse mammary glands. 2A: Numbers correspond to the case numbers which successfully transplanted. Representative immunofluorescent staining of human CK 5 (K5) or 19 (K19), smooth muscle actin (SMA), and DNA. K5 and K19 are conjugated to Alexa-fluor 594, shown in red, and SMA is conjugated to Alexa-fluor 488, shown in green. Nuclei are counterstained with DAPI. 2B: Representation fluorescent in situ hybridization (FISH) analysis of a MIND xenograft using probes labeled for mouse DNA, human DNA, and DAPI. The human DNA specific probe was conjugated to Alexa-fluor 594, shown in red, and the mouse DNA specific probe was conjugated to Alexa-fluor 488, shown in green. Images Bi-ii are with only the 488 and 594 signal on, while images Biii-iv additionally have the DAPI and DIC signals on.
Table 3.
Summary of successful MIND xenotransplantation characteristics. Cases are grouped based on the success of engraftment and strain of mouse used for transplantation. Data is presented as mean ± standard error
| Case no. | 2,3,4,7,8,10,12,14 | 5,9,11 | 1 | 6,13 |
|---|---|---|---|---|
| Pathology | DCIS | DCIS | DCIS | Hyperplasia |
| No. of cases | 8/11 (73%) | 3/11 (25%) | 0/1 (0%) | 2/2 (100%) |
| No. recovered cells/mg tissue | 3489± 1653 | 827±631 | 1159 | 5166±1806 |
| Mouse Strain | NSG | NSG | SCID-Beige | NSG |
| Growth | Yes | No | No | Yes |
Biomarker expression
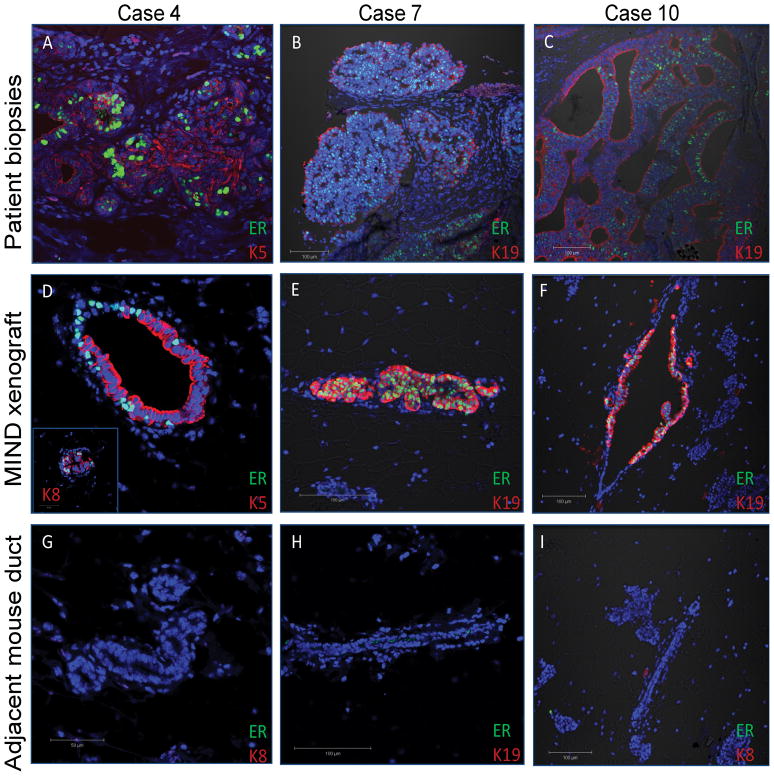
The MIND xenografts recapitulated the patient’s original DCIS as evidenced by IF staining and immunoFISH for the biomarkers ER and HER2 in the primary human biopsies and MIND xenografts (figures 3 and 4). Overall, 3/7 ER+ and 1/3 HER2+ cases which did successfully transplant had expression of these biomarkers in the MIND transplanted mammary glands. For Case 4, in which 10% of cells in the original patient biopsy were ER+ (figure 3A), expression was found in the MIND xenograft in approximately 20% of the human cytokeratin expressing cells (figure 3A,D,G). ER expressing cells in this xenograft did not express human cytokeratin 5, as many of the transplanted cells did, but did co-stain with human cytokeratin 8 (figure 3D, inset image). In xenografts from cases 7 and 10, of which 98% and 99% of cells were ER+ in the patient biopsies (figure 3B–C), nearly 100% of transplanted cells retained ER expression (figure 3E–F). These cells also co-stained with human cytokeratin 19. Normal mouse ducts that did not contain human cells within each of the xenotransplanted glands did not stain for ER, demonstrating the specificity of the antibody for human ER (figure 3G–I).
