Abstract
Several mutant strains of Synechocystis sp. PCC 6803 with large deletions in the D-E loop of the photosystem II (PSII) reaction center polypeptide D1 were subjected to high light to investigate the role of this hydrophilic loop in the photoinhibition cascade of PSII. The tolerance of PSII to photoinhibition in the autotrophic mutant ΔR225-F239 (PD), when oxygen evolution was monitored with 2,6-dichloro-p-benzoquinone and the equal susceptibility compared with control when monitored with bicarbonate, suggested an inactivation of the QB-binding niche as the first event in the photoinhibition cascade in vivo. This step in PD was largely reversible at low light without the need for protein synthesis. Only the next event, inactivation of QA reduction, was irreversible and gave a signal for D1 polypeptide degradation. The heterotrophic deletion mutants ΔG240-V249 and ΔR225-V249 had severely modified QB pockets, yet exhibited high rates of 2,6-dichloro-p-benzoquinone-mediated oxygen evolution and less tolerance to photoinhibition than PD. Moreover, the protein-synthesis-dependent recovery of PSII from photoinhibition was impaired in the ΔG240-V249 and ΔR225-V249 mutants because of the effects of the mutations on the expression of the psbA-2 gene. No specific sequences in the D-E loop were found to be essential for high rates of D1 polypeptide degradation.
Prolonged strong irradiation impairs the photochemical efficiency of all oxygen-evolving photosynthetic organisms. PSII, an intricate thylakoid membrane complex with a light-driven water-plastoquinone-oxidoreductase activity, is the primary target of this phenomenon, which is known as photoinhibition (see Prasil et al., 1992; Aro et al., 1993). Despite intense research, the molecular mechanism of PSII photoinhibition in vivo still remains unclear. On the basis of in vitro studies, two major pathways have been implicated in the photodamage of PSII. The acceptor-side photoinhibition, typical under strong illumination, occurs at the level of the primary quinone electron acceptor QA, which leaves its site in the D2 polypeptide after being double reduced (Styring et al., 1990; Vass et al., 1992). Such conditions lead to the recombination of the primary radical pair P680+ Pheo− and to the formation of chlorophyll triplets (Vass et al., 1992). Triplet chlorophyll may then react with molecular oxygen to produce toxic singlet oxygen, which damages PSII. The other mechanism, donor-side photoinhibition, takes place when the donation of electrons to PSII occurs more slowly than their removal to the acceptor side, leading to the formation of long-lived, highly oxidizing radicals such as Tyr Z+ and P680+ (Blubaugh and Cheniae, 1990; Jegerschöld et al., 1990; Bumann and Oesterhelt, 1995). These species have the capacity to extract electrons from their surroundings, causing irreversible damage to PSII. Depending on environmental conditions, both of these photoinhibition mechanisms have been implicated in in vivo photoinhibition as well (Wang et al., 1992; Ohad et al., 1994). Studies conducted with intact cells suggest an initial role for the QB pocket in the induction of PSII photoinhibition (Kyle et al., 1984; Kirilovsky et al., 1988; Ohad et al., 1990).
It is generally agreed that light-induced damage to PSII renders the D1 polypeptide ready for degradation. Detrimental oxidative species produced by PSII may induce a conformational change in D1, thereby making it susceptible to proteolysis (see Prasil et al., 1992; Aro et al., 1993). Restoration of PSII function following photoinhibitory damage and subsequent degradation of the damaged D1 polypeptide requires de novo synthesis and the incorporation of a new D1 copy into the PSII complex (Prasil et al., 1992; Aro et al., 1993).
Although the mechanism of D1 degradation is not yet known, the D-E loop of this polypeptide is thought to have an important regulatory role in the process of D1 polypeptide degradation. The D-E loop not only forms an essential structural component of the QB pocket (Trebst, 1986), but also has specific amino acid sequences, the PEST-like sequence and the putative cleavage region (Greenberg et al., 1987), which are postulated to carry information for D1 polypeptide degradation. We have recently constructed Synechocystis sp. PCC 6803 mutants with large deletions in the D-E loop of the D1 polypeptide, and have shown that the electron transfer on the acceptor side of PSII is severely disturbed in these mutants (Mulo et al., 1997). The deletion of the PEST-like sequence in the autotrophic strain PD impaired the QA-to-QB electron transfer in PSII, whereas deletions in the putative cleavage region of the D1 polypeptide in CD and PCD also resulted in perturbation of the quinone-exchange reaction in the QB pocket, preventing photosynthetic growth (Mulo et al., 1997).
In the present study the mutants described above were employed to investigate how the severe modifications of the QB pocket and consequent perturbations in the function of the acceptor side of PSII modulate the susceptibility of PSII to photoinactivation and how they exert their effect on the subsequent repair process.
MATERIALS AND METHODS
Bacterial Strains and Growth Conditions
D1 polypeptide mutants, which are derivatives of the Glc-tolerant strain of Synechocystis sp. PCC 6803 (Williams, 1988), were constructed as described by Mulo et al. (1997). The mutant strains PD, CD, and PCD were maintained on BG-11 plates supplemented with antibiotics (see Mäenpää et al., 1993), 5 mm Glc, and 10 μm DCMU to prevent any selective advantage of possible revertants or secondary mutants. The only functional psbA gene in all mutants was the mutated psbA-2, whereas psbA-1 and psbA-3 genes were inactivated by interruption with antibiotic-resistance cassettes (Mohamed and Jansson, 1989).
All strains were grown under a continuous PPFD of 40 μmol m−2 s−1 at 32°C. The AR control, which contains the same antibiotic-resistance cassettes as the mutants but no mutations in the psbA-2 gene, and the autotrophic mutant PD were grown as described in Mäenpää et al. (1993), except that 5 mm Glc was included during growth on the plates. Glc was omitted from the liquid medium to allow photoautotrophic growth (the growth mode of the cells did not affect the results obtained). The heterotrophic strains CD and PCD were grown on plates like those used for AR and PD, but the liquid growth medium was supplemented with 5 mm Glc.
