Abstract
Summary
Symporters are membrane proteins that couple energy stored in electrochemical potential gradients to drive the cotransport of molecules and ions into cells. Traditionally, proteins are classified into gene families based on sequence homology and functional properties, e.g. the sodium glucose (SLC5 or Sodium Solute Symporter Family, SSS or SSF) and GABA (SLC6 or Neurotransmitter Sodium Symporter Family, NSS or SNF) symporter families [1-4]. Recently, it has been established that four Na+-symporter proteins with unrelated sequences have a common structural core containing an inverted repeat of 5 transmembrane (TM) helices [5-8]. Analysis of these four structures reveals that they reside in different conformations along the transport cycle providing atomic insight into the mechanism of sodium solute cotransport.
Introduction
While membrane proteins account for about a quarter of the genes in organisms, they constitute less than 1% of the structures in the protein databases (http://blanco.biomol.uci.edu/Membrane_proteins_xtal.html). An important class of membrane proteins is the secondary active transporters which utilize electrochemical potential gradients to drive a diverse group of substrates into cells. Na+-symporters are a subclass responsible for the accumulation of sugars, amino acids and other ions and molecules in cells. Many hundreds of the Na+-symporters in both prokaryotes and eukaryotes have been identified and classified into ten major families, including the Na+-glucose (SGLTs, SSS, T.C.2.21) and Na+-amino acid (NSS, T.C.2.A.22) symporters (Transporter classification database, http://www.tcdb.org/tcdb/ and PFam, http://pfam.sanger.ac.uk). In humans the SSS and NSS transporters play critical roles in the physiology of the brain, intestine, kidneys, and thyroid, where mutations in their genes are responsible for severe congenital diseases, and furthermore are the designated targets for drugs in the treatment of depression, diabetes and obesity [4,9-12]. Elucidating the structural basis for symport will greatly facilitate our understanding of these diseases and aid in the development of novel drugs.
A central concept in membrane transport is the alternating access mechanism proposed by Jardetzky in 1966 [13]. This model states that substrate binds to one face of the membrane protein and an energy input (e.g. Δμs, ΔV, Δ-μx +/-) then drives a conformational change to expose the substrate to the opposite face of the membrane where it is released. In the case of Na+-symporters, the sodium motive force (Δ-μNa, the Na+ electrochemical potential gradient) drives the conformational changes [9,10,14]. For almost 40 years alternating access has been the model of secondary active transport, but the structural basis has not yet been established. Recently, we solved the crystal structure of a prokaryotic member of the SSS family, the Na+-galactose symporter, vSGLT [6], and were surprised to find that the architecture of the core domain closely resembles that of the Na+-leucine symporter from the NSS family, LeuT [5]. More recently, it has been revealed that the crystal structures of the nucleobase (Mhp1 [7]) and the glycine-betaine/proline-betaine (BetP [8]) symporters from two different gene families (NSS1, T.C.2.A.39, and BCCT, T.C.2.A.15) have the same core architecture.
In this review, we will discuss this highly conserved protein fold in terms of the common features of the structures of vSGLT, LeuT, Mhp1 and BetP. In particular, we will address the binding of various substrates and the driver ion, sodium. We will further highlight the conformational differences between these structures and identify rearrangements required to achieve the binding and release of substrate and sodium.
vSGLT, LeuT, Mhp1 and BetP share the same core domain
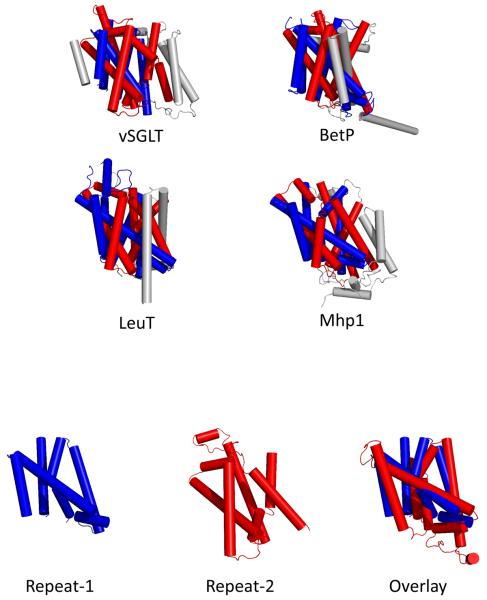
All four symporters belong to different gene families but maintain a common core structure of 10 transmembrane helices (TM1-10) (RMSDs 3.8-4.5 Ǻ, Figure 3A) formed through the association of an unanticipated internal structural repeat of a 5 TM helical bundle (Red TM1-TM5 and Blue TM6-TM10, Figures 1-2). Although there is no sequence similarity between the internal repeats, they have a high degree of structural conservation permitting superimposition with each other (RMSD 3.9Ǻ). They have an inverted topology where TM1-TM5 is related to TM6-TM10 by an apparent two-fold symmetry around an axis through the center of the membrane plane. The transmembrane helices are intertwined giving rise to a group of seven central helices (TM1, TM2, TM3, TM6, TM7, TM8 and TM10) which house the substrate as well as ion binding sites. Although additional helices on the N- and/or C-termini may be present and be important for function (Figure 1), it would appear that the core domain represents a structural hallmark for many sodium driven transporters and thus an alternative classification scheme not solely based on primary sequence alignment is required. Along these lines, Lolkema and Slotbloom used hydrophathy profile alignments [15] to identify a structural similarity between the vSGLT and LeuT. They further predict that amino acid uniporters, symporters and antiporters in the APC (Amino acid-Polyamine-Organocation) superfamily (T.C. 2.A 3) share the inverted repeat core structure. It is interesting that the APC family includes both Na+ and H+ symporters. In the Pfam classification the APC superfamily (Pfam Clan CL0062) contains 14 families including those with transporters for sugars and amino acids, e.g. T.C. 2.A.3, T.C.2.A.18, T.C.A.21, T.C.A.25, T.C.2.A.26, but not those for the LeuT, Mph1 or BetP families. As more structures of transporters are resolved, there is a greater probability for being able to identify these signature motifs and develop better classification tools.