Figure 3.
Immunofluorescent staining of human ER, human cytokeratin 5 or 19, and DNA in patient biopsies (A–C) and MIND xenograft transplanted with DCIS cells (D–I) from the corresponding patient after 8 weeks. Human K5 and K19 were conjugated to Alexa-fluor 595, shown in red, and ER and HER2 were conjugated to Alexa-fluor 488, shown in green. Nuclei are counterstained with DAPI. G–I are mouse mammary ducts from the transplanted gland which did not stain with the human specific antibodies demonstrating the human specificity of the cytokeratin antibodies.
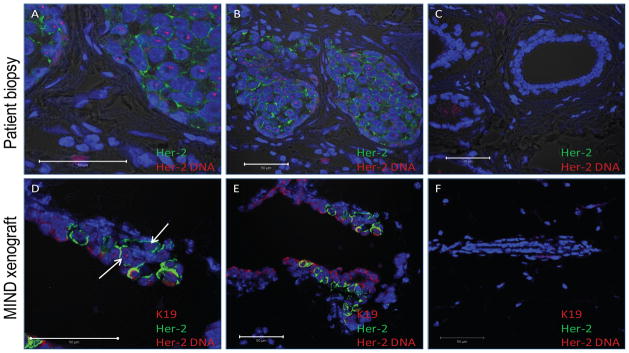
Figure 4.
ImmunoFISH staining of human HER-2 (FISH and immuno) and human cytokeratin 19 (IF), and DNA in the patient biopsy (A–C) and MIND xenograft transplanted with DCIS cells (D–F) from Case 4 after 8 weeks. All images are from Case 4 with high magnification in panels A and D. Human K19 was conjugated to Alexa-fluor 595, shown in red, and HER-2 was conjugated to Alexa-fluor 488, shown in green. The Texas Red-labelled DNA probe covering a 218 kb region of human chromosome 17 including HER2 appears as red (indicated by white arrows) in the nuclei of cells with amplified HER2 gene. Nuclei are counterstained with DAPI. Shown in panel C is a normal duct within the same tissue section as panel A and B, and shown in panel F is a mouse mammary duct from the same tissue section as panel D and E, neither of which stained with the human specific antibodies or the FISH HER2 probe, demonstrating the human specificity of the antibodies and probe.
MIND xenografts of the HER2+ case 4 exhibited cell membrane staining of HER2+ and HER2 gene amplification in approximately 25% of engrafted human cells. In the patient biopsy sections both HER2 protein expression on the cell membrane and nuclear HER2 gene over-expression within the nuclei is restricted to the DCIS lesions and is not found in the surrounding stroma (figures 4A–B) or normal ducts within the same tissue section (figure 4C). Similarly, in the MIND xenograft the HER2+ cells, which also co-stained for cytoplasmic human cytokeratin 19, were not found in normal mouse ducts within the transplanted gland. The HER2 FISH data on the xenografted DCIS case 4 is a strong indication that the cells were truly DCIS. Genomic DNA analysis (by Affymetrix’s Oncoscan) is currently being performed to confirm that the remaining non-HER2 positive cells are also truly DCIS and not hyperplastic or normal human cells.