In Vivo High-Light Treatments
Synechocystis sp. PCC 6803 cells were harvested (4500g, 10 min, room temperature) at the logarithmic growth phase and resuspended in BG-11 medium to a final concentration of 10 μg chlorophyll mL−1. The cells were illuminated at a PPFD of 500, 1000, or 1500 μmol photons m−2 s−1 as indicated, using a slide projector as a light source. Temperature was controlled so as not to exceed 32°C, and aliquots for various measurements were withdrawn every 15 min. After 60 min of incubation at high light, cells were transferred back to growth light (40 μmol photons m−2 s−1) and the recovery of the cells from photoinhibition was monitored by measuring the oxygen-evolving capacity after 0, 1, 2, 3, and 4 h. In some experiments, as indicated, the translation inhibitor lincomycin (400 μg mL−1) was added immediately after the high-light treatment, prior to the transfer of the cells to recovery conditions.
In Vivo Pulse-Chase Experiments and Determination of D1 Protein Half-Life
In vivo pulse-chase experiments were performed as described previously (Tyystjärvi et al., 1994). Before high-light treatment, l-[35S]Met (in vivo cell-labeling grade, 1000 Ci mmol−1, Amersham) was added to the cell suspension (10 μg chlorophyll mL−1) to a final concentration of 1.2 μm. The cells were pulse-labeled for 75 min at 1500 μmol photons m−2 s−1, unlabeled Met (1 mm) was added, and thereafter the cells were washed and resuspended in fresh BG-11 medium supplemented with 1 mm cold Met. Radioactivity was chased for 0, 15, 30, 45, and 60 min at high light (1500 μmol photons m−2 s−1).
Photosynthetic membranes were isolated and polypeptides were separated as described previously (Tyystjärvi et al., 1994) by using 12% SDS-polyacrylamide gels including 4 m urea (Laemmli, 1970). Solubilized membranes containing 2 μg of chlorophyll were loaded into each well. After electrophoresis, gels were treated with Amplify (Amersham), dried, and exposed to radiographic film. Fluorograms were scanned with a laser densitometer, and the half-life of the D1 polypeptide was determined by fitting the data to the first-order equation.
In Vivo Measurement of Oxygen Evolution
Oxygen evolution was measured under saturating red light with a Clark-type oxygen electrode (Hansatech, Kings Lynn, UK) at 32°C with a slide projector as the light source. Before measurements, 1 mL of cell suspension corresponding to 10 μg chlorophyll mL−1 was harvested and resuspended in fresh BG-11 medium. Either 0.5 mm DCBQ or 1.0 mm DMBQ was used as the artificial electron acceptor, and 0.25 mm ferricyanide was added to keep the quinones in oxidized form. The light-saturated rate of oxygen evolution was also measured by supplementing the growth medium with 0.6 mm bicarbonate.
Isolation of RNA and Northern Blotting
Cell samples were collected after 0, 15, 30, 45, and 60 min of high-light treatment (1500 μmol photons m−2 s−1), and the total RNA was isolated using the hot phenol method in the presence of 60 mm EDTA and 0.5% SDS (Sherrer and Darnell, 1962). Northern analysis was performed using the standard methods (Sambrook et al., 1989) described in detail by Tyystjärvi et al. (1996). Membranes were first probed with the entire psbA-2 gene of Synechocystis sp. PCC 6803, and then the membrane was stripped and reprobed with rrn genes of Anacystis nidulans (Tomioka et al., 1981) to verify equal loading of the gel and even transfer of RNA in blotting.
Chlorophyll Determination
The chlorophyll a content of the cell suspensions and isolated membranes was determined according to the methods of Bennett and Bogorad (1973) and Arnon (1949), respectively.
RESULTS
Oxygen Evolution Rates and Inhibition of Electron Transport by DCMU in the Mutant Strains
The D-E loop deletion mutants of the D1 polypeptide, PD, CD, and PCD, showed striking differences in the oxygen-evolving capacity with various electron acceptors (Table I). The autotrophic PD and control AR strains, as well as the heterotrophic strains CD and PCD, were all capable of electron transfer from water to DCBQ, which has been suggested to accept electrons directly from QA (Graan and Ort, 1986). In a slight variance from our earlier results (Mulo et al., 1997), all D-E loop mutants, including PD, were severely perturbed in electron transfer to DMBQ, which accepts electrons from QB, probably by interacting with the QB site (Graan and Ort, 1986). Despite low-DMBQ-mediated oxygen-evolution activity, the PD mutant exhibited nearly as high a rate of oxygen evolution as the control strain AR when no artificial electron acceptors were present.
Table I.
Oxygen-evolution rates in Synechocystis sp. PCC 6803 control and mutant strains measured with different electron acceptors, and the inhibition of oxygen evolution by DCMU
| Strain | Oxygen Evolution
|
||
|---|---|---|---|
| DCBQ | DMBQ | Bicarbonate | |
| μmol O2 mg−1 Chlah−1 | |||
| AR | 231 ± 18 (+) | 260 ± 39 (+) | 160 ± 15 (+) |
| PD | 91 ± 8 (−) | 34 ± 2 (+) | 136 ± 9 (+) |
| CD | 146 ± 15 (−) | 32 ± 2 (+) | Trace (+) |
| PCD | 148 ± 15 (−) | 40 ± 4 (+/−) | Trace (+) |
Susceptibility of oxygen evolution to inhibition by 1 μm DCMU is given in parentheses: +, complete inhibition of oxygen evolution; +/−, partial inhibition of oxygen evolution, and −, no or only a minor effect of 1 μm DCMU. Results are the means ± se of 3 to 10 independent experiments from different cell cultures.
Chl, Chlorophyll.