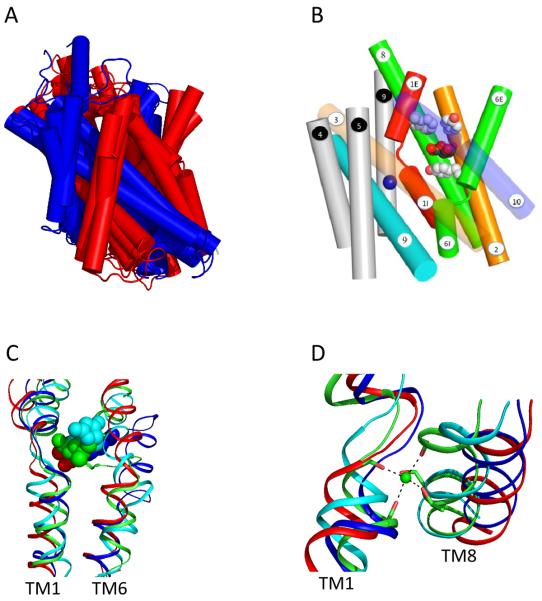
Figure 3. The location of substrate and the common sodium binding site.
A) Structural alignment of the core domains from vSGLT, BetP, LeuT and Mhp1 yield an RMSD between 3.8 Å and 4.5 Å. TM1-TM5 and TM6-TM10 are colored red and blue respectively. B) Side view of vSGLT showing the location of the sugar binding site, the extracellular (M73, Y87 and F424) and intracellular (Y263) gates. The numbering of the TMs is according to the convention shown in Figure 1. C) A structural alignment of TM1, TM6 and the substrate-binding site for vSGLT (red), BetP (blue), LeuT (green) and Mhp1 (cyan). D) A structural alignment of TM1, TM8 and the conserved sodium-binding site for vSGLT (red), BetP (blue), LeuT (green) and Mhp1 (cyan).
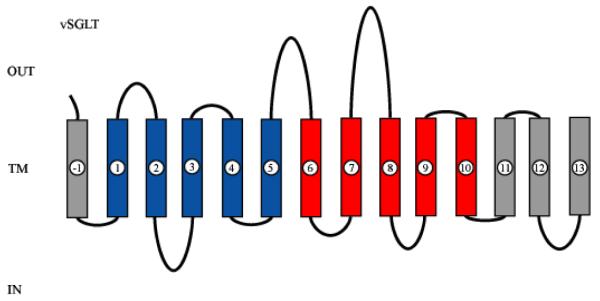
Figure 1. 2-D structure of vSGLT.
A 2D model of vSGLT highlighting the core domain of 10 TMs composed of inverted repeats of 5TMs. vSGLT is a 543 amino acid protein containing 14 TMs with N- and C-termini on the extracellular face of the membrane [41,42]. To avoid confusion, we number the TMs in the inverted repeats as in LeuT ([5], TM1-TM5 (Blue) and TM6-TM10 (red). As members of families contain up to 15 TMs, addition N-terminal TMs are numbered TM(-1) (vSGLT), TM(-1) and TM(-2) (BetP) and additional C-terminal TMs are numbered TM11 and TM12 (LeuT, Mhp1) and TM11, TM12, and TM13 (vSGLT). Note: the N-terminus of the first repeat is intracellular while that for the inverted repeat is extracellular.
Figure 2. The 3D structures of the sodium symporters.