Discussion
We have demonstrated the creation of a novel model of DCIS that closely mimics the human disease. Our results indicate that these human DCIS cells are capable of growing within the mouse mammary ducts and retain the pattern of biomarker expression observed in original human biopsy. Immunofluorescent staining with antibody specific to human cytokeratins and FISH analysis indicates the presence of human DCIS cells within the mammary ducts. Our success in view of past failed attempts to transplant DCIS cells was likely due to several different factors, including the strain of the recipient mice and the method of isolation of the primary human cells. The use of the NSG mice appears to be essential for successful MIND xenotransplantation of primary DCIS cells. Both in our hands and others (personal correspondence, DM) use of other immunocompromised mice, including SCID-beige and NOD-SCID has not provided a suitable microenvironment for the growth of these cells. The NSG mice have targeted mutations in the common cytokine-receptor interleukin-2 receptor (IL-2R), which appears essential for the development of a permissive environment for engraftment of primary human DCIS cells. The NSG mice have also successfully been used to develop an assay for quantification of the mammary stem cell population [30]. However, to our knowledge, we are the first to engraft human cells directly into the mammary ducts of NSG mice.
In the past, human DCIS research has been limited due to the absence of a suitable model system to study morphological and biological qualities of the DCIS lesions. Previous DCIS studies have utilized a method in which DCIS cell lines or tissue fragments are implanted subcutaneously or into cleared mammary fat pad in nude mice, and thus do not effectively mimic the microenvironment [25–27, 31, 32]. In the current study, primary human DCIS cells were injected through the nipple into the network of mammary ducts, based on the concept that DCIS initiates inside the ducts and therefore this will provide a natural microenvironment for DCIS cell growth. Thus, the MIND model mimics human DCIS in the mouse to the closest condition that it is found in vivo and has the potential to allow examination of processes involving the progression of human breast cancers from the initial growth as carcinoma in situ to invasion through the myoepithelial cell layer and basement membrane.
None of the MIND xenografts of primary human DCIS in this study appeared to become invasive after 8 weeks, a time point that was chosen based on our previous experience with immortal DCIS cell line transplantations to allow enough time for detectable engraftment [29]. Future studies will allow growth to continue over an extended time course up to 1 year, and we predict that at least a subset of the MIND xenografts will become invasive over time. Additionally, using the methods established by Farnie et al [33], mammosphere culture will be used to enrich for the stem cell population prior to intraductal xenotransplantation, allowing the characterization of these cell and their role in DCIS progression.
The potential of the MIND model for facilitating the elucidation of the mechanisms underlying DCIS progression is tremendous. While there is obvious clinical and genetic heterogeneity among patients with respect to risk for malignant progression of DCIS lesions, as well as response to treatment, currently we are unable to identify the subset of premalignant lesions that will progress to invasion [34, 35]. Therefore currently all patients diagnosed with DCIS are treated similarly. If left untreated, it is predicted that only a small subset of patients with premalignant changes will evolve to invasive breast cancer [34–36]. The long-term goal of our research is to develop xenograft lines which can be used to characterize and classify various subtypes of DCIS by immunoassay, gene profiling, expression of markers of stem and progenitor cells, histopathology, and invasive potential. Additionally, the role of the microenvironment and factors influence cell growth and apoptosis can be examined. The molecular and cellular characterization of DCIS subtypes, as the temporal events leading to invasive progression unique to an individual patient’s tumor type, may offer new tailored preventive strategies and spare many patients from the development of invasive and metastatic breast.
Supplementary Material
Acknowledgments
We thank Drs. Carol Conner and Marilee McGinness, University of Kansas Medical Center, and the University of Kansas Medical Center Biospecimen Shared Resources for tissue collection, Dr. Arindam Paul and Yan Hong, University of Kansas Medical Center, for technical assistance, and Dr. Mike Lewis, Baylor College of Medicine, for valuable guidance. This work was supported by NIH 5 R00 CA127462 to FB.
List of abbreviations
- DCIS
Ductal carcinoma in situ
- MIND
Mouse intraductal
- NSG
NOD-SCID IL2Rgammanull
- ER
Estrogen receptor α
- PR
Progesterone receptor
- IBC
Invasive breast cancer
- SAGE
Serial analysis of gene expression
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KV participated in the design of the study, carried out the processing of primary biopsies, animal surgeries, immunofluorescent staining, FISH analysis, imaging, and drafted the manuscript. FF performed the histopathological analysis of the human biopsies. WS was responsible for identifying potential study subjects. DM and CA participated in the conception and design of the study. FB conceived of the study, and participated in its design and coordination. All authors read and approved the final manuscript.