We also tested the ability of DCMU to replace different electron acceptors in the QB pocket. The addition of 1 μm DCMU completely inhibited the DCBQ-dependent oxygen-evolution activity in the control AR cells, but had no effect in the mutants PD, CD, and PCD. However, a minor inhibition of DCBQ-dependent oxygen evolution was observed in the mutants if the DCMU concentration was increased to 10 μm. DMBQ- and bicarbonate-mediated oxygen evolution, by contrast, were completely blocked by 1 μm DCMU, both in the control AR and in the mutants PD and CD. Only in the PCD strain was a higher DCMU concentration (10 μm) required to totally inhibit the oxygen evolution from water to DMBQ (Table I).
Susceptibility of PSII to Photoinhibition in the D-E Loop- Deletion Strains of Synechocystis sp. PCC 6803
Upon illumination at a PPFD of 1500 μmol photons m−2 s−1, the autotrophic PEST-deletion strain PD was much more resistant to photoinhibition of PSII than the control strain AR or the heterotrophic strains CD and PCD when assayed with DCBQ after the high-light treatment (Fig. 1A). Light treatment at 1000 μmol photons m−2 s−1 resulted in an even more drastic difference between AR and PD; more than 80% of activity was lost in AR, indicating that the PPFD of 1000 μmol m−2 s−1 was still strong enough to induce severe inactivation of PSII oxygen evolution, whereas only a 30% loss was observed in PD after 60 min of illumination (Fig. 1B). To exclude the possibility that the PD cells with low PSII activity were already partially photoinactivated during the growth period at 40 μmol photons m−2 s−1, both AR and PD cells were grown at much lower light intensity, below 10 μmol photons m−2 s−1, and no variance to the results above was observed (data not shown).
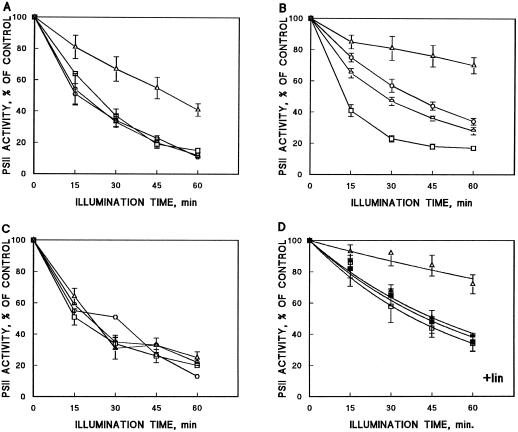
Figure 1.
Photoinactivation of PSII oxygen evolution during illumination of AR (□) and mutant cells (PD, ▵; CD, ○; and PCD, ) of Synechocystis sp. PCC 6803. Results are the means of three to five independent experiments from different cell cultures, and are expressed as a percentage of oxygen-evolution activity measured before the high-light treatment. Bars denote se. A, Cells were illuminated at a PPFD of 1500 μmol m−2 s−1 and oxygen evolution was measured in vivo using DCBQ as the electron acceptor. B, Cells were illuminated at a PPFD of 1000 μmol photons m−2 s−1 and oxygen evolution was assayed in vivo with DCBQ as the electron acceptor. C, Cells were illuminated at a PPFD of 1000 μmol photons m−2 s−1 and oxygen evolution was assayed in vivo with DMBQ as the electron acceptor. D, The PD and AR cells were illuminated at a PPFD of 500 μmol photons m−2 s−1 in the presence of lincomycin (400 μg/mL) and oxygen evolution was monitored with DCBQ (□, ○) or with bicarbonate (▪, •) as the electron acceptor.
The cleavage-region mutants CD and PCD, which exhibited similar kinetics of photoinactivation under a PPFD of 1500 μmol m−2 s−1 (as did the control strain AR [Fig. 1A]), were somewhat more resistant to photoinhibition than AR under a PPFD of 1000 μmol m−2 s−1, but were still clearly more susceptible than PD (Fig. 1B). Notably, however, when PSII activity was measured with DMBQ (Fig. 1C) or bicarbonate (Fig. 2B) as the electron acceptor, the mutants were as susceptible to photoinhibition as AR. Unlike the mutant strains, no significant differences were detected in the susceptibility of the AR cells to photoinhibition whether DCBQ or DMBQ was used as the artificial quinone electron acceptor to monitor the progress of photoinhibition (Fig. 1, B and C).
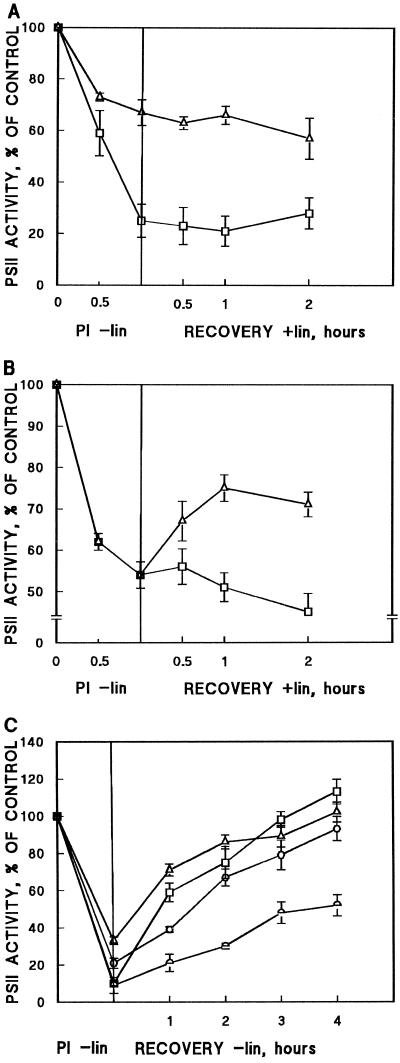
Figure 2.