A) Cylinder representation of vSGLT, BetP, LeuT and Mhp1 structures. All four structures have a core domain of 10 transmembrane (TM) helices formed from an internal structural repeat. B) The two structural repeats have an inverted topology where TM1-TM5 is related to TM6-TM10 by an apparent two-fold symmetry around an axis through the center of the membrane plane permitting superimposition. A superposition of vSGLT's TM1-TM5 and inverted TM6-TM10 yields an RMSD of 3.9 Å for 131 Cα atoms. TM1-TM5 and TM6-TM10 are colored red and blue respectively with additional helices on the N- and C-termini colored gray.
Substrate binding site
The substrates in the crystal structures (PDB's: 2A65, 3DH4, 2JLO and 2W8A) have a common location, Site-1, approximately halfway across the membrane bilayer in the center of the core domain (Figure 3B). In LeuT and Mhp1, a vestibule from the extracellular surface to the substrate binding site is formed from TM1, TM3, TM6, TM8 and TM10. The exit pathway from the substrate binding site into the intracellular compartment was identified in the vSGLT structure formed from TM1, TM2, TM3, TM6, TM8 and TM10. A pronounced feature of the 5×5 inverted repeat motif is the position of two discontinuous helices, TM1 and the symmetrically related TM6 located roughly halfway across the membrane bilayer at the interface of the internal repeats. These unwound segments are thought to generate a local polar environment for the binding of substrates and ions, providing a possible link for coupled transport in these and other transport proteins [16,17].
Contrary to what was observed in the structure of the secondary active transporter lactose permease [18], the bound substrates are occluded from both the extracellular and intracellular environment (for vSGLT see Figure 3B). In general, large hydrophobic residues enclose the substrate within the binding pocket preventing access from the surrounding solutes. Thus, a prerequisite for the binding and release of substrate is a conformational change that opens the substrate-binding site to either the extracellular or intracellular milieu.
Although the relative location of the substrate-binding sites may be shared amongst transporters (Figure 3C), the specific interactions vary in order to accommodate the diverse range of substrates. As seen for vSGLT, all OH-moieties of galactose are coordinated by hydrogen bonds from polar side chains in the central helices (TM1, TM6, TM7 and TM10). BetP maintains its substrate, betaine, within a tryptophan box defined by three tryptophan and one tyrosine residues (TM2 and TM6). For Mhp1, benzyl-hydantoin is placed between two tryptophan residues (TM3 and TM6) and coordinated by conserved glutamine and asparagine residues (TM1 and TM8). Finally, LeuT coordinates its substrate, leucine, primarily through main chain atoms in the unwound segments of TM1 and TM6. Thus, the 5×5 inverted repeat topology appears to act as a common scaffold and it is capable of accommodating a wide range of substrates by substituting key amino acids. Mutation of residues coordinating the substrates markedly reduces transport [6-8].
Structures of LeuT, co-crystallized in the presence of antidepressants, revealed the position of an inhibitory binding site (Site-2) located near the extracellular surface, ~11 Å above the Site-1 [19-21]. These noncompetitive inhibitors appear to lock the extracellular gate preventing substrate translocation. Subsequently, steered molecular dynamics simulations in conjunction with binding and flux assays revealed Site-2 to be a secondary binding site whereupon substrate binding induces release of sodium and substrate from Site-1 [22]. To date, there have been no crystal structures showing two substrates bound simultaneously indicating, at least in the occluded conformation, a secondary site has a significantly lower affinity. However, Quick and colleagues have shown that octyl glucoside (the detergent used for crystallization) is a potent inhibitor of LeuT and may prevent the binding of substrate to Site-2 [23]. So far, only single substrate binding sites have been found in the other symporter structures. However, functional data suggests a similar architecture for the inhibitor binding sites in human SGLT1 (SSS) and GAT1 (NSS) [24]. The large hydrophobic domains of competitive inhibitors bind at sites are within 8 Ǻ of the substrate binding site.
Sodium binding site(s)
Sodium is the ion driving substrate translocation in LeuT, vSGLT, Mhp1 and BetP, however the sodium to substrate stoichiometry may differ amongst these sodium dependent transporters: LeuT and BetP have a 2:1 stoichiometry [8,22,23]; and vSGLT and Mhp1 have a 1:1 stoichiometry [7,25] [7]. In the founding structure of LeuT (at 1.65Å), two likely sodium sites (Na1 and Na2) were identified [5]. The positions of both bound ions are in the vicinity of the unwound segments of TM1 and TM6 [26]. In both LeuT and BetP the sodium ion at the Na1 site is directly coupled to the carboxy oxygen of their corresponding substrates. For both vSGLT and Mhp1, there is no Na1 site; however, members of the SSS and NSS families with a 2:1 stoichiometry [27,28] may harbor a sodium ion at position Na1.
The Na2 sodium site appears to be present in all four structures and plays an essential role for substrate translocation (Figure 3D). For LeuT, vSGLT and Mhp1, the site is located at the intersection of TM1 and TM8 ~8Å away from the substrate binding site. There appears to be a conserved coordination pattern made by carbonyl oxygens in the unwound segment of TM1 and hydroxyl groups from serine and/or threonine residues in TM8 [26]. A slightly different conformation is suggested in BetP where TM5 fulfills the role of TM8. However, Thr467 and Ser468 in TM8 of BetP correspond to the proposed Na2 coordinating residues from vSGLT, LeuT and Mhp1. Functional evidence for Na2 in vSGLT is provided by the finding that mutation of S365 abolishes transport [6].