List of online supporting
Supplementary table 1. List of antibodies, suppliers, and dilutions used.
Contributor Information
Kelli Elizabeth Valdez, Email: kvaldez@kumc.edu.
Fan Fang, Email: ffan@kumc.edu.
William Smith, Email: wsmith2@kumc.edu.
D. Craig Allred, Email: dcallred@path.wustl.edu.
Daniel Medina, Email: dmedina@bcm.edu.
References
- 1.Boeker W. Preneoplasia of the Breast. Elservier GmbH; Munich: 2006. [Google Scholar]
- 2.Gudjonsson T, Adriance MC, Sternlicht MD, et al. Myoepithelial cells: their origin and function in breast morphogenesis and neoplasia. J Mammary Gland Biol Neoplasia. 2005;10:261–272. doi: 10.1007/s10911-005-9586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tavassoli F. In: Tumors of the Breast and Female Genital Organs. Tavassoli FaD P, editor. IARC-Press; Lyon: 2003. pp. 63–73. [Google Scholar]
- 4.Allred C. Biological characteristics of ductal carcinoma in situ. In: Silverstein MJ, editor. Ductal Carcinoma In Situ of the Breast. 2. Lipincott, Williams, and Wilkins; Philadelphia: 2002. pp. 37–48. [Google Scholar]
- 5.Allred C. Biological features of human premalignant breast disease and the progression to cancer. In: Harris JRLM, Morrow M, Obsorne CK, editors. Diseases of the Breast. 4. Lippincott, Williams, and Wilkins; Philidelphia: 2009. pp. 323–334. [Google Scholar]
- 6.Wellings SR, Jensen HM. On the origin and progression of ductal carcinoma in the human breast. J Natl Cancer Inst. 1973;50:1111–1118. doi: 10.1093/jnci/50.5.1111. [DOI] [PubMed] [Google Scholar]
- 7.Shamliyan T, Wang SY, Virnig BA, et al. Association between patient and tumor characteristics with clinical outcomes in women with ductal carcinoma in situ. J Natl Cancer Inst Monogr. 2010:121–129. doi: 10.1093/jncimonographs/lgq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen M, Jensen J, Andersen J. Precancerous and cancerous breast lesions during lifetime and at autopsy. A study of 83 women. Cancer. 1984;54:612–615. doi: 10.1002/1097-0142(1984)54:4<612::aid-cncr2820540403>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 9.Leonard GD, Swain SM. Ductal carcinoma in situ, complexities and challenges. J Natl Cancer Inst. 2004;96:906–920. doi: 10.1093/jnci/djh164. [DOI] [PubMed] [Google Scholar]
- 10.Polyak K. Molecular markers for the diagnosis and management of ductal carcinoma in situ. J Natl Cancer Inst Monogr. 2010:210–213. doi: 10.1093/jncimonographs/lgq019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bombonati A, Sgroi DC. The molecular pathology of breast cancer progression. J Pathol. 223:307–317. doi: 10.1002/path.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 13.Allred DC, Wu Y, Mao S, et al. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution. Clin Cancer Res. 2008;14:370–378. doi: 10.1158/1078-0432.CCR-07-1127. [DOI] [PubMed] [Google Scholar]
- 14.Ma XJ, Salunga R, Tuggle JT, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci U S A. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porter D, Lahti-Domenici J, Keshaviah A, et al. Molecular markers in ductal carcinoma in situ of the breast. Mol Cancer Res. 2003;1:362–375. [PubMed] [Google Scholar]
- 16.Porter DA, Krop IE, Nasser S, et al. A SAGE (serial analysis of gene expression) view of breast tumor progression. Cancer Res. 2001;61:5697–5702. [PubMed] [Google Scholar]
- 17.Vincent-Salomon A, Lucchesi C, Gruel N, et al. Integrated genomic and transcriptomic analysis of ductal carcinoma in situ of the breast. Clin Cancer Res. 2008;14:1956–1965. doi: 10.1158/1078-0432.CCR-07-1465. [DOI] [PubMed] [Google Scholar]
- 18.Wulfkuhle JD, Sgroi DC, Krutzsch H, et al. Proteomics of human breast ductal carcinoma in situ. Cancer Res. 2002;62:6740–6749. [PubMed] [Google Scholar]