Recovery of PSII from photoinhibition. Protein-synthesis-independent (A and B) and protein-synthesis-dependent (C) restoration of PSII oxygen evolution in Synechocystis PCC 6803 strains after photoinhibitory illumination. Results are means of three independent experiments from different cell cultures and bars denote se. A, AR (□) and PD (▵) cells were illuminated at 1000 μmol photons m−2 s−1 for 60 min (PI) in the absence of lincomycin. The subsequent recovery of oxygen evolution was followed in the presence of lincomycin at 40 μmol photons m−2 s−1 and was monitored with DCBQ as the electron acceptor. B, The AR (□) and PD (▵) cells were illuminated at 1000 μmol photons m−2 s−1 for 60 min (PI) in the absence of lincomycin. The subsequent recovery of oxygen evolution at 40 μmol photons m−2 s−1 in the presence of lincomycin was monitored using bicarbonate as the electron acceptor. C, AR (□), PD (▵), CD (○), and PCD ( ) strains were illuminated at 1500 μmol photons m−2 s−1 for 60 min (PI), and the restoration of DCBQ-dependent oxygen evolution was followed at 40 μmol photons m−2 s−1. All treatments were performed in the absence of lincomycin.
Because of rapid photoinactivation of the AR cells at 1000 μmol photons m−2 s−1, we further lowered the light intensity to measure the intrinsic susceptibility of PSII photoinactivation in the presence of the protein-synthesis inhibitor lincomycin, which prevents the concurrent repair of PSII. Distinct differences prevailed in the susceptibility of PSII to photoinhibition between AR and PD, even when protein synthesis was blocked; PD was clearly more tolerant to photoinhibition than the control strain AR when measured with DCBQ (Fig. 1D). Under similar conditions, however, no difference could be detected in the rate of photoinhibition between the AR and PD strains if oxygen evolution was assayed with bicarbonate as the final electron acceptor; a severe photoinhibition of PSII was now also seen in the PD strain (Fig. 1D).
Recovery from Photoinhibition
The results above indicate that PSII in the PD cells is intrinsically more tolerant against photoinhibition when assayed with DCBQ, whereas a very similar sensitivity of the AR and PD cells was revealed if oxygen evolution was monitored with bicarbonate (Fig. 1D). To elucidate this apparent controversy, we illuminated the AR and PD cells at 1000 μmol photons m−2 s−1 and then transferred the cells to low recovery light with the simultaneous addition of the protein-synthesis inhibitor lincomycin to prevent protein-synthesis-dependent repair of PSII centers. The differential susceptibility of PSII to high light in the AR and PD cells was evident when oxygen evolution was monitored with DCBQ (Fig. 2A), whereas equal susceptibility was recorded when bicarbonate-dependent oxygen evolution was monitored (Fig. 2B).
After the transfer of cells to low light in the presence of lincomycin, practically no changes in oxygen-evolution activity took place in the PD or AR cells when monitored with DCBQ (Fig. 2A) or in AR cells monitored without the addition of artificial electron acceptor (Fig. 2B). In sharp contrast to AR, nearly 50% restoration of bicarbonate-dependent oxygen evolution, with no concurrent protein synthesis, took place in the PD cells during 1 h of incubation at low light (Fig. 2B). Taken together, these results strongly suggest that photoinhibition in the PD strain, when monitored with bicarbonate, was comprised of two components, one irreversible and one reversible, whereas DCBQ-mediated oxygen-evolution measurements only recorded the irreversible photoinhibitory component of PSII.
To follow the protein-synthesis-dependent restoration of PSII oxygen evolution (water–DCBQ) in photoinhibited cells, all strains were first illuminated for 60 min at a PPFD of 1500 μmol m−2 s−1. This illumination induced severe (about 80%) photoinhibition in all strains except PD; the PSII activity of the PD cells remained somewhat higher despite several trials to enhance photoinhibition either by prolonged incubation of the cells at 1500 μmol photons m−2 s−1 or by increasing the light intensity up to 2500 μmol photons m−2 s−1 (data not shown). Despite severe photoinhibition, the transfer of the control AR and the mutant strains PD and CD back to growth-light conditions induced full restoration of their PSII function within 4 h (Fig. 2C). This restoration of PSII function was fully dependent on protein synthesis in all strains (data not shown). It should be noted that during the 1st h of recovery, more than 50% restoration of the PSII activity occurred in AR and PD cells, whereas the cleavage-region mutants recovered more slowly, with less than 20% restoration occurring within the 1st h. The overall capacity of the PCD strain to recover from photoinhibition during 4 h of incubation was clearly suppressed compared with the other strains (Fig. 2C).
Degradation Rates of the D1 Protein at High Light
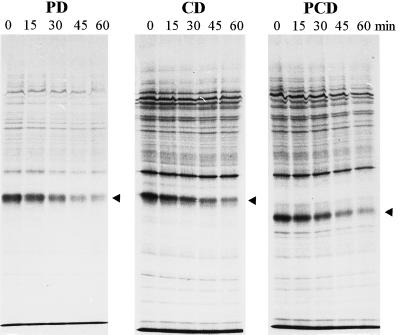
The half-lives of the D1 protein in AR and the mutant strains at a PPFD of 1500 μmol m−2 s−1 measured with the pulse-chase technique are given in Table II. Half-lives at growth conditions (Mulo et al., 1997) are also given for comparison. In AR the half-life of the D1 polypeptide at high light was 26 min, whereas in all mutants the half-life was clearly longer: 45 min for PD, 56 min for CD, and 53 min for PCD. Figure 3 shows examples of pulse-chase fluorograms for the mutant strains PD, CD, and PCD, the two latter strains lacking the putative cleavage region (Greenberg et al., 1987). Thylakoid proteins of the heterotrophic strains CD and PCD were more heavily labeled than those of the autotrophic strain PD because of the faster growth rate of the cells, which was induced by Glc in the growth media of CD and PCD. In addition to pulse-chase experiments, monitoring the degradation of the D1 polypeptide by immunoblotting during the high-light treatment of cells in the presence of the protein-synthesis inhibitor lincomycin also revealed a significantly longer half-life for all mutant strains compared with the AR control strain (data not shown).
Table II.
Half-life of the D1 polypeptide in the site-specific D-E loop mutants of Synechocystis sp. PCC 6803 at high-light and growth-light conditions
| Strain | Half-Life
|
|
|---|---|---|
| High light | Growth lighta | |
| min | ||
| AR | 26 ± 3 | 198 ± 18 |
| PD | 45 ± 3 | 120 ± 6 |
| CD | 56 ± 2 | 108 ± 18 |
| PCD | 53 ± 5 | 162 ± 18 |
Cells were pulse labeled with l-[35S]Met and the radioactivity was chased at either high-light (1500 μmol photons m−2 s−1) or growth-light (40 μmol photons m−2 s−1) conditions. Fluorograms were analyzed with a laser densitometer, and the results were fitted to first-order kinetics to determine the half-life (t½) of the D1 polypeptide. Results are the means ± se of two to four independent experiments.
Data were taken from Mulo et al. (1997).
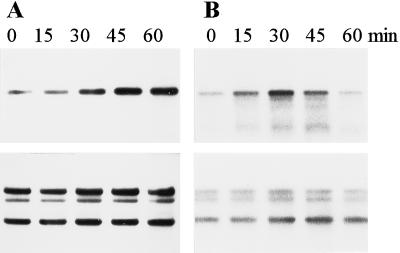
Figure 3.
Chase of radioactivity in membrane polypeptides of Synechocystis sp. PCC 6803 mutants. The cells were first pulse labeled for 75 min in the presence of l-[35S]Met, and then radioactivity was chased for 0, 15, 30, 45, and 60 min at 1500 μmol photons m−2 s−1. Arrowheads indicate the location of the mutated D1 polypeptides. Correct identification of the radiolabeled D1 polypeptide was confirmed by western blotting.
Variation of the psbA-2 Transcript Level during Photoinhibitory Illumination
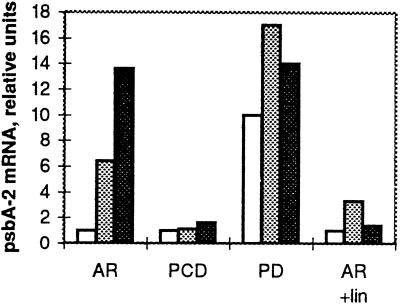
The fluctuation in the amount of the psbA-2 mRNA, the product of the only active psbA gene in all of the strains, was followed in the course of photoinhibitory illumination. In the AR strain, the amount of the psbA-2 mRNA at growth-light conditions was low, but the high-light treatment induced a strong accumulation of the psbA-2 mRNA (Figs. 4 and 5). PCD also had a low level of the psbA-2 mRNA at growth-light conditions, but, unlike the AR strain, no drastic accumulation of the psbA-2 mRNA occurred during high-light illumination. In the PD and CD strains the steady-state level of the psbA-2 mRNA under growth-light conditions was about 10-fold higher than that in the AR and PCD strains (Mulo et al., 1997), and remained at a high level during the high-light illumination of 60 min. In the presence of the protein-synthesis inhibitors lincomycin (Figs. 4 and 5) and chloramphenicol (data not shown), only a slight increase in the amount of the psbA-2 transcripts was detected at the beginning of the high-light illumination. However, this declined back to the growth-light level during the 60-min high-light treatment.
Figure 4.
Accumulation of the psbA-2 mRNA in the AR, PCD, and PD cells in the course of high-light illumination (1500 μmol photons m−2 s−1) in the absence or presence of lincomycin, as indicated. The amount of RNA is given in arbitrary units comparable between the strains. White bars, 0 min of illumination; stippled bars, 30 min of illumination; and black bars, 60 min of illumination.
Figure 5.
Representative northern blots showing the psbA-2 mRNA levels in the AR strain during photoinhibitory illumination of 1500 μmol photons m−2 s−1 in the absence (A) and presence (B) of the protein-synthesis inhibitor lincomycin. The bottom panels present the same membranes probed with rrn genes of A. nidulans. Five micrograms of total RNA was loaded in each well.
DISCUSSION
We have constructed a unique autotrophic PSII mutant with a deletion in the D-E loop of the D1 polypeptide (Mulo et al., 1997). This deletion has significantly modified the conformation of the QB pocket, as evidenced by a strongly modified oxygen-evolution capacity with different artificial electron acceptors and the incapability of DCMU to inhibit DCBQ-dependent oxygen evolution (Table I). Apparently because of these drastic modifications in the QB site, the PD mutant showed specific tolerance against photoinactivation of PSII when assayed with DCBQ, which has been suggested to accept electrons directly from QA (Graan and Ort, 1986; Satoh et al., 1993, 1995) (Figs. 1, A, B, and D, and 2A). Since the yield of variable fluorescence was similar in the presence and absence of DCBQ, it can be concluded that in the PD mutant, reduced QA also serves as the electron donor to DCBQ (E. Tyystjärvi, personal communication), thereby excluding the possibility that the mutation had created new avenues for the reduction of DCBQ. No specific tolerance of the PD cells to high light was evident when photoinhibition of oxygen evolution was assayed with DMBQ (Fig. 1C) (there were, however, extremely low activities; Table I) or with bicarbonate as the final electron acceptor (Fig. 2B). Nevertheless, even this inhibition differed from that of the control AR cells in being largely reversible, without the need for de novo protein synthesis, when the cells were transferred to low-light conditions (Fig. 2B).
The photoinhibitory behavior of the PD mutant has important consequences for our understanding of the mechanism of PSII photoinhibition in vivo. These results imply that the process of PSII photoinhibition can be dissected into at least two relatively independent but probably sequential phases. The initial light-induced modification of the QB-binding site in the PD mutant appeared to be a reversible event, with no requirement for protein synthesis, as far as the inhibition had not progressed to an inhibition of QA reduction (Fig. 2B). The latter event, in contrast, was irreversible in all strains (Fig. 2A, data not shown for CD and PCD). Such a reversible, high-light-induced initial modification of the QB pocket could not be trapped in the AR control strain, probably because of a short lifetime.
The initial role of the QB pocket in PSII photoinactivation in vivo was suggested in very early photoinhibition studies (Kyle et al., 1984) and later with the ΔPEST (ΔE226-Q233; Nixon et al., 1995) and ΔpsbH (Komenda and Barber, 1995) mutants of PSII. The reversibility of this reaction in vivo was deduced from fluorescence and thermoluminescence experiments (Kirilovsky et al., 1990; Ohad et al., 1990), but a clear biochemical demonstration for such a reversible conformational change in the QB pocket under physiological conditions has been missing until now. It is conceivable that the initial modification in the QB-binding site renders the cells susceptible to subsequent irreversible events: an inhibition of QA reduction and a consequent irreversible damage to the D1 protein. Generally, the reversible phase is difficult to trap (Vavilin et al., 1995) because the subsequent irreversible phases rapidly follow the initial conformational change in the QB pocket. This model of early photoinhibitory events in the QB pocket agrees with the reduced rate of PSII photoinactivation at low temperatures reported in several systems both in vivo and in vitro (Aro et al., 1990; Kirilovsky et al., 1990), presumably because the initial conformational changes in the QB pocket slow down with declining temperatures.
What could be the molecular basis for such a unique response of the PD cells to photoinhibitory illumination? The ΔPEST mutant (Nixon et al., 1995), which has a somewhat smaller deletion in the same region of the D-E loop compared with that of PD, showed no such tolerance against high-light-induced photoinhibition. Detailed comparison of these two autotrophic mutants ΔPEST and PD, both showing perturbations in the function of the acceptor side of PSII, also revealed an important difference. Both thermoluminescence and fluorescence-decay measurements point to a very small driving force for the electron transfer from QA− to QB in the PD mutant (Mulo et al., 1997), whereas ΔPEST is much less severely affected in this respect (Nixon et al., 1995). It is conceivable that the large deletion in PD, including amino acids of the possible contact site between the D1 and D2 proteins (Trebst and Soll-Bracht, 1996), also drastically modifies the QA-binding site in the D2 protein, rendering QA− less accessible to reoxidation.
Generally, the mutants with modified acceptor-side function of PSII, including herbicide-resistant mutants carrying point mutations in the QB-binding site (Kirilovsky et al., 1989; Constant et al., 1996), the psbH deletion mutant (Mayes et al., 1993; Komenda and Barber, 1995), the ΔPEST mutant (Nixon et al., 1995), and the strains with point mutations or small deletions in the PEST and cleavage-site regions (Tyystjärvi et al., 1994), have been reported to be intrinsically either equally or more sensitive to light-induced damage of PSII than the wild type. No simple correlation, however, seems to exist between the efficiency of electron-transfer reactions on the acceptor side of PSII and the susceptibility of PSII to photoinhibition. Nevertheless, a picture is now emerging suggesting that mutations that mainly alter the QB-binding properties may render the cells more susceptible to photoinhibition, whereas mutations that lead to an isolation of QA− from QB may protect PSII centers against irreversible photodamage. PD belongs to the latter group, whereas the heterotrophic CD and PCD strains also suffer from extremely poor exchange of plastoquinone in the QB pocket (Table I; Mulo et al., 1997), which exacerbates the irreversible photoinhibitory damage to PSII.
PSII photoinactivation was accompanied by D1 protein degradation in all strains. Although the amino acid stretches in the PEST-like sequence and in the putative cleavage region probably carry no specific information for light-dependent D1 protein degradation, the overall changes in the conformation of the loop are likely to modulate the degradation of the D1 protein. Thus, all mutant strains exhibited somewhat slower D1 degradation at high light than the AR control strain (Table II). In the PD mutant this is probably related to slower irreversible inactivation of PSII centers, whereas a distorted conformation of the D-E loop is likely to retard D1 degradation in CD and PCD compared with the AR cells. Our results also question a signal from the QB site to the degradation of the D1 protein upon illumination of the cells (Bracht and Trebst, 1994; Komenda and Barber, 1995).
As demonstrated with the PD mutant, the degradation of the D1 protein is not directly related to the initial conformational change in the QB-binding site that renders the cells incapable to reduce plastoquinone. This reaction indeed seems to be a reversible event with no need for D1 protein replacement (Fig. 2B). Only the subsequent event, inactivation of electron transfer from water to QA, was clearly also an irreversible process in PD (Fig. 2A), and this inhibition is likely to give a signal for degradation of the D1 polypeptide.
The retarded D1 degradation in the D-E loop deletion mutants under strong illumination seems to contradict results obtained under growth-light conditions, which showed accelerated D1 protein turnover compared with the AR control strain (Table II). However, it is conceivable that under growth-light conditions the mutated and misfolded D1 polypeptides are efficiently degraded by nonspecific, “household” protease(s), whereas under high irradiances such “cleaning” is masked by an efficient function of a still unknown D1-specific proteinase.
Slow initial protein-synthesis-dependent recovery of the heterotrophic mutants CD and PCD from photoinhibition (water to DCBQ) compared with the AR and PD strains (Fig. 2C) agrees with the measured low degradation capacity of the photodamaged D1 polypeptide in these mutants (Table II). The final recovery of the CD cells, however, was as complete as that of the control AR strain, whereas the PCD mutant, with almost similar susceptibility to photoinhibition as the CD cells, showed only poor capacity for PSII repair (Fig. 2C). Since degradation of the D1 polypeptide from photoinhibited centers is equally retarded in the CD and PCD mutants, this difference in the recovery capacity possibly points to impaired D1 synthesis in PCD.
The wild type (Mohamed and Jansson, 1989; Kanervo et al., 1993) and control AR and the mutants PD and CD possessed high amounts of psbA-2 transcripts at high light, whereas PCD was the only mutant not able to efficiently accumulate psbA-2 mRNA at high light (Figs. 4 and 5). A similar situation could be mimicked in the AR cells by blocking the protein synthesis with lincomycin during the high-light treatment. This might infer a link between regulation of the psbA-2 gene transcription and the D1 protein synthesis and suggests that there are interacting factors in the regulation of efficient D1 turnover rather than a single determinant such as the light intensity.
Copyright Clearence Center: 0032-0889/98/117/0483/08.
ACKNOWLEDGMENTS
We thank Drs. T. Tyystjärvi and E. Tyystjärvi for discussions and critical reading of the manuscript.
Abbreviations:
- AR
antibiotic-resistant strain
- CD
ΔG240-V249 mutant
- DCBQ
2,6-dichloro-p-benzoquinone
- DMBQ
2,5-dimethyl-p-benzoquinone
- PCD
ΔR225-V249 mutant
- PD
ΔR225-F239 mutant
Footnotes
This study was financially supported by the Academy of Finland (Helsinki).
LITERATURE CITED
- Arnon DI. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24:1–5. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro E-M, Hundal T, Carlberg I, Andersson B. In vitro studies on light-induced inhibition of Photosystem II and D1-protein degradation at low temperatures. Biochim Biophys Acta. 1990;1019:269–275. [Google Scholar]
- Aro E-M, Virgin I, Andersson B. Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta. 1993;1143:113–134. doi: 10.1016/0005-2728(93)90134-2. [DOI] [PubMed] [Google Scholar]
- Bennett A, Bogorad L. Complementary chromatic adaptation in a filamentous blue-green alga. J Cell Biol. 1973;58:419–435. doi: 10.1083/jcb.58.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blubaugh DJ, Cheniae GM. Kinetics of photoinhibition in hydroxylamine-extracted photosystem II membranes: relevance to photoactivation and sites of electron donation. Biochemistry. 1990;29:5109–5118. doi: 10.1021/bi00473a016. [DOI] [PubMed] [Google Scholar]
- Bracht E, Trebst A. Hypothesis on the control of D1 protein turnover by nuclear coded proteins in Chlamydomonas reinhardtii. Z Naturforsch. 1994;49c:439–446. [Google Scholar]
- Bumann D, Oesterhelt D. Destruction of a single chlorophyll is correlated with the photoinhibition of photosystem II with a transiently inactive donor side. Proc Natl Acad Sci USA. 1995;92:12195–12199. doi: 10.1073/pnas.92.26.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constant S, Perewoska I, Nedbal L, Miranda T, Etienne A-L, Kirilovsky D. A new phenotype for a herbicide resistant mutant of Synechocystis6714 with a high sensitivity to photoinhibition. Plant Sci. 1996;115:165–174. [Google Scholar]
- Graan T, Ort DR. Detection of oxygen-evolving photosystem II centers inactive in plastoquinone reduction. Biochim Biophys Acta. 1986;852:320–330. [Google Scholar]
- Greenberg BM, Gaba V, Mattoo EK, Edelman M. Identification of a primary in vivodegradation product of the rapidly-turning-over 32 kd protein of photosystem II. EMBO J. 1987;6:2865–2869. doi: 10.1002/j.1460-2075.1987.tb02588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegerschöld C, Virgin I, Styring S. Light-dependent degradation of the D1 protein in photosystem II is accelerated after inhibition of the water splitting reaction. Biochemistry. 1990;29:6179–6186. doi: 10.1021/bi00478a010. [DOI] [PubMed] [Google Scholar]
- Kanervo E, Mäenpää P, Aro E-M. D1 protein degradation and psbA transcript levels in Synechocystis PCC 6803 during photoinhibition in vivo. J Plant Physiol. 1993;142:669–675. [Google Scholar]
- Kirilovsky D, Ajlani G, Picaud M, Etienne A-L. Mutations responsible for high light sensitivity in an atrazine-resistant mutant of Synechocystis6714. Plant Mol Biol. 1989;13:355–363. doi: 10.1007/BF00015547. [DOI] [PubMed] [Google Scholar]
- Kirilovsky D, Vernotte C, Astier C, Etienne A-L. Reversible and irreversible photoinhibition in herbicide-resistant mutants of Synechocystis6714. Biochim Biophys Acta. 1988;933:124–131. [Google Scholar]
- Kirilovsky DL, Vernotte C, Etienne A-L. Protection from photoinhibition by low temperature in Synechocystis 6714 and in Chlamydomonas reinhardtii: detection of an intermediary state. Biochemistry. 1990;29:8100–8106. doi: 10.1021/bi00487a016. [DOI] [PubMed] [Google Scholar]
- Komenda J, Barber J. Comparison of psbO and psbH deletion mutants of Synechocystis PCC 6803 indicates that degradation of D1 protein is regulated by the QBsite and dependent on protein synthesis. Biochemistry. 1995;34:9625–9631. doi: 10.1021/bi00029a040. [DOI] [PubMed] [Google Scholar]
- Kyle DJ, Ohad I, Arntzen CJ. Membrane protein damage and repair: selective loss of a quinone-protein function in chloroplast membranes. Proc Natl Acad Sci USA. 1984;81:4070–4074. doi: 10.1073/pnas.81.13.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mäenpää P, Kallio T, Mulo P, Salih G, Aro E-M, Tyystjärvi E, Jansson C. Site-specific mutations in the D1 polypeptide affect the susceptibility of Synechocystis6803 cells to photoinhibition. Plant Mol Biol. 1993;22:1–12. doi: 10.1007/BF00038991. [DOI] [PubMed] [Google Scholar]
- Mayes SR, Dubbs JM, Vass I, Hideg E, Nagy L, Barber J. Further characterization of the psbH locus of Synechocystis sp. PCC 6803: inactivation of psbH impairs QA to QBelectron transport in photosystem II. Biochemistry. 1993;32:1454–1465. doi: 10.1021/bi00057a008. [DOI] [PubMed] [Google Scholar]
- Mohamed A, Jansson C. Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis6803. Plant Mol Biol. 1989;13:693–700. doi: 10.1007/BF00016024. [DOI] [PubMed] [Google Scholar]
- Mulo P, Tyystjärvi T, Tyystjärvi E, Govindjee, Mäenpää P, Aro E-M. Mutagenesis of the D-E loop of photosystem II reaction centre protein D1: function and assembly of photosystem II. Plant Mol Biol. 1997;33:1059–1071. doi: 10.1023/a:1005765305956. [DOI] [PubMed] [Google Scholar]
- Nixon PJ, Komenda J, Barber J, Deak Z, Vass I, Diner BA. Deletion of the PEST-like region of photosystem two modifies the QB-binding pocket but does not prevent rapid turnover of D1. J Biol Chem. 1995;270:14919–14927. doi: 10.1074/jbc.270.25.14919. [DOI] [PubMed] [Google Scholar]
- Ohad I, Adir N, Koike H, Kyle DJ, Inoue Y. Mechanisms of photoinhibition in vivo: a reversible light-induced conformational change of reaction center II is related to an irreversible modification of the D1 protein. J Biol Chem. 1990;265:1972–1979. [PubMed] [Google Scholar]
- Ohad I, Keren N, Zer H, Gong H, Mor TS, Gal A, Tal S, Domovich Y. Light-induced degradation of the photosystem II reaction centre D1 protein in vivo: an integrative approach. In: Baker NR, Bowyer JR, editors. Photoinhibition of Photosynthesis from Molecular Mechanisms to the Field. Oxford, UK: Bios Scientific Publishers; 1994. pp. 161–177. [Google Scholar]
- Prasil O, Adir N, Ohad I. Dynamics of photosystem II. In: Barber J, editor. Topics in Photosynthesis, Vol 11. Amsterdam, The Netherlands: Elsevier; 1992. pp. 293–348. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Satoh K, Kashino Y, Koike H. Electron transfer from QA to thymoquinone in a Synechococcus oxygen-evolving photosystem II preparation: role of QB and binding affinity of thymoquinone to the QBsite. Z Naturforsch. 1993;48c:174–178. [Google Scholar]
- Satoh K, Oh-hashi M, Kashino Y, Koike H. Mechanism of electron flow through the QB site in photosystem II. 1. Kinetics of the reduction of electron acceptors at the QB and plastoquinone sites in photosystem II particles from the cyanobacterium Synechococcus vulcanus. Plant Cell Physiol. 1995;36:597–605. [Google Scholar]
- Sherrer K, Darnell JE. Sedimentation characteristics of rapidly labelled RNA from HeLa cells. Biochem Biophys Res Commun. 1962;7:486–490. doi: 10.1016/0006-291x(62)90341-8. [DOI] [PubMed] [Google Scholar]
- Styring S, Virgin I, Ehrenberg A, Andersson B. Strong light photoinhibition of electrontransport in photosystem II: impairment of the function of the first quinone acceptor, QA. Biochim Biophys Acta. 1990;1015:269–278. [Google Scholar]
- Tomioka N, Shinozaki K, Sugiura M. Molecular cloning and characterization of ribosomal RNA genes from a blue-green alga, Anacystis nidulans. Mol Gen Genet. 1981;184:359–363. [Google Scholar]
- Trebst A. The topology of the plastoquinone and herbicide binding peptides of photosystem II in the thylakoid membrane. Z Naturforsch. 1986;41c:240–245. [Google Scholar]
- Trebst A, Soll-Bracht E. Cycloheximide retards high light driven D1 protein degradation in Chlamydomonas reinhardtii. Plant Sci. 1996;115:191–197. [Google Scholar]
- Tyystjärvi T, Aro E-M, Jansson C, Mäenpää P. Changes of amino acid sequence in PEST-like area and QEEET motif affect degradation rate of D1 polypeptide in photosystem II. Plant Mol Biol. 1994;25:517–526. doi: 10.1007/BF00043879. [DOI] [PubMed] [Google Scholar]
- Tyystjärvi T, Mulo P, Mäenpää P, Aro E-M. D1 polypeptide degradation may regulate psbA gene expression at transcriptional and translational levels in Synechocystissp. PCC 6803. Photosynth Res. 1996;47:111–120. doi: 10.1007/BF00016174. [DOI] [PubMed] [Google Scholar]
- Vass I, Styring S, Hundal T, Koivuniemi A, Aro E-M, Andersson B. Reversible and irreversible intermediates during photoinhibition of photosystem II: stable reduced QAspecies promote chlorophyll triplet formation. Proc Natl Acad Sci USA. 1992;89:1408–1412. doi: 10.1073/pnas.89.4.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavilin DV, Tyystjärvi E, Aro E-M. In search of a reversible stage of photoinhibition in a higher plant: no changes in the amount of functional photosystem II accompany relaxation of variable fluorescence after exposure of lincomycin-treated Cucurbita pepoleaves to high light. Photosynth Res. 1995;45:239–247. doi: 10.1007/BF00015564. [DOI] [PubMed] [Google Scholar]
- Wang W-Q, Chapman DJ, Barber J. Inhibition of water splitting increases the susceptibility of photosystem II to photoinhibition. Plant Physiol. 1992;99:16–20. doi: 10.1104/pp.99.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JGK. Constructions of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis6803. Methods Enzymol. 1988;167:766–778. [Google Scholar]