A closer inspection of the Na2 site reveals some distinct alterations in TM8 that appear to be dependent on the conformation of the transporter. In LeuT and Mhp1, which reside in the outward and/or the outward-occluded conformations (C2 or C3, Figure 4), TM1 and TM8 align nicely with one another. Conversely, BetP and vSGLT, which reside in an intermediate (C4) and the inward-occluded conformations (C5), TM1 and TM8 align well with one another but are shifted ~3 Å away from those in LeuT and Mhp1. This can further be observed by a comparison of the sodium coordinating distances, where both LeuT (2.2 to 2.5 Å) and Mhp1 (2.1 to 2.7 Å) are tighter than those for vSGLT (3.1 to 3.8 Å). The more open environment around the vSGLT Na2 site raises the possibility that sodium is not bound in the vSGLT crystal. Molecular Dynamic simulations on the Na2 site in LeuT indicate a tightly bound sodium [29], whereas in vSGLT sodium escapes from Na2 site into the inner aqueous vestibule and, after a transient interaction with D186, into the cytoplasm (E Tajkhorshid, personal communication). The conserved aspartate has been shown to be involved in cation selectivity in human SGLT1 [30].
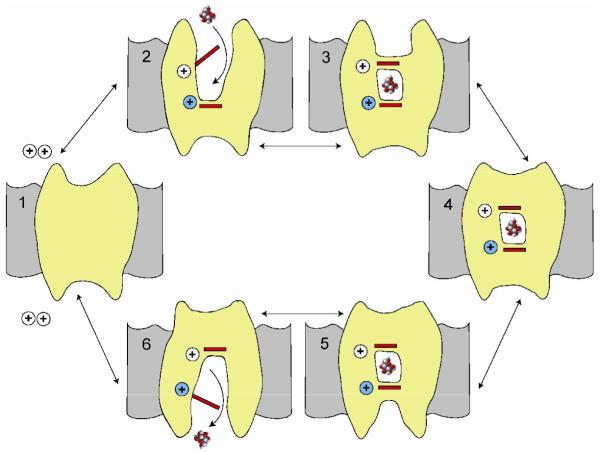
Figure 4. A Simple 6-state model for sodium-substrate symport.
In this model the starting point is state 1 (C1) with no ligands bound. In the presence of extracellular sodium, one (or two) Na+ ions bind first to open the external gates to the substrate binding site (C2). External substrate is then able to bind to its binding site and this in turn closes the external gate (C3). The next step is closure of the external vestibule (C4) and this is followed by opening of the internal vestibule (C5). Opening of the internal gate permits substrate and sodium ion(s) to dissociate and exit at the intracellular face of the membrane (C6). The cycle is completed by closure of the internal vestibule and the return to C1. The complete cycle results in the transport of sodium and substrate across the membrane in a fixed stoichiometry (1/1, 2/1). The direction of transport is determined by the ligand concentrations on each side of the membrane and the membrane potential [14,25,27,31-33,43]. The crystal structures of Mhp1 appear to correspond to C2 and C3, LeuT to C3, BetP to C4 and vSGLT to C5. At least in the case of vSGLT the monomer is completely functional [41].
Alternating access
According to kinetic models for members of the SSS and NSS families [14,27,31-34] the rate and direction of symport is a function of the ligand concentrations on each side of the membrane and the voltage.The transport cycle occurs through at least 6 states (see Figure 4). The common core structure of the four symporters and the fact that the crystal structures reside in distinct conformations, provides insight into the structural basis for sodium/solute symport.
In these kinetic models, sodium in the extracellular compartment binds first and facilitates substrate binding [14,27,31-34]. In the sodium bound open-out conformation (C2) of Mhp1 (PDB: 2JLN) a clear pathway leading to the substrate binding site is observed between the extracellular halves of TM1, TM3, TM6, TM8 and TM10. A similar pathway is evident in the C3 conformation of LeuT (PDB: 2A65) and this becomes more apparent in the structure with the bound competitive inhibitor tryptophan (PDB: 3F3A, [19]). Upon substrate binding, the external gates close to occlude substrate from the extracellular environment (C3). LeuT achieves this through stacking of large bulky residues F253 and Y108 directly above the substrate-binding site (toward extracellular side) yet the pathway above this obstruction facing the extracellular milieu remains intact. For Mhp1 and human SGLT1 [35], the extracellular portion of TM10 (residues 355 to 370) kinks in toward the extracellular facing cavity preventing accessibility to the substrate binding site. Although the mechanism by which the occlusion occurs differs between LeuT and Mhp1 these structures demonstrate an outward facing occluded conformation (C3, Figure 4).
The corresponding conversion from the outward- to inward-facing conformation appears to transition through an intermediate state (C4, Figure 4)) where the transporter, in complex with substrate and sodium, has neither a clear extracellular nor intracellular-facing cavity. BetP (PDB: 2W8A) shows the makings of an intracellular pathway formed from TM1, TM5, TM6 and TM8, but there is insufficient space to facilitate substrate release into the intracellular compartment. The transporters proceed through the cycle where an inward occluded conformation is formed (C5, Figure 4), as represented by the structure of vSGLT (PDB: 3DH4), where galactose is sandwiched between hydrophobic residues (intracellular side Y263; extracellular side M73, Y87 and F424) and is inaccessible to the extracellular and intracellular compartments. The intracellular exit pathway has a large hydrophilic cavity formed from portions of TM1, TM2, TM3, TM6, TM8 and TM10. To date, there are no structures of an inward-facing conformation (IC) devoid of substrate, but based on the structure of vSGLT it would require the rotation of the intracellular gating residue Y263.
How then does the transition from the outward- to inward facing conformation occur? With multiple structures in hand, movements of isolated segments- of the core structure become apparent as the transporters proceed from substrate uptake to release. The Gouaux group suggested [5] a conformation change in the unwound segments TM1 and TM6, where the extracellular portions would flip `out' and the intracellular portions would flip `in' forming the C3 conformation and the reverse would occur to achieve the C5 conformation. With the vSGLT structure, we were able to capitalize on the C3 (LeuT) and the C5 (vSGLT) states to reveal large conformational changes in TM2, TM6 and TM10 with additional displacements TM3, TM7, and TM8. Concurrently, Forrest and colleagues suggested that a 4TM bundle composed of TM1, TM2, TM6 and TM7 can `rock' between the C3 and C5 conformations by simply switching the tilt angle of the bundle [36]. The Iwata group [7] also compared their structures of Mhp1 in the C2 (PDB: 2JLN) and the C3 (PDB: 2JLO) conformations with that for vSGLT in the C5 conformation to develop a simpler rocker bundle model composed of TM3 and TM8. Similarly, Ressl and colleagues [8] combined their BetP structure in the C4 conformation and the other structural data to postulate an iris type mechanism composed primarily of TM1, TM2, TM6 and TM7. Finally, it should be noted that in confronting kinetic models with these four structures it is difficult to predict the dwell time of any given structure during the transport cycle.
Conclusions
The structures obtained for the four sodium symporters from different gene families provide new perspective on the classification of membrane proteins based on gene sequences: First, it is difficult to predict internal structural repeats in a given membrane protein from the primary and/or secondary structures alone; Second, members of different genes families with similar functions may have homologous 3D structures; and Third, a common structural architecture implies a common transport mechanism. In addition, differences in conformation of different transporters with the same core structure provide a tantalizing view of the structural basis of the conformational changes underlying the transport mechanism. Nevertheless, the goal now is to solve the structure of a single transporter in each step of the transport cycle. Finally, we anticipate that other sodium and proton symporters will have the same core structure. Proton symporters are kinetically indistinguishable from the sodium symporters, and in some transport can be driven by either proton or sodium electrochemical potential gradients [37].
Update
While the manuscript was under review the prediction made by Lolkema & Slotbloom [15] transporters in the APC superfamily (T.C. 2.A 3) also share the LeuT structural fold. Two groups [38,39] have solved the structure of the arginine/agmatine antiporter AdiC and found that it too has the 5 TM inverted repeat with disrupted TM 1 and 6.and a central cavity opening to the extracellular surface. In both cases the central cavity, 20 × 10 Å, is substrate free and no sodium binding sites are identified. Since this antiporter and the sodium symporters share the same structure this points to a common mechanism of alternating access. In addition, the Gouaux group [40] has recently published an insightful review of the structure and function of sodium-coupled transporters.
Acknowledgements
We are indebted to Salam Faham, Akira Watanabe. Gabriel Besserer, Duilio Cascio, Alexandre Specht, Bruce Hirayama, Peter Zwart for their contributions towards solving the vSGLT structure and to Vincent Chaptal, Bruce Hirayama and Don Loo for their critical comments on the manuscript. This work was supported by grants from the NIH (JA GM07884; EMW DK19567).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nelson N. The Family of Na+/C1- Neuortransmitter Transporters. Journal of Neurochemistry. 199871:1786–1803. doi: 10.1046/j.1471-4159.1998.71051785.x. [DOI] [PubMed] [Google Scholar]
- 2.Wright EMaT E. The Sodium Glucose Cotransport Family SLC5. Pflugers Arch. 2004:510–518. doi: 10.1007/s00424-003-1063-6. [DOI] [PubMed] [Google Scholar]
- 3.Turk E, Wright EM. Membrane topology motifs in the SGLT cotransporter family. J Membr Biol. 1997;159:1–20. doi: 10.1007/s002329900264. [DOI] [PubMed] [Google Scholar]
- 4.Chen NH, Reith ME, Quick MW. Synaptic uptake and beyond: the sodium-and chloride-dependent neurotransmitter transporter family SLC6. Pflugers Arch. 2004;447:519–531. doi: 10.1007/s00424-003-1064-5. [DOI] [PubMed] [Google Scholar]
- 5**.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]; The first high resolution crystal structure of a sodium symporter with the inverted 5TM motif containing a pair of transmembrane discontinuous helices. The substrate, leucine, and one sodium was bound in a occluded site halfway across the membrane close to these two discontinuous helices. A route for substrate entry to the binding site from the external solution was identified and this conformation of the protein was termed the outward facing substrate occluded conformation.
- 6**.Faham S, Watanabe A, Besserer GM, Cascio D, Specht A, Hirayama BA, Wright EM, Abramson J. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science. 2008;321:810–814. doi: 10.1126/science.1160406. [DOI] [PMC free article] [PubMed] [Google Scholar]; The second sodium symporter with the inverted 5TM motif with a pair of discontinuous transmembrane helices. As for LeuT the substrate was bound in an occluded binding site devoid of water, but in this case the protein was in an inward facing conformation where a hyrophobic gate blocked sugar exit into a hydrophobic channel leading to cytoplasmic side of the membrane. The outward facing LeuT and inward facing vSGLT structures enabled modeling of the structural changes that underlie alternating access transport mechanism.
- 7**.Weyand S, Shimamura T, Yajima S, Suzuki S, Mirza O, Krusong K, Carpenter EP, Rutherford NG, Hadden JM, O'Reilly J, et al. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science. 2008;322:709–713. doi: 10.1126/science.1164440. [DOI] [PMC free article] [PubMed] [Google Scholar]; The third report of a symporter with the LeuT structural motif, i.e. the inverted 5TM repeat with a pair of discontinuous transmembrane helices. One structure was solved in the absence of substrate, an outward facing open conformation. Another structure with substrate bound was solved at a lower resolution, outward facing occluded conformation. This permitted modeling of the substrate induced These together with the structure modeled on the inward occluded facing conformation of vSGLT resulted in a more complete structural model for transport. It is not yet fully clear whether or not Mhp1 is a sodium or a proton symporter.
- 8**.Ressl S, Terwisscha van Scheltinga AC, Vonrhein C, Ott V, Ziegler C. Molecular basis of transport and regulation in the Na(+)/betaine symporter BetP. Nature. 2009;458:47–52. doi: 10.1038/nature07819. [DOI] [PubMed] [Google Scholar]; The most recent report of a sodium symporter with the LeuT structural motif where the structure is an intermediate between the outward and inward facing occluded conformations. As for LeuT, two putative sodium binding sites were identified, Na1 and Na2. Only Na2 sites were identified in vSGLT and Mhp1. The BetP family of proteins play an important role in osmoregulation in cells.
- 9.Wright EM, Hirayama BA, Loo DF. Active sugar transport in health and disease. J Intern Med. 2007;261:32–43. doi: 10.1111/j.1365-2796.2006.01746.x. [DOI] [PubMed] [Google Scholar]
- 10.Dohan O, De La Vieja A, Paroder V, Riedel C, Artani M, Reed M, Ginter CS, Carrasco N. The sodium/iodide Symporter (NIS): characterization, regulation, and medical significance. Endocr Rev. 2003;24:48–77. doi: 10.1210/er.2001-0029. [DOI] [PubMed] [Google Scholar]
- 11.Isaji M. Sodium-glucose cotransporter inhibitors for diabetes. Current Opinion in Investigational Drugs. 2007;8:285–292. [PubMed] [Google Scholar]
- 12.Bakris GL, Fonseca VA, Sharma K, Wright EM. Renal sodium-glucose transport: role in diabetes mellitus and potential clinical implications. Kidney Int. 2009 doi: 10.1038/ki.2009.87. [DOI] [PubMed] [Google Scholar]
- 13.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 14.Kanner BI. Structure and function of sodium-coupled GABA and glutamate transporters. J Membr Biol. 2006;213:89–100. doi: 10.1007/s00232-006-0877-5. [DOI] [PubMed] [Google Scholar]
- 15.Lolkema JS, Slotboom DJ. The major amino acid transporter superfamily has a similar core structure as Na+-galactose and Na+-leucine transporters. Mol Membr Biol. 2008;25:567–570. doi: 10.1080/09687680802541177. [DOI] [PubMed] [Google Scholar]
- 16**.Hunte C, Screpanti E, Venturi M, Rimon A, Padan E, Michel H. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;435:1197–1202. doi: 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]; A highlight of this 12 TM transporter protein is a pair of transmembrane discontinuous helices. The crossed extended chains create a balanced electrostatic environment and the authors proposed that the binding of charged substrates at this site causes a charge imbalance that promotes a rapid alternating access transport mechanism.
- 17.Yernool D, Boudker O, Jin Y, Gouaux E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature. 2004;431:811–818. doi: 10.1038/nature03018. [DOI] [PubMed] [Google Scholar]
- 18.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 19*.Singh SKPC, Yamashita A, Gouaux E. A competitive inhibitor traps LeuT in an open-to-out conformation. Science. 2008;322:1655–1661. doi: 10.1126/science.1166777. 2008 Dec 12;(5908):1655-61. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is this biochemical and crystallographic study of LeuT it was shown that several amino acid substrates are substrates and bind in the same conformation as leucine, i.e. outward facing occluded, but with local differences in the binding site. In contrast, tryptophan is a non-transported, competitive inhibitor that traps LeuT in an open-to-out conformation with solvent accessibility to the substrate binding pocket. In addition, a second low affinity substrate binding site was postulated to exist at the base of the extracellular vestibule. On the basis of the substrate and competitive inhibitor bound structures of LeuT a structural model for transport and inhibition was proposed.
- 20*.Singh SK, Yamashita A, Gouaux E. Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature. 2007;448:952–956. doi: 10.1038/nature06038. [DOI] [PubMed] [Google Scholar]; Antidepressants block the transport of neurotransmitters by the NSS symporters. In this paper it was shown that the bacterial homolog LeuT the antidepressants also block amino acid transport and bind to an extracellular facing vestibule about 11 Å above the leucine binding site. It is concluded that the inhibitor stabilizes the protein in an outward facing conformation with a closed extracellular gate to the substrate binding site.
- 21*.Zhou Z, Zhen J, Karpowich NK, Goetz RM, Law CJ, Reith ME, Wang DN. LeuT-desipramine structure reveals how antidepressants block neurotransmitter reuptake. Science. 2007;317:1390–1393. doi: 10.1126/science.1147614. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is an independent report on the binding of antidepressants to the LeuT symporter. These authors also find that the antidepressants bind to the inner end of an extracellular cavity separated from the leucine binding site by an extracellular gate. In addition, a mutagenesis analysis of the human NSS symporters also indicates that the inhibitor binding sites are probably conserved in the human transporters.
- 22*.Shi L, Quick M, Zhao Y, Weinstein H, Javitch JA. The mechanism of a neurotransmitter:sodium symporter--inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol Cell. 2008;30:667–677. doi: 10.1016/j.molcel.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; Molecular dynamic studied of LeuT suggested a second substrate binding site in the proximity of the inhibitor binding site. This conclusion was strengthened by biochemical experiments indicating that both substrate sites were occupied simultaneously. The authors proposed that substrate binding to the second site triggers the intracellular release of sodium and substrate, and that when antidepressants bind the is no release of ligands to the cytoplasm.
- 23*.Quick M, Winther AM, Shi L, Nissen P, Weinstein H, Javitch JA. Binding of an octylglucoside detergent molecule in the second substrate (S2) site of LeuT establishes an inhibitor-bound conformation. Proc Natl Acad Sci U S A. 2009;106:5563–5568. doi: 10.1073/pnas.0811322106. [DOI] [PMC free article] [PubMed] [Google Scholar]; N-octyl-β-D-glucopyranoside is a popular detergent used for solubilizing membrane proteins and it was used in the successful crystallization of LeuT. Here it was shown that this detergent is an inhibit transport and occupies the second substrate (and antidepressant) binding site in LeuT.
- 24.Hirayama BA, Diez-Sampedro A, Wright EM. Common mechanisms of inhibition for the Na+/glucose (hSGLT1) and Na+/Cl-/GABA (hGAT1) cotransporters. Br J Pharmacol. 2001;134:484–495. doi: 10.1038/sj.bjp.0704274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Veenstra M, Lanza S, Hirayama BA, Turk E, Wright EM. Local conformational changes in the Vibrio Na+/galactose cotransporter. Biochemistry. 2004;43:3620–3627. doi: 10.1021/bi0357210. [DOI] [PubMed] [Google Scholar]
- 26*.Noskov SY, Roux B. Control of Ion Selectivity in LeuT:Two Na+ Binding Sites with Two Different Mechanisms. Journal Mol. Biol. 2008;377:804–818. doi: 10.1016/j.jmb.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; This contains a most valuable analysis of Na+ selectivity in LeuT and other sodium binding proteins in the PDB using molecular dynamic simulations.
- 27.Eskandari S, Loo DD, Dai G, Levy O, Wright EM, Carrasco N. Thyroid Na+/I-symporter. Mechanism, stoichiometry, and specificity. J Biol Chem. 1997;272:27230–27238. doi: 10.1074/jbc.272.43.27230. [DOI] [PubMed] [Google Scholar]
- 28.Mackenzie B, Loo DD, Wright EM. Relationships between Na+/glucose cotransporter (SGLT1) currents and fluxes. J Membr Biol. 1998;162:101–106. doi: 10.1007/s002329900347. [DOI] [PubMed] [Google Scholar]
- 29.Celik L, Schiott B, Tajkhorshid E. Substrate binding and formation of an occluded state in the leucine transporter. Biophys J. 2008;94:1600–1612. doi: 10.1529/biophysj.107.117580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quick M, Loo DD, Wright EM. Neutralization of a conserved amino acid residue in the human Na+/glucose transporter (hSGLT1) generates a glucose-gated H+ channel. J Biol Chem. 2001;276:1728–1734. doi: 10.1074/jbc.M005521200. [DOI] [PubMed] [Google Scholar]
- 31.Eskandari S, Wright EM, Loo DD. Kinetics of the reverse mode of the Na+/glucose cotransporter. J Membr Biol. 2005;204:23–32. doi: 10.1007/s00232-005-0743-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32*.Loo D, Hirayama BA, Sala-Rabanal M, Wright EM. How drugs interact with transporters; SGLT1 as a model. J.Membrane Biology. 2008;223:87–106. doi: 10.1007/s00232-008-9116-6. [DOI] [PubMed] [Google Scholar]; This paper provides an entre into the most current approach to modeling symporter kinetics.
- 33.Loo DDF, Hirayama BA, Karakossian MH, Meinmild A-K, Wright EM. Conformational Dynamics of hSGLT1 during Na+/glucose cotransport. J. General Physiology. 2006;128:701–720. doi: 10.1085/jgp.200609643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung H. The sodium/substrate symporter family: structural and functional features. FEBS Lett. 2002;2:73–77. doi: 10.1016/s0014-5793(02)03184-8. [DOI] [PubMed] [Google Scholar]
- 35.Hirayama BA, Loo DD, Diez-Sampedro A, Leung DW, Meinild AK, Lai-Bing M, Turk E, Wright EM. Sodium-dependent reorganization of the sugar-binding site of SGLT1. Biochemistry. 2007;46:13391–13406. doi: 10.1021/bi701562k. [DOI] [PubMed] [Google Scholar]
- 36*.Forrest LR, Zhang YW, Jacobs MT, Gesmonde J, Xie L, Honig BH, Rudnick G. Mechanism for alternating access in neurotransmitter transporters. Proc Natl Acad Sci U S A. 2008;105:10338–10343. doi: 10.1073/pnas.0804659105. [DOI] [PMC free article] [PubMed] [Google Scholar]; These authors independently proposed a structural model for alternating access based on molecular dynamic simulations of LeuT.
- 37.Hirayama BA, Hirayama LD, Wright EM. Cation effects on protein conformation and transport in the Na+/glucose cotransporter. J Biol Chem. 1997 Jan 24;272(4) doi: 10.1074/jbc.272.4.2110. 1997. [DOI] [PubMed] [Google Scholar]
- 38**.Gao X, Lu F, Zhou L, Dang S, Sun L, Li X, Wang J, Shi Y. Structure and Mechanism of an Amino Acid Antiporter. Science. 2009 doi: 10.1126/science.1173654. [DOI] [PubMed] [Google Scholar]; The structure of the arginine:agmatine antiporter AdiC has the same LeuT fold as the sodium symporters.
- 39**.Fang Y, Jayaram H, Shane T, Kolmakova-Partensky L, Wu F, Williams C, Xiong Y, Miller C. Structure of a Virtual Proton Pump. Nature. 2009 doi: 10.1038/nature08201. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]; An independent report that AdiC has the same fold as LeuT
- 40**.Krishnamurthy H, Piscitelli CL, Gouaux E. Unlocking the molecular secrets of sodium-coupled transporters. Nature. 2009;459:347–355. doi: 10.1038/nature08143. [DOI] [PMC free article] [PubMed] [Google Scholar]; A timely review of the structure and function of sodium symporters.
- 41.Turk E, Kim O, le Coutre J, Whitelegge JP, Eskandari S, Lam JT, Kreman M, Zampighi G, Faull KF, Wright EM. Molecular characterization of Vibrio parahaemolyticus vSGLT: a model for sodium-coupled sugar cotransporters. J Biol Chem. 2000;275:25711–25716. doi: 10.1074/jbc.M003127200. [DOI] [PubMed] [Google Scholar]
- 42.Turk E, Gasymov OK, Lanza S, Horwitz J, Wright EM. A reinvestigation of the secondary structure of functionally active vSGLT, the vibrio sodium/galactose cotransporter. Biochemistry. 2006;45:1470–1479. doi: 10.1021/bi052160z. [DOI] [PubMed] [Google Scholar]
- 43.Parent L, Supplisson S, Loo DD, Wright EM. Electrogenic properties of the cloned Na+/glucose cotransporter: II. A transport model under nonrapid equilibrium conditions. J Membr Biol. 1992;125:63–79. doi: 10.1007/BF00235798. [DOI] [PubMed] [Google Scholar]