- 19.Ma XJ, Dahiya S, Richardson E, et al. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 2009;11:R7. doi: 10.1186/bcr2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park SY, Gonen M, Kim HJ, et al. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest. 120:636–644. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerlikowske K, Molinaro AM, Gauthier ML, et al. Biomarker expression and risk of subsequent tumors after initial ductal carcinoma in situ diagnosis. J Natl Cancer Inst. 102:627–637. doi: 10.1093/jnci/djq101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Espina V, Liotta LA. What is the malignant nature of human ductal carcinoma in situ? Nat Rev Cancer. 11:68–75. doi: 10.1038/nrc2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCaskill-Stevens W. National Institutes of Health State-of-the-Science Conference on the Management and Diagnosis of Ductal Carcinoma in Situ: a call to action. J Natl Cancer Inst Monogr. 2010:111–112. doi: 10.1093/jncimonographs/lgq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boland GP, Knox WF, Bundred NJ. Molecular markers and therapeutic targets in ductal carcinoma in situ. Microsc Res Tech. 2002;59:3–11. doi: 10.1002/jemt.10172. [DOI] [PubMed] [Google Scholar]
- 25.Hu M, Yao J, Carroll DK, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 2008;13:394–406. doi: 10.1016/j.ccr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller FR. Xenograft models of premalignant breast disease. J Mammary Gland Biol Neoplasia. 2000;5:379–391. doi: 10.1023/a:1009577811584. [DOI] [PubMed] [Google Scholar]
- 27.Miller FR, Santner SJ, Tait L, et al. MCF10DCIS. com xenograft model of human comedo ductal carcinoma in situ. J Natl Cancer Inst. 2000;92:1185–1186. doi: 10.1093/jnci/92.14.1185a. [DOI] [PubMed] [Google Scholar]
- 28.Tait LR, Pauley RJ, Santner SJ, et al. Dynamic stromal-epithelial interactions during progression of MCF10DCIS. com xenografts. Int J Cancer. 2007;120:2127–2134. doi: 10.1002/ijc.22572. [DOI] [PubMed] [Google Scholar]
- 29.Behbod F, Kittrell FS, LaMarca H, et al. An intraductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast Cancer Res. 2009;11:R66. doi: 10.1186/bcr2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eirew P, Stingl J, Raouf A, et al. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat Med. 2008;14:1384–1389. doi: 10.1038/nm.1791. [DOI] [PubMed] [Google Scholar]
- 31.Hu M, Peluffo G, Chen H, et al. Role of COX-2 in epithelial-stromal cell interactions and progression of ductal carcinoma in situ of the breast. Proc Natl Acad Sci U S A. 2009;106:3372–3377. doi: 10.1073/pnas.0813306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deome KB, Faulkin LJ, Jr, Bern HA, et al. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 1959;19:515–520. [PubMed] [Google Scholar]
- 33.Farnie G, Clarke RB, Spence K, et al. Novel cell culture technique for primary ductal carcinoma in situ: role of Notch and epidermal growth factor receptor signaling pathways. J Natl Cancer Inst. 2007;99:616–627. doi: 10.1093/jnci/djk133. [DOI] [PubMed] [Google Scholar]
- 34.Silverstein MJ, Barth A, Poller DN, et al. Ten-year results comparing mastectomy to excision and radiation therapy for ductal carcinoma in situ of the breast. Eur J Cancer. 1995;31A:1425–1427. doi: 10.1016/0959-8049(95)00283-o. [DOI] [PubMed] [Google Scholar]
- 35.Schulze-Garg C, Lohler J, Gocht A, et al. A transgenic mouse model for the ductal carcinoma in situ (DCIS) of the mammary gland. Oncogene. 2000;19:1028–1037. doi: 10.1038/sj.onc.1203281. [DOI] [PubMed] [Google Scholar]
- 36.Solin LJ, Kurtz J, Fourquet A, et al. Fifteen-year results of breast-conserving surgery and definitive breast irradiation for the treatment of ductal carcinoma in situ of the breast. J Clin Oncol. 1996;14:754–763. doi: 10.1200/JCO.1996.14.3.754